 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Exploring the sensitivities of experimental techniques to various types of membrane asymmetry using atomistic simulations
Frederick A.
Heberle
 a and
Milka
Doktorova
a and
Milka
Doktorova
 *b
*b
aDepartment of Chemistry, University of Tennessee Knoxville, Knoxville, TN 37916, USA
bDepartment of Biochemistry and Biophysics, Stockholm University, Science for Life Laboratory, Solna 171 65, Sweden. E-mail: milka.doktorova@dbb.su.se
First published on 16th January 2025
Abstract
Biological membranes have two leaflets that can differ in both lipid composition and total lipid abundance. These different types of asymmetries play a major role in determining the biophysical properties of the membrane; however, they have proven challenging to assay experimentally even in simpler model systems. Molecular dynamics simulations offer the means for detailed computational investigation of systematically varied interleaflet lipid distributions, but opportunities for critical validation with wet lab experiments are scarce. To help address this problem, here we use atomistic simulations of asymmetric bilayers to generate synthetic experimental data and thus investigate the sensitivity of various approaches to changes in relative lipid composition, number, and cholesterol distribution. Contrary to trends in symmetric bilayers, the simulations showed a decrease in lipid packing with increasing cholesterol in differentially stressed asymmetric bilayers, with more pronounced changes in the more loosely packed leaflet. Representative experimental data computed from the simulation trajectories indicated that the detection of asymmetry-induced changes in leaflet properties should be possible with environment-sensitive fluorescent probes and NMR observables, but may require optimization of sample preparation conditions. On the other hand, small-angle scattering data are already experimentally accessible and can reveal differential leaflet packing densities through a model-free analysis. We further show that computationally generated cryo-EM intensity profiles are highly sensitive to phospholipid imbalance between membrane leaflets. Together, these findings provide a roadmap for developing targeted applications of the in vitro techniques and obtaining experimental data critical for validating computationally derived principles related to membrane asymmetry.
Introduction
As biological interfaces, lipid bilayers have evolved to support a large number of functions and processes. This is achieved by a rich palette of biophysical properties created in part by the bilayer lipid composition.1 Extensive studies of the physio-chemical behavior of single component lipid bilayers have linked lipid structure (e.g., chain length, head group type) to membrane permeability, fluidity and elasticity.2–6 Further analyses of the thermodynamics of lipid mixing have revealed the ability of membranes to compartmentalize their protein constituents via lateral heterogeneities characterized by in-plane (within leaflet) co-existence of regions with different lipid compositions and biophysical profiles.7–9 These versatile structure–property relationships have been investigated with a large suite of experimental and computational techniques,10–13 and formulate the basis for current models of membrane structure and function.In addition to their in-plane complexity, lipid bilayers can exhibit non-random organization with respect to the two leaflets. For example, the existence and prevalence of compositional (and by extension biophysical) asymmetry in biological membranes has been recognized since the 1970s.14–16 Cells use valuable resources to actively maintain gradients in lipid saturation and charge across the bilayer midplane, which lead to contrasting fluidity and electrostatic surface potentials in the two leaflets.16–20 Lipids also have different spontaneous curvatures, and their interleaflet distributions may induce curvature stress in the bilayer.2,21–23 Cholesterol, with its unique structure and dynamics, further contributes to these effects via its (re)distribution.21,24 The overall compositional asymmetry of model and cell membranes has been assayed with various approaches including fluorescence quenching, peptide–membrane interactions, and NMR and mass spectrometry, although in many cases the exact quantification of the lipid make-up of both leaflets has not been possible.16,25–29
Lipid composition however, may not be the only (or even the main) determinant of the properties of a membrane. Lipid bilayers can tolerate a range of imbalances in the relative abundance of lipids between their leaflets.22,30,31 This is realized by adjustments in the lipid packing in each monolayer to conform to the area imposed by the closed membrane surface while minimizing hydrocarbon–water contacts. The result is a build-up of tension in one leaflet and pressure in the other (termed differential, or asymmetry, stress) with concomitant opposite effects on the structure and dynamics of the two leaflets.32 Analogously to externally applied net bilayer tension, differential stress can be tolerated up to a certain threshold before the membrane ruptures or changes morphology.33 For example, the addition of lipids to one leaflet of pre-formed membranes has been used to induce excessive asymmetry stress and budding or membrane shape changes in both synthetic and living systems.34–37 At the same time, theory and experiment indicate that substantial resting differential stress is likely present in giant synthetic vesicles with engineered compositional asymmetry as well as unperturbed cell plasma membranes, ‘silently’ affecting their measurable properties without any clear morphological signatures.22,38–40
Differential stress can be quantified from the lateral pressure distribution of simulated lipid bilayers.22,30,41 However, the ability to unambiguously detect and measure it in experiments remains a formidable challenge.42 To begin to address this problem, here we examine the sensitivities of various experimental techniques to different types of interleaflet membrane asymmetries including in phospholipid composition, number and cholesterol distribution. To this end, we employ molecular dynamics simulations to analyze the properties and dynamics of detailed atomistic models, then use them to generate representative experimental data from liposomes with the corresponding bilayer structures. The experiments we focus on include fluorescence lifetime measurements of environment-sensitive probes, NMR observables, small-angle scattering form factors, and cryo-EM images and intensity profiles. Our goal is to examine the differences in biophysical properties induced by the interplay of the various types of asymmetries and evaluate the capabilities of the in vitro approaches to detect them. Our results present a framework for identifying the conditions and controls for each technique that are best suited for analysis of the multifaceted asymmetry of lipid membranes, thus providing a roadmap to help guide future research efforts in these directions.
Methods
Atomistic bilayer models
Solvated lipid bilayer models were constructed with CHARMM-GUI43–45 and were composed of 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC), 1,2-diarachidonoyl-sn-glycero-3-phosphocholine (DAPC) and cholesterol (Chol). In particular, eight symmetric bilayers had either DPPC or DAPC with 0, 10, 30 and 50 mol% Chol, and one symmetric bilayer (referred to as ‘scramble’) was composed of DPPC/DAPC/Chol 35/35/30 mol%. All bilayers contained 100 lipids per leaflet and were hydrated with 50 waters per lipid and no ions. Two additional symmetric bilayers with DPPC, 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) and Chol, were constructed, with DPPC/DOPC/Chol in molar percents of 56/9/35 (liquid-ordered, Lo) and 23/67/10 (liquid-disordered, Ld).In addition, six asymmetric bilayers were taken from ref. 46 for further analysis. The bilayers had 70 DPPC lipids in one leaflet, a varying number of DAPC lipids in the opposite leaflet, and Chol at 30 mol% of all lipids distributed according to its chemical potential (as characterized from initial coarse-grained simulations, see ref. 46). Table 1 lists the number of lipid and cholesterol molecules in each leaflet. All bilayers were similarly constructed with CHARMM-GUI and hydrated with 50 waters per lipid and no ions.
| Bilayer (PL imb.) | Composition, # lipids (DPPC/DAPC/Chol) | Chol leaflet (mol%) | Area per lipid (Å2) | Leaflet thickness (Å), phosphate to midplane | Leaflet tension (mN m−1) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Top | Bot | Top | Bot | Top | Bot | Top | Bot | Top | Bot | |
| A.1 (0.91) | 70/0/56 | 0/77/7 | 44 | 8 | 42.0 (0.36) | 63.0 (0.54) | 23.5 (0.26) | 20.2 (0.29) | 14.8 (0.9) | −14.8 (0.9) |
| A.2 (1.00) | 70/0/52 | 0/70/8 | 43 | 10 | 41.3 (0.30) | 64.7 (0.46) | 23.7 (0.22) | 19.8 (0.28) | 7.2 (0.9) | −7.2 (0.9) |
| A.3 (1.11) | 70/0/47 | 0/63/10 | 40 | 14 | 40.8 (0.28) | 65.4 (0.45) | 23.9 (0.21) | 19.5 (0.27) | −0.1 (1.0) | 0.1 (1.0) |
| A.4 (1.25) | 70/0/43 | 0/56/11 | 38 | 16 | 40.3 (0.19) | 67.9 (0.33) | 24.1 (0.18) | 18.9 (0.26) | −11.1 (1.0) | 11.1 (1.0) |
| A.5 (1.43) | 70/0/37 | 0/49/14 | 35 | 22 | 40.1 (0.23) | 68.1 (0.39) | 24.1 (0.20) | 18.7 (0.28) | −14.5 (1.0) | 14.5 (1.0) |
| A.6 (2.00) | 70/0/29 | 0/35/16 | 29 | 31 | 39.7 (0.20) | 77.0 (0.39) | 24.1 (0.17) | 16.8 (0.28) | −31.7 (1.0) | 31.7 (1.0) |
Molecular dynamics simulations
All bilayers were simulated with NAMD![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 47 and the CHARMM36 force field for lipids,48 as described in ref. 46. Briefly, the systems were first equilibrated with CHARMM-GUI’s 6-step equilibration protocol, then simulated with a 2 fs timestep and the length of all hydrogen bonds constrained (rigidbonds parameter set to all). Nonbonded interactions were modeled with a 10–12 Å Lennard-Jones potential function using force switching via the vdwforceswitching option in NAMD. The particle mesh Ewald method with a grid spacing of 1 Å was used for electrostatic interactions. The simulations were performed with semi-isotropic pressure coupling at a constant temperature of 40 °C (or 25 °C for the Lo and Ld bilayers) and pressure at 1 atm using Langevin thermostat (damping coefficient of 5 ps−1) and barostat (period 200 fs and decay 50 fs). The symmetric bilayers were run for 600–1180 ns. All asymmetric bilayers were run for over 1 µs with the last 400 ns used for analysis, as detailed in Table S4 in ref. 46.
47 and the CHARMM36 force field for lipids,48 as described in ref. 46. Briefly, the systems were first equilibrated with CHARMM-GUI’s 6-step equilibration protocol, then simulated with a 2 fs timestep and the length of all hydrogen bonds constrained (rigidbonds parameter set to all). Nonbonded interactions were modeled with a 10–12 Å Lennard-Jones potential function using force switching via the vdwforceswitching option in NAMD. The particle mesh Ewald method with a grid spacing of 1 Å was used for electrostatic interactions. The simulations were performed with semi-isotropic pressure coupling at a constant temperature of 40 °C (or 25 °C for the Lo and Ld bilayers) and pressure at 1 atm using Langevin thermostat (damping coefficient of 5 ps−1) and barostat (period 200 fs and decay 50 fs). The symmetric bilayers were run for 600–1180 ns. All asymmetric bilayers were run for over 1 µs with the last 400 ns used for analysis, as detailed in Table S4 in ref. 46.
Calculation of bilayer structural parameters
Simulations were first centered so that the average position of all terminal methyl carbons was at (x,y,z) = (0,0,0) in every frame. Average area per lipid (APL) was calculated by dividing the lateral area of the simulation box by the number of lipids (including Chol) in each leaflet. Lateral pressure profiles were obtained with NAMD as described in detail in ref. 30. The differential stress and bilayer torque density were calculated from the integral and first moment of the pressure profiles, respectively, as outlined in ref. 46. The order parameters and relaxation rates of the carbon–hydrogen bonds were calculated following the protocols in ref. 49. Errors on APL and order parameters were obtained by block averaging, while errors on differential stress and bilayer torque density were calculated with bootstrapping analysis. Number and charge density profiles were computed for the whole systems in thin slabs of thickness 0.2 Å with the density profile plugin in VMD.50 The bilayer dipole potential was calculated from the charge density distribution following the approach in ref. 51.Calculation of small-angle scattering intensity
Scattering intensity was calculated from the time-averaged atomic number density distributions projected onto the bilayer normal, ni(z), where z denotes position along the bilayer normal and i indexes the individual lipid and water atoms. Each distribution was scaled by the appropriate atomic scattering factor (i.e., the X-ray atomic form factor or the coherent neutron scattering length) and then summed to produce a scattering length density profile, ρ(z). The scattering intensity as a function of scattering vector q is given by:52 | (1) |
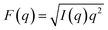 . The center-of-mass of the form factor was calculated as:
. The center-of-mass of the form factor was calculated as: | (2a) |
 | (2b) |
Calculation of cryo-EM intensity profiles
In a projection image of a liposome, the azimuthally averaged intensity profile (IP) contains information about the bilayer structure. IPs were calculated as previously described53 and briefly summarized here. First, the electrostatic potential within the lipid bilayer, Φ(z), was approximated as a sum of contributions from individual lipid and water atoms with an additional contribution from the membrane dipole potential Φd, i.e., | (3) |
| g(z) = σeΦ(z), | (4) |
 | (5) |
To mimic experimental images, Γ(r) is convolved with a CTF, c(s), and associated phase perturbation factor, χ(s):
c(s) = [sin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) χ(s) − Q χ(s) − Q![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cos cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) χ(s)]exp(−B|s|2), χ(s)]exp(−B|s|2), | (6) |
 | (7) |
 | (8) |
 is the Fourier transform of the projected 2D phase shift image Γ(r) and m is an arbitrary scale factor that adjusts the intensity contrast in the spatial domain. The corresponding real-space image I(r) is the inverse Fourier transform of I(s),
is the Fourier transform of the projected 2D phase shift image Γ(r) and m is an arbitrary scale factor that adjusts the intensity contrast in the spatial domain. The corresponding real-space image I(r) is the inverse Fourier transform of I(s), | (9) |
For the calculations in this work, we used the following cryo-EM parameter values: R = 50 nm, B = 300 Å2, Q = 0.075, λ = 1.97 pm (corresponding to 300 keV electrons), ΔZ = 2.5 µm, Cs = 2.0 mm, m = 400.
Results and discussion
Asymmetry-mediated changes in leaflet composition and properties
To investigate the effects of biologically relevant asymmetries on membrane properties, we analyzed a series of molecular dynamics (MD) simulation trajectories of all-atom bilayers where one leaflet contained the saturated lipid DPPC, the other contained the unsaturated lipid DAPC, and cholesterol was at a fixed overall concentration and free to redistribute between the leaflets. This simplified lipid composition aims to capture the asymmetry in lipid saturation reported for red blood cell membranes,16 which also plays a key role in determining the chemical potential of cholesterol in the leaflets.24 The bilayers had a fixed number of DPPC lipids (NDPPC) in the top leaflet but varying numbers of DAPC lipids (NDAPC) in the bottom leaflet (Table 1). Initial coarse-grained simulations showed that while such bilayers with more DPPC than DAPC lipids were stable up to relatively large imbalances, deviations in the opposite direction (more DAPC lipids) were not tolerated as well and produced unphysical bilayer morphologies even at smaller imbalances.46 The phospholipid (PL) imbalance, NDPPC/NDAPC, in the systems we studied here thus ranged from 0.91 to 2.0.All systems had cholesterol at 30 mol% of all PLs. In real membranes, cholesterol can quickly flip between leaflets and redistribute according to its chemical potential. Since proper sampling of this movement is challenging in all-atom representation, the equilibrium cholesterol distribution in the models was determined from initial long coarse-grained trajectories, and did not change during the all-atom simulations (Table 1).46 In all but one of the bilayers, cholesterol showed a strong preference for the saturated DPPC leaflet (Fig. 1A). Its strong pairwise interactions with DPPC competed with other factors including the overabundance of the saturated lipid (relative to DAPC), as predicted from theory.24 Cholesterol’s enrichment in the top leaflet thus decreased gradually across the systems until it completely disappeared in the A.6 bilayer which had twice as many DPPC than DAPC lipids (Fig. 1A and Table 1).
 | ||
Fig. 1 Biophysical properties of simulated asymmetric bilayers. (A) Cholesterol asymmetry (cholesterol mol% in the top vs. the bottom leaflet) as a function of interleaflet PL imbalance. Solid gray lines indicate the conditions of no PL imbalance and no cholesterol asymmetry. (B) Leaflet area per lipid (APL) as a function of cholesterol mol% in the individual leaflets of symmetric DPPC or DAPC bilayers (open squares, ‘sym’) and the asymmetric DPPC/DAPC bilayers from Table 1 (filled triangles, ‘asym’). Shown for comparison is a symmetric ‘scramble’ bilayer with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DPPC 1 DPPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DAPC and 30 mol% cholesterol (gray square). (C) Bilayer torque density as a function of PL imbalance in the asymmetric bilayers from Table 1. DAPC and 30 mol% cholesterol (gray square). (C) Bilayer torque density as a function of PL imbalance in the asymmetric bilayers from Table 1. | ||
Cholesterol has a well characterized condensing effect on lipid packing in both saturated and unsaturated membranes.55–57 We confirmed this trend in symmetric DPPC and DAPC bilayers which showed a systematic decrease in the average area per lipid (APL) with increasing amounts of cholesterol (Fig. 1B, open symbols). Since the individual leaflets of the asymmetric bilayers are binary mixtures with varying concentrations of cholesterol, we can similarly analyze the effect of the sterol on their APLs. In contrast to the expected trend observed in the symmetric membranes, cholesterol had an exactly opposite effect in the asymmetric bilayers, increasing rather than decreasing the leaflet APL (Fig. 1B, closed symbols). This counter-intuitive trend can be related to the internal stresses in the leaflets, i.e. differential stress, resulting from the interleaflet PL and cholesterol distributions (Table 1). In most bilayers, one leaflet had tension while the lipids in the other leaflet were compressed (i.e., a negative tension). These stresses constrained the effects of cholesterol, making them deviate from the trends observed in symmetric bilayers which by definition have zero leaflet tension.30 Only the A.3 bilayer, which had a PL imbalance of 1.11 and about 3-fold more cholesterol in the DPPC leaflet, had no differential stress. Interestingly, in that bilayer, the APL of the DAPC leaflet containing 14 mol% cholesterol (Table 1) was lower than the APL of a symmetric DAPC bilayer with similar cholesterol concentration, while the condensing effect of cholesterol in the DPPC leaflet was similar to its symmetric counterpart (Fig. 1B). These results reveal that even in the absence of internal stresses, compositional asymmetry can influence bilayer properties in ways that cannot be fully recapitulated in symmetric bilayers, which is a manifestation of interleaflet coupling.58–60 How the specific lipid compositions of the leaflets affect the observed relationships between PL imbalance and differential stress, and APL and cholesterol concentration, remains to be investigated.
The high resolution of the simulated bilayers also enables analysis of the propensity of the bilayers to curve, which in experiments can be monitored via changes in membrane morphology.34,61,62 Depending on its lipid composition, each leaflet has a spontaneous curvature that determines the curvature stress experienced by the leaflet in a given configuration (e.g., when flat).63,64 The balance between curvature stress and differential stress governs the energy cost of changes in the bilayer morphology.22 In particular, the first moment of the lateral pressure profile (whose integral gives the leaflet stresses) is the bilayer torque density.41 Its deviation from zero provides an estimate of the bilayer tendency to curve, with positive and negative values indicating curvature towards the top and bottom leaflets, respectively. Analysis of the asymmetric bilayers shows that this parameter is highly sensitive to their molecular organization, with a consistent preference for curvature towards the compressed leaflet (Fig. 1C). Only in the tensionless A.3 bilayer is a flat morphology the most energetically favorable. Since this bilayer has no area strain, the result implies that it has vanishing curvature stress (i.e., a spontaneous curvature of zero)41 despite its highly asymmetric profile (Table 1). This is likely due to the modulation of the leaflets’ spontaneous curvature by cholesterol.21 However, vanishing differential stress is not a pre-requisite for a stable flat configuration, as the latter can exist even when the leaflets experience significant tension and compression that counterbalances curvature stress.46 These findings demonstrate that the absence of curvature in experimental asymmetric membranes cannot be used to infer the presence or absence of both differential and curvature stresses.
Sensitivity of environment-sensitive probes
One commonly applied approach for analyzing the structural properties of lipid membranes in both model systems and cells, is measuring changes in their lipid packing via environment-sensitive probes.65 A typical experiment relies on incorporation of the probe in one or both bilayer leaflets and recording its fluorescence lifetime (for e.g. Di4-ANEPPDHQ,66 Flipper67) or generalized polarization (laurdan,68 NR12S![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 69). While each of these probes likely senses an interplay of multiple leaflet properties,70 they are often used as reporters of lipid packing. This has been shown to be a good approximation for Di4, whose lifetime in symmetric giant unilamellar vesicles is very strongly correlated with the average area per lipid calculated from corresponding simulations of flat bilayer patches for a wide range of simple 1- and 2-component lipid mixtures.46 To verify that the relationship holds for 3-component bilayers, we simulated the two timeline endpoints of DPPC/DOPC/Chol 40/40/20 representing its coexisting Lo and Ld compositions (see Methods). The bilayers were thus symmetric and uniform, and we compared the APLs from the simulations to the experimentally measured Di4 lifetimes in the mixtures from ref. 16. To account for any differences in the experimental conditions between ref. 16 and 46, we used the Di4 lifetime of DOPC reported in both studies to normalize the results. The Lo and Ld compositions both fell precisely on the calibration curve as shown in Fig. 2A, confirming that even in more complex multi-component mixtures, Di4 lifetime is highly correlated with simulated APLs.
69). While each of these probes likely senses an interplay of multiple leaflet properties,70 they are often used as reporters of lipid packing. This has been shown to be a good approximation for Di4, whose lifetime in symmetric giant unilamellar vesicles is very strongly correlated with the average area per lipid calculated from corresponding simulations of flat bilayer patches for a wide range of simple 1- and 2-component lipid mixtures.46 To verify that the relationship holds for 3-component bilayers, we simulated the two timeline endpoints of DPPC/DOPC/Chol 40/40/20 representing its coexisting Lo and Ld compositions (see Methods). The bilayers were thus symmetric and uniform, and we compared the APLs from the simulations to the experimentally measured Di4 lifetimes in the mixtures from ref. 16. To account for any differences in the experimental conditions between ref. 16 and 46, we used the Di4 lifetime of DOPC reported in both studies to normalize the results. The Lo and Ld compositions both fell precisely on the calibration curve as shown in Fig. 2A, confirming that even in more complex multi-component mixtures, Di4 lifetime is highly correlated with simulated APLs.
 | ||
| Fig. 2 Predicted sensitivity of Di4 lifetime to membrane asymmetries. (A) Fluorescence lifetime of Di4 calculated from leaflet APLs in simulated bilayers using the experimentally derived calibration curve from ref. 46. Shown is data for the simulated top DPPC leaflets (blue triangles pointing up), bottom DAPC leaflets (red triangles pointing down) and symmetric scrambled bilayer (green diamond). Also shown are the results for symmetric 3-component Lo and Ld compositions of DPPC/DOPC/Chol as explained in the text (pink and purple stars). The solid black curve shows the linear relationship between simulation and experiment derived in ref. 46 from model membranes, while the dotted lines indicate regions outside of the calibration range. (B) Calculated Di4 lifetime as a function of PL imbalance for the top and bottom leaflets in the asymmetric bilayers (same symbols as in (A)). Also shown is the difference between the two (gray squares, right y-axis) which shows the largest sensitivity to the transverse lipid organization in the membranes. | ||
Using the experimentally derived linear relationship between the two parameters, we then converted the leaflet APLs in our other simulated bilayers to Di4 lifetimes and compared the sensitivity of this parameter to the asymmetries in the membranes (Fig. 2). Since Di4 is charged and does not easily flip between leaflets,16 we analyzed the expected lifetime measured when the probe is added to the bilayer from the outside, labeling only the top leaflet, or selectively incorporated exclusively in the bottom leaflet. Thus, while the simulations do not contain the probe itself, the structural properties of the leaflets are used to estimate the leaflet-specific fluorescence lifetime read-out from Di4 in an experiment. In experiments, adding Di4 to outer membrane leaflets is straightforward since in aqueous solutions the probe partitions strongly into lipid bilayers due to its hydrophobic groups. Adding Di4 to selectively label the inner (cytosolic) leaflet is more challenging but has been accomplished experimentally by microinjection of cells.16 This was confirmed by the difference in Di4 lifetime of the cytoplasmic leaflet relative to the exoplasmic leaflet, and the lack of BSA back-extraction from the PM (see Fig. S7 in ref. 16). In model membranes, selective labeling of the inner leaflet of liposomes has been accomplished using cyclodextrin to exchange the outer leaflet probe with lipid from unlabelled donor vesicles,71 although this has not been specifically demonstrated for Di4. The selective incorporation of a lipid dye in the inner leaflet can be verified with externally added quenchers, and other environment-sensitive probes can in principle be used in analogous ways (see Introduction).
The first noticeable result from our computational analysis was that two of the data points—corresponding to two DAPC-rich leaflets—fell outside of the calibration range between simulation and experiment (Fig. 2A).46 One of them is a leaflet in bilayer A.6 that has the most extreme PL imbalance of 2.0 and substantial differential stress (Table 1) and bilayer torque density (Fig. 1C). This highly stressed bilayer state is tolerated in simulations due to the relatively small system size and applied periodic boundary conditions but may exceed the tolerance threshold of liposomes in vitro, where the stress can be relieved instead by mechanisms such as extreme curvature and budding.34,72 The calibration range could be extended by analyzing more disordered symmetric bilayers under net bilayer tension or at higher temperatures (both of which can be applied in simulations and experiments to increase leaflet APL). However, the tolerance threshold for bilayer stress in the experiments, as well as any potential changes to the correspondence between the simulations and Di4 measurements from external perturbations, must be investigated and accounted for.
Comparison between the calculated Di4 lifetimes of the outer and inner leaflets across the series of simulated bilayers reveals differences in the sensitivity of this parameter to the properties of the respective lipid compositions (Fig. 2B). The relatively tightly packed DPPC leaflet, whose APL varies only from 39.7 ± 0.2 Å2 to 42 ± 0.36 Å2 between the two extremes (bilayers A.1 and A.6), shows marginal variation in Di4 lifetime. In contrast, the predicted changes in the DAPC leaflet, whose APL increases by 22% (Table 1), are more pronounced. Since the trends in the leaflet APLs are inversely correlated, where the smaller APL becomes smaller while the larger APL increases, most sensitive to the asymmetries is the difference between outer and inner leaflet Di4 lifetimes (Fig. 2B, gray squares). Measuring this difference in vitro would require concurrent experiments of membranes with either their top or bottom leaflets labelled with Di4, or calibrating and incorporating in the two leaflets different environment-sensitive probes with non-overlapping fluorescence spectra that can be imaged and analyzed simultaneously.
Sensitivity of NMR structural parameters
In addition to APL, bilayer structure is often characterized by the degree of order of the lipid acyl chains. This property is quantified from the dynamic reorientation of the carbon–hydrogen (CH) bonds along the lipid chains relative to the bilayer normal. In particular, the average order parameter (SCD) characterizing the mean of the fluctuations, as well as their relaxation rate (R1Z), can be measured with NMR and also calculated from simulations (e.g. ref. 49 and 73). The experiments are often done with deuterium NMR which allows for better spectral resolution and identification of chain segments.74 The measurements are typically performed on oriented lipid stacks or multilamellar lipid dispersions,75 but can also be done on unilamellar liposomes,76,77 thus making them suitable for asymmetry studies using model membranes. Since deuterated lipids with saturated chains are in general more readily available from commercial vendors, we analyzed the CH bond dynamics for DPPC in the simulated bilayers as representative experimental data (Fig. 3). Similar comparisons of the order at specific carbon segments can also be made with electron spin resonance (ESR) experiments via spin probes selectively incorporated into one of the leaflets.71,78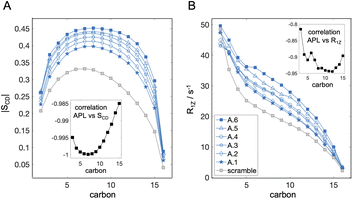 | ||
Fig. 3 Variation in simulation parameters corresponding to properties measured with NMR. (A) Acyl chain order parameter profiles for carbons 2 through 16 averaged over the sn-1 and sn-2 chains of DPPC in the asymmetric bilayers A.1 to A.6 from Table 1, and the scramble analog with DPPC/DAPC 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 30 mol% cholesterol. Errors are less than 10−2 and are omitted for clarity. (B) Spin-lattice relaxation rates of the corresponding CH bond fluctuations analyzed in (A). Insets in both (A) and (B) show the correlation between SCD or R1Z and leaflet APL (Table 1) calculated from the plotted data for the asymmetric bilayers. 1 and 30 mol% cholesterol. Errors are less than 10−2 and are omitted for clarity. (B) Spin-lattice relaxation rates of the corresponding CH bond fluctuations analyzed in (A). Insets in both (A) and (B) show the correlation between SCD or R1Z and leaflet APL (Table 1) calculated from the plotted data for the asymmetric bilayers. | ||
The resulting order parameter profiles show an overall increase in chain order (i.e., higher |SCD|) with a decrease in leaflet tension and APL (Fig. 3A and Table 1), as expected. There is a strong negative correlation between APL and SCD for all carbons (inset in Fig. 3A) but the best sensitivity to the asymmetry-mediated properties (i.e., the largest separation between all conditions) is achieved in the bottom parts of the chains, at carbons 12 through 15 (Fig. 3A). This is not surprising since this is the part of the leaflet in closest contact with the opposing leaflet. In contrast, the order parameter profiles for some bilayers (e.g. A.5 and A.4) overlap in the plateau region (carbons 5 through 10). Interestingly, in the same bilayers, these carbon segments have detectable differences in their relaxation rates even though the correlation between APL and R1Z is slightly weaker (Fig. 3B), indicating that the structural information from SCD and R1Z is complementary. Thus, if available, data from both parameters can maximize the ability to distinguish the leaflet states in experiments. Most challenging in this endeavour would be optimizing the size and hydration level of the asymmetric vesicles for the measurements. While the simulations revealed overall strong correlation between the NMR parameters and APL (Fig. 3), experimental conditions may be key for determining the consistency of the measured bilayer structure with other techniques.73 The relationship between the order parameters and relaxation rates in the region of leaflet–leaflet interaction has also been connected to bilayer elasticity via the membrane thickness in symmetric membranes.49 Further research is needed to test and develop this dependence in asymmetric bilayers.
Sensitivity of small-angle scattering data
Small-angle X-ray and neutron scattering (SAXS and SANS, respectively) are robust and well-established techniques for the precise measurement of bilayer structural parameters including area per lipid and bilayer thickness.79,80 Using atomic number density profiles from simulated bilayers as input, we calculated form factors for SAXS (Fig. 4A) and SANS (Fig. 4B–D), the latter using different combinations of protiated and perdeuterated lipids to explore the effects of internal bilayer contrast. Whereas symmetric bilayers produce clearly delineated scattering lobes separated by minima that approach zero intensity, asymmetry results in a characteristic “liftoff” of the minima that is particularly pronounced in cases of extreme transbilayer scattering length density contrast (e.g., protiated DAPC in the inner leaflet and DPPC-d62 in the outer leaflet as in Fig. 4C and D).81–83 Such liftoff is also visible (albeit to a smaller extent) in the first minimum following the first peak at the lowest q in the SAXS form factors (Fig. 4A). In principle, structural parameters can be extracted from the data by fitting to a suitable structural model; however, this is a daunting task for an asymmetric membrane where each leaflet has its own variables thus requiring a multitude of free parameters in the fitting routine.84,85 Furthermore, as illustrated in the fully protiated condition in Fig. 4B, bilayers with very different PL and cholesterol asymmetries can lead to almost indistinguishable scattering curves, making it difficult to unambiguously determine the underlying lipid organization.An alternative approach to analyzing such data uses model-free quantities that correlate with structural features. For example, increasing phospholipid imbalance results in systematic changes in the intensity profiles that are particularly prominent in the first two scattering lobes for SAXS (Fig. 4A) and internally contrasted SANS (Fig. 4C and D) data. These changes are readily apparent in the centers-of-mass of the F(q) curves, calculated as described in the Methods and plotted as small filled symbols (highlighted by solid arrows) in Fig. 4A–D. Fig. 4E shows the corresponding trends and reveals that the q-coordinate of the center-of-mass increases nearly linearly with phospholipid imbalance in all but the fully protiated SANS data. A useful aspect of this analysis is that it is insensitive to sample-to-sample differences in lipid concentration. Furthermore, the q-range over which it is calculated can be adjusted to include only data with the highest signal-to-noise in the experiments. Importantly, this quantification demonstrates that both SAXS and SANS with at least one asymmetrically distributed perdeuterated lipid are sensitive to compositional and number imbalances, in contrast to SANS of fully protiated samples. Joint analysis of these various conditions may thus provide the best approach for identifying a bilayer model with the correct structure.
Sensitivity of cryo-EM imaging and analysis
Cryo-EM enables the direct visualization of lipid bilayers at sub-nanometer resolution.86,87 When spherical liposomes are imaged in projection, the defining characteristic is a pair of dark concentric circles roughly corresponding to the electron dense lipid headgroup layers (Fig. 5A). For laterally homogeneous bilayers, images of individual vesicles can be reduced to 1D intensity profiles normal to the bilayer by azimuthal integration and then ensemble averaged to produce data suitable for quantitative analysis.88,89 As described in Methods, the cryo-EM image formation process for liposomes can also be accurately mimicked computationally using atomic number density profiles from MD simulations as input.90Fig. 5B shows cryo-EM intensity profiles calculated from the simulated asymmetric bilayers with varying phospholipid imbalance, as well as the symmetric scrambled bilayer with 30 mol% Chol.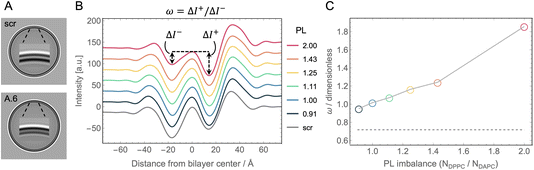 | ||
| Fig. 5 Cryo-EM intensity profiles calculated from simulation trajectories. (A) Synthetic cryo-EM images of 100 nm-diameter vesicles corresponding to the scramble (top) and asymmetric A.6 (bottom, see Table 1) simulated bilayers. Shown in the middle of each vesicle is an enlargement of the intensity close to and across the bilayer, showing how the contrast between the outer and inner troughs in the two images is reversed, consistent with the change in PL imbalance in the bilayer leaflets. (B) Intensity profiles for the asymmetric bilayers (color-coded according to their PL imbalance), and the scrambled bilayer (gray) computationally derived from the simulated atomic number density profiles and membrane dipole potential as described in Methods. The outer (ΔI+) and inner (ΔI−) trough depths relative to the central peak which define the parameter ω, are illustrated for the curve corresponding to a PL imbalance of 2.0. Profiles have been shifted vertically for better visualization. (C) ω as a function of PL imbalance for all asymmetric bilayers (symbols). ω for the scramble membrane is also shown for comparison (dashed line). | ||
Because the intensity profiles arise from electron density variation across the bilayer, their features are sensitive to details of the bilayer structure. For example, the distance between the two deep troughs has been shown to be strongly correlated with bilayer thickness in both simulated and experimental data.91 Similarly, the depth of the troughs can reveal differences in lipid packing density within the leaflets.53 To explore this further, we defined a model-free parameter, ω = ΔI+/ΔI−, calculated as the ratio of the outer (ΔI+) and inner (ΔI−) trough depths relative to the central peak as depicted in Fig. 5B. Fig. 5C shows that ω nearly doubled (from 0.95 to 1.86) when the phospholipid imbalance was varied from 0.9 to 2.0. It is important to note that ω for perfectly symmetric bilayers is not unity but instead ranges from ∼0.65–0.8 (i.e., the outer trough is shallower than the inner trough) due to the spherical geometry of the liposome and the nature of projection imaging, as shown by the scrambled bilayer in Fig. 5B (gray curve). These results demonstrate that a simple model-free analysis can be used to accurately estimate the phospholipid imbalance of liposomes, provided that a sufficient number of liposomes are analyzed to achieve high signal-to-noise in the intensity profile. Whether the trends in ω for asymmetric bilayers can be affected by variations in lipid composition, cholesterol concentration and electron contrast-enhancers (such as substitutions of hydrogen in the lipid molecules with halogen atoms) needs to be further explored.
Conclusion
While the compositional asymmetry of biological membranes has been known for over half a century, it has only recently been recognized that an asymmetric membrane cannot solely be defined as just having leaflets with different lipid compositions. Imbalances in the total lipid abundances of the two monolayers can accompany interleaflet compositional variations and have profound effects on the bilayer biophysical properties. This makes the detection and characterization of both compositional and number asymmetries critical for analysis of model and cell membranes. Existing biophysical techniques have different sensitivities to the precise interleaflet lipid organization in a bilayer, that can be maximized by optimizing experimental conditions and data analysis protocols. Coupling the in vitro approaches with simulations of corresponding atomistic models holds great promise for refining these methodologies, fully defining the state of the experimental systems and uncovering the full potential of membrane asymmetry for shaping and regulating bilayer properties.Data availability
The data corresponding to this article have been stored at https://zenodo.org/records/14626488, https://zenodo.org/records/14655810, and https://zenodo.org/records/14627064.Author contributions
MD: conceptualization, data curation, formal analysis, investigation, project administration, writing – original draft, writing – review & editing. FAH: formal analysis, methodology, writing – original draft, writing – review & editing.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
This research was supported by NIH grant R01GM138887 (to FAH) and the SciLifeLab & Wallenberg Data Driven Life Science Program grant: KAW 2024.0159 (to MD). The table of contents entry graphic was created in https://BioRender.com.References
- G. van Meer, D. R. Voelker and G. W. Feigenson, Membrane lipids: where they are and how they behave, Nat. Rev. Mol. Cell Biol., 2008, 9(2), 112–124 CrossRef CAS PubMed.
- R. M. Venable, F. L. Brown and R. W. Pastor, Mechanical properties of lipid bilayers from molecular dynamics simulation, Chem. Phys. Lipids, 2015, 192, 60–74 CrossRef CAS PubMed.
- S. D. Guler, D. D. Ghosh, J. Pan, J. C. Mathai, M. L. Zeidel, J. F. Nagle and S. Tristram-Nagle, Effects of ether vs. ester linkage on lipid bilayer structure and water permeability, Chem. Phys. Lipids, 2009, 160(1), 33–44 CrossRef CAS PubMed.
- J. C. Mathai, S. Tristram-Nagle, J. F. Nagle and M. L. Zeidel, Structural determinants of water permeability through the lipid membrane, J. Gen. Physiol., 2008, 131(1), 69–76 CrossRef CAS PubMed.
- N. Kucerka, F. A. Heberle, J. Pan and J. Katsaras, Structural Significance of Lipid Diversity as Studied by Small Angle Neutron and X-ray Scattering, Membranes, 2015, 5(3), 454–472 Search PubMed.
- D. Marsh Handbook of Lipid Bilayers; CRC Press, 2013 Search PubMed.
- I. Levental, K. R. Levental and F. A. Heberle, Lipid Rafts: Controversies Resolved, Mysteries Remain, Trends Cell Biol., 2020, 30(5), 341–353 Search PubMed.
- E. Sezgin, I. Levental, S. Mayor and C. Eggeling, The mystery of membrane organization: composition, regulation and roles of lipid rafts, Nat. Rev. Mol. Cell Biol., 2017, 18(6), 361–374 CrossRef CAS PubMed.
- I. Levental and E. Lyman, Regulation of membrane protein structure and function by their lipid nano-environment, Nat. Rev. Mol. Cell Biol., 2023, 24(2), 107–122 Search PubMed.
- H. A. Faizi, A. Tsui, R. Dimova and P. M. Vlahovska, Bending Rigidity, Capacitance, and Shear Viscosity of Giant Vesicle Membranes Prepared by Spontaneous Swelling, Electroformation, Gel-Assisted, and Phase Transfer Methods: A Comparative Study, Langmuir, 2022, 38(34), 10548–10557 CrossRef CAS PubMed.
- G. Enkavi, M. Javanainen, W. Kulig, T. Rog and I. Vattulainen, Multiscale Simulations of Biological Membranes: The Challenge To Understand Biological Phenomena in a Living Substance, Chem. Rev., 2019, 119(9), 5607–5774 CrossRef CAS PubMed.
- R. Ashkar, H. Bilheux, H. Bordallo, R. Briber, D. J. Callaway, X. Cheng, X.-Q. Chu, J. E. Curtis, M. Dadmun and P. Fenimore, Neutron scattering in the biological sciences: Progress and prospects, Acta Crystallogr., Sect. D:Struct. Biol., 2018, 74(12), 1129–1168 Search PubMed.
- K. J. Mallikarjunaiah, J. J. Kinnun, H. I. Petrache and M. F. Brown, Flexible lipid nanomaterials studied by NMR spectroscopy, Phys. Chem. Chem. Phys., 2019, 21(34), 18422–18457 Search PubMed.
- A. J. Verkleij, R. F. Zwaal, B. Roelofsen, P. Comfurius, D. Kastelijn and L. L. van Deenen, The asymmetric distribution of phospholipids in the human red cell membrane. A combined study using phospholipases and freeze-etch electron microscopy, Biochim. Biophys. Acta, Biomembr., 1973, 323(2), 178–193 CrossRef CAS PubMed.
- P. F. Devaux, Static and dynamic lipid asymmetry in cell membranes, Biochemistry, 1991, 30(5), 1163–1173 CrossRef CAS PubMed.
- J. H. Lorent, K. R. Levental, L. Ganesan, G. Rivera-Longsworth, E. Sezgin, M. Doktorova, E. Lyman and I. Levental, Plasma membranes are asymmetric in lipid unsaturation, packing and protein shape, Nat. Chem. Biol., 2020, 16(6), 644–652 CrossRef CAS PubMed.
- T. Yeung, G. E. Gilbert, J. Shi, J. Silvius, A. Kapus and S. Grinstein, Membrane phosphatidylserine regulates surface charge and protein localization, Science, 2008, 319(5860), 210–213 CrossRef CAS PubMed.
- A. Gupta, T. Korte, A. Herrmann and T. Wohland, Plasma membrane asymmetry of lipid organization: fluorescence lifetime microscopy and correlation spectroscopy analysis, J. Lipid Res., 2020, 61(2), 252–266 Search PubMed.
- C. Patra, M. Samad, T. S. van Zanten, M. Singh, R. Vishwakarma, P. Pal Singh and S. Mayor, Solvatochromic reporter to image plasma membrane order leaflet by leaflet reveals a highly asymmetric bilayer locally modulated by transbilayer interactions, bioRxiv, 2024, preprint, DOI:10.1101/2024.07.23.604763.
- A. A. Gurtovenko and I. Vattulainen, Membrane potential and electrostatics of phospholipid bilayers with asymmetric transmembrane distribution of anionic lipids, J. Phys. Chem. B, 2008, 112(15), 4629–4634 Search PubMed.
- D. W. Allender, A. J. Sodt and M. Schick, Cholesterol-Dependent Bending Energy Is Important in Cholesterol Distribution of the Plasma Membrane, Biophys. J., 2019, 116(12), 2356–2366 CrossRef CAS PubMed.
- A. Hossein and M. Deserno, Spontaneous Curvature, Differential Stress, and Bending Modulus of Asymmetric Lipid Membranes, Biophys. J., 2020, 118(3), 624–642 CrossRef CAS PubMed.
- B. Kollmitzer, P. Heftberger, M. Rappolt and G. Pabst, Monolayer spontaneous curvature of raft-forming membrane lipids, Soft Matter, 2013, 9(45), 10877–10884 Search PubMed.
- M. Varma and M. Deserno, Distribution of cholesterol in asymmetric membranes driven by composition and differential stress, Biophys. J., 2022, 121(20), 4001–4018 Search PubMed.
- H. L. Scott, K. B. Kennison, T. A. Enoki, M. Doktorova, J. J. Kinnun, F. A. Heberle and J. Katsaras, Model Membrane Systems Used to Study Plasma Membrane Lipid Asymmetry, Symmetry, 2021, 13(8), 1356 Search PubMed.
- J. Niu, Y. Ma, Y. Yang, H. Lv, J. Wang, T. Wang, F. Liu, S. Xu, Z. Jiang and W. Lin, Lighting up the changes of plasma membranes during apoptosis with fluorescent probes, Coord. Chem. Rev., 2023, 476, 214926 CrossRef CAS.
- J. R. St Clair, Q. Wang, G. Li and E. London, Preparation and Physical Properties of Asymmetric Model Membrane Vesicles, in The Biophysics of Cell Membranes, ed. Epand, R. and Ruysschaert, J. M., Springer, 2017, vol. 19, pp. 1–27 Search PubMed.
- M. Doktorova, J. L. Symons and I. Levental, Structural and functional consequences of reversible lipid asymmetry in living membranes, Nat. Chem. Biol., 2020, 16(12), 1321–1330 Search PubMed.
- M. Doktorova, F. A. Heberle, B. Eicher, R. F. Standaert, J. Katsaras, E. London, G. Pabst and D. Marquardt, Preparation of asymmetric phospholipid vesicles for use as cell membrane models, Nat. Protoc., 2018, 13(9), 2086–2101 Search PubMed.
- M. Doktorova and H. Weinstein, Accurate in silico modeling of asymmetric bilayers based on biophysical principles, Biophys. J., 2018, 115(9), 1638–1643 CrossRef CAS PubMed.
- S. Park, A. H. Beaven, J. B. Klauda and W. Im, How Tolerant are Membrane Simulations with Mismatch in Area per Lipid between Leaflets?, J. Chem. Theory Comput., 2015, 11(7), 3466–3477 CrossRef CAS PubMed.
- S. L. Foley, M. Varma, A. Hossein and M. Deserno, Elastic and thermodynamic consequences of lipid membrane asymmetry, Emerging Top. Life Sci., 2023, 7(1), 95–110 Search PubMed.
- A. Sreekumari and R. Lipowsky, Large stress asymmetries of lipid bilayers and nanovesicles generate lipid flip-flops and bilayer instabilities, Soft Matter, 2022, 18(32), 6066–6078 Search PubMed.
- L. Hua, M. Kaiser, I. Carabadjac, A. Meister, G. Hause and H. Heerklotz, Vesicle budding caused by lysolipid-induced asymmetry stress, Biophys. J., 2023, 122(20), 4011–4022 CrossRef CAS PubMed.
- N. Bettache, L. Baisamy, S. Baghdiguian, B. Payrastre, P. Mangeat and A. Bienvenue, Mechanical constraint imposed on plasma membrane through transverse phospholipid imbalance induces reversible actin polymerization via phosphoinositide 3-kinase activation, J. Cell Sci., 2003, 116(11), 2277–2284 CrossRef CAS PubMed.
- H. Heerklotz, Membrane stress and permeabilization induced by asymmetric incorporation of compounds, Biophys. J., 2001, 81(1), 184–195 CrossRef CAS PubMed.
- T. Fujii and A. Tamura, Shape change of human erythrocytes induced by phosphatidylcholine and lysophosphatidylcholine species with various acyl chain lengths, Cell Biochem. Funct., 1984, 2(3), 171–176 Search PubMed.
- Y. Elani, S. Purushothaman, P. J. Booth, J. M. Seddon, N. J. Brooks, R. V. Law and O. Ces, Measurements of the effect of membrane asymmetry on the mechanical properties of lipid bilayers, Chem. Commun., 2015, 51(32), 6976–6979 Search PubMed.
- B. W. Rickeard, M. H. L. Nguyen, M. DiPasquale, C. G. Yip, H. Baker, F. A. Heberle, X. Zuo, E. G. Kelley, M. Nagao and D. Marquardt, Transverse lipid organization dictates bending fluctuations in model plasma membranes, Nanoscale, 2020, 12(3), 1438–1447 Search PubMed.
- M. P. K. Frewein, P. Piller, E. F. Semeraro, O. Czakkel, Y. Gerelli, L. Porcar and G. Pabst, Distributing aminophospholipids asymmetrically across leaflets causes anomalous membrane stiffening, Biophys. J., 2023, 122(12), 2445–2455 CrossRef CAS PubMed.
- S. Park, W. Im and R. W. Pastor, Developing initial conditions for simulations of asymmetric membranes: a practical recommendation, Biophys. J., 2021, 120(22), 5041–5059 Search PubMed.
- M. Doktorova, I. Levental and F. A. Heberle, Seeing the Membrane from Both Sides Now: Lipid Asymmetry and Its Strange Consequences, Cold Spring Harbor Perspect. Biol., 2023, a041393 CrossRef CAS PubMed.
- S. Jo, T. Kim, V. G. Iyer and W. Im, CHARMM-GUI: a web-based graphical user interface for CHARMM, J. Comput. Chem., 2008, 29(11), 1859–1865 CrossRef CAS PubMed.
- J. Lee, X. Cheng, J. M. Swails, M. S. Yeom, P. K. Eastman, J. A. Lemkul, S. Wei, J. Buckner, J. C. Jeong and Y. Qi, et al., CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field, J. Chem. Theory Comput., 2016, 12(1), 405–413 CrossRef CAS PubMed.
- B. R. Brooks, C. L. Brooks, A. D. Mackerell, L. Nilsson, R. J. Petrella, B. Roux, Y. Won, G. Archontis, C. Bartels and S. Boresch, et al., CHARMM: The Biomolecular Simulation Program, J. Comput. Chem., 2009, 30(10), 1545–1614 CrossRef CAS PubMed.
- M. Doktorova, J. L. Symons, X. Zhang, H.-Y. Wang, J. Schlegel, J. H. Lorent, F. A. Heberle, E. Sezgin, E. Lyman, K. R. Levental and I. Levental, Cell Membranes Sustain Phospholipid Imbalance Via Cholesterol Asymmetry, Cell, 2025 DOI:10.1016/j.cell.2025.02.034.
- J. C. Phillips, D. J. Hardy, J. D. C. Maia, J. E. Stone, J. V. Ribeiro, R. C. Bernardi, R. Buch, G. Fiorin, J. Henin and W. Jiang, et al., Scalable molecular dynamics on CPU and GPU architectures with NAMD, J. Chem. Phys., 2020, 153(4), 044130 CrossRef CAS PubMed.
- J. B. Klauda, R. M. Venable, J. A. Freites, J. W. O’Connor, D. J. Tobias, C. Mondragon-Ramirez, I. Vorobyov, A. D. MacKerell Jr and R. W. Pastor, Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types, J. Phys. Chem. B, 2010, 114(23), 7830–7843 CrossRef CAS PubMed.
- M. Doktorova, G. Khelashvili, R. Ashkar and M. F. Brown, Molecular simulations and NMR reveal how lipid fluctuations affect membrane mechanics, Biophys. J., 2023, 122(6), 984–1002 CrossRef CAS PubMed.
- W. Humphrey, A. Dalke and K. Schulten, VMD: visual molecular dynamics, J. Mol. Graphics, 1996, 14(1), 33–38 CrossRef CAS PubMed.
- J. N. Sachs, P. S. Crozier and T. B. Woolf, Atomistic simulations of biologically realistic transmembrane potential gradients, J. Chem. Phys., 2004, 121(22), 10847–10851 CrossRef CAS PubMed.
- N. Kucerka, J. Pencer, J. N. Sachs, J. F. Nagle and J. Katsaras, Curvature effect on the structure of phospholipid bilayers, Langmuir, 2007, 23(3), 1292–1299 CrossRef CAS PubMed.
- K. D. Sharma, M. Doktorova, M. N. Waxham and F. A. Heberle, Cryo-EM images of phase-separated lipid bilayer vesicles analyzed with a machine-learning approach, Biophys. J., 2024, 123(17), 2877–2891 CrossRef CAS PubMed.
- E. J. Kirkland, Advanced Computing in Electron Microscopy, Springer, 1998 Search PubMed.
- F. Leeb and L. Maibaum, Spatially Resolving the Condensing Effect of Cholesterol in Lipid Bilayers, Biophys. J., 2018, 115(11), 2179–2188 CrossRef CAS PubMed.
- W. C. Hung, M. T. Lee, F. Y. Chen and H. W. Huang, The condensing effect of cholesterol in lipid bilayers, Biophys. J., 2007, 92(11), 3960–3967 CrossRef CAS PubMed.
- M. Alwarawrah, J. Dai and J. Huang, A molecular view of the cholesterol condensing effect in DOPC lipid bilayers, J. Phys. Chem. B, 2010, 114(22), 7516–7523 CrossRef CAS PubMed.
- H. T. Cheng and E. London, Preparation and properties of asymmetric large unilamellar vesicles: interleaflet coupling in asymmetric vesicles is dependent on temperature but not curvature, Biophys. J., 2011, 100(11), 2671–2678 CrossRef CAS PubMed.
- B. Eicher, D. Marquardt, F. A. Heberle, I. Letofsky-Papst, G. N. Rechberger, M. S. Appavou, J. Katsaras and G. Pabst, Intrinsic Curvature-Mediated Transbilayer Coupling in Asymmetric Lipid Vesicles, Biophys. J., 2018, 114(1), 146–157 CrossRef CAS PubMed.
- M. A. Perez and P. A. Beales, Dynamics of asymmetric membranes and interleaflet coupling as intermediates in membrane fusion, Biophys. J., 2023, 122(11), 1985–1995 CrossRef PubMed.
- B. L. Mui, H. G. Dobereiner, T. D. Madden and P. R. Cullis, Influence of transbilayer area asymmetry on the morphology of large unilamellar vesicles, Biophys. J., 1995, 69(3), 930–941 CrossRef CAS PubMed.
- R. Dimova, Giant vesicles and their use in assays for assessing membrane phase state, curvature, mechanics, and electrical properties, Annu. Rev. Biophys., 2019, 48(1), 93–119 CrossRef CAS PubMed.
- A. J. Sodt, R. M. Venable, E. Lyman and R. W. Pastor, Nonadditive Compositional Curvature Energetics of Lipid Bilayers, Phys. Rev. Lett., 2016, 117(13), 138104 CrossRef CAS PubMed.
- M. Deserno, Fluid lipid membranes: from differential geometry to curvature stresses, Chem. Phys. Lipids, 2015, 185, 11–45 CrossRef CAS PubMed.
- F. Ragaller, E. Sjule, Y. B. Urem, J. Schlegel, R. El, D. Urbancic, I. Urbancic, H. Blom and E. Sezgin, Quantifying Fluorescence Lifetime Responsiveness of Environment-Sensitive Probes for Membrane Fluidity Measurements, J. Phys. Chem. B, 2024, 128(9), 2154–2167 CrossRef CAS PubMed.
- D. M. Owen, P. M. Lanigan, C. Dunsby, I. Munro, D. Grant, M. A. Neil, P. M. French and A. I. Magee, Fluorescence lifetime imaging provides enhanced contrast when imaging the phase-sensitive dye di-4-ANEPPDHQ in model membranes and live cells, Biophys. J., 2006, 90(11), L80–L82 CrossRef CAS PubMed.
- K. Strakova, J. Lopez-Andarias, N. Jimenez-Rojo, J. E. Chambers, S. J. Marciniak, H. Riezman, N. Sakai and S. Matile, HaloFlippers: A General Tool for the Fluorescence Imaging of Precisely Localized Membrane Tension Changes in Living Cells, ACS Cent. Sci., 2020, 6(8), 1376–1385 Search PubMed.
- G. Gunther, L. Malacrida, D. M. Jameson, E. Gratton and S. A. Sánchez, LAURDAN since Weber: the quest for visualizing membrane heterogeneity, Acc. Chem. Res., 2021, 54(4), 976–987 CrossRef CAS PubMed.
- F. Ragaller, L. Andronico, J. Sykora, W. Kulig, T. Rog, Y. B. Urem, Abhinav, D. I. Danylchuk, M. Hof and A. Klymchenko, et al., Dissecting the mechanisms of environment sensitivity of smart probes for quantitative assessment of membrane properties, Open Biol., 2022, 12(9), 220175 CrossRef CAS PubMed.
- M. Amaro, F. Reina, M. Hof, C. Eggeling and E. Sezgin, Laurdan and Di-4-ANEPPDHQ probe different properties of the membrane, J. Phys. D: Appl. Phys., 2017, 50(13), 134004 CrossRef PubMed.
- M. N. Doktorova, Biophysics of Asymmetric Membranes: Protocols and Revelations, Weill Medical College of Cornell University, 2018 Search PubMed.
- R. Dasgupta, M. S. Miettinen, N. Fricke, R. Lipowsky and R. Dimova, The glycolipid GM1 reshapes asymmetric biomembranes and giant vesicles by curvature generation, Proc. Natl. Acad. Sci. U.S.A., 2018, 115(22), 5756–5761 CrossRef CAS PubMed.
- M. Doktorova, N. Kucerka, J. J. Kinnun, J. Pan, D. Marquardt, H. L. Scott, R. M. Venable, R. W. Pastor, S. R. Wassall and J. Katsaras, et al., Molecular Structure of Sphingomyelin in Fluid Phase Bilayers Determined by the Joint Analysis of Small-Angle Neutron and X-ray Scattering Data, J. Phys. Chem. B, 2020, 124(25), 5186–5200 CrossRef CAS PubMed.
- T. R. Molugu, X. Xu, A. Leftin, S. Lope-Piedrafita, G. V. Martinez, H. I. Petrache and M. F. Brown, Solid-state deuterium NMR spectroscopy of membranes, Mod. Magn. Reson., 2017, 1–23 Search PubMed.
- A. A. Nevzorov, T. P. Trouard and M. F. Brown, Lipid bilayer dynamics from simultaneous analysis of orientation and frequency dependence of deuterium spin-lattice and quadrupolar order relaxation, Phys. Rev. E:Stat. Phys., Plasmas, Fluids, Relat. Interdiscip. Top., 1998, 58(2), 2259 CrossRef CAS.
- D. B. Fenske and P. R. Cullis, Acyl chain orientational order in large unilamellar vesicles: comparison with multilamellar liposomes: a 2H and 31P nuclear magnetic resonance study, Biophys. J., 1993, 64(5), 1482–1491 CrossRef CAS PubMed.
- O. Cruciani, L. Mannina, A. P. Sobolev, C. Cametti and A. Segre, An improved NMR study of liposomes using 1-palmitoyl-2-oleoyl-sn-glycero-3-phospatidylcholine as model, Molecules, 2006, 11(5), 334–344 Search PubMed.
- W. K. Subczynski, J. Widomska and J. B. Feix, Physical properties of lipid bilayers from EPR spin labeling and their influence on chemical reactions in a membrane environment, Free Radical Biol. Med., 2009, 46(6), 707–718 CrossRef CAS PubMed.
- F. A. Heberle, J. Pan, R. F. Standaert, P. Drazba, N. Kucerka and J. Katsaras, Model-based approaches for the determination of lipid bilayer structure from small-angle neutron and X-ray scattering data, Eur. Biophys. J., 2012, 41(10), 875–890 CrossRef CAS PubMed.
- E. F. Semeraro, L. Marx, M. P. Frewein and G. Pabst, Increasing complexity in small-angle X-ray and neutron scattering experiments: from biological membrane mimics to live cells, Soft Matter, 2021, 17(2), 222–232 RSC.
- M. R. Brzustowicz and A. T. Brunger, X-ray scattering from unilamellar lipid vesicles, J. Appl. Crystallogr., 2005, 38(1), 126–131 CrossRef CAS.
- F. A. Heberle, D. Marquardt, M. Doktorova, B. Geier, R. F. Standaert, P. Heftberger, B. Kollmitzer, J. D. Nickels, R. A. Dick and G. W. Feigenson, et al., Subnanometer Structure of an Asymmetric Model Membrane: Interleaflet CouplingInfluences Domain Properties, Langmuir, 2016, 32(20), 5195–5200 CrossRef CAS PubMed.
- M. P. K. Frewein, P. Piller, E. F. Semeraro, K. C. Batchu, F. A. Heberle, H. L. Scott, Y. Gerelli, L. Porcar and G. Pabst, Interdigitation-Induced Order and Disorder in Asymmetric Membranes, J. Membr. Biol., 2022, 255(4–5), 407–421 CrossRef CAS PubMed.
- B. Eicher, F. A. Heberle, D. Marquardt, G. N. Rechberger, J. Katsaras and G. Pabst, Joint small-angle X-ray and neutron scattering data analysis of asymmetric lipid vesicles, J. Appl. Crystallogr., 2017, 50(2), 419–429 CrossRef CAS PubMed.
- E. F. Semeraro, M. P. Frewein and G. Pabst, Structure of symmetric and asymmetric lipid membranes from joint SAXS/SANS, Methods Enzymol., 2024, 700, 349–383 Search PubMed.
- Y. Tahara and Y. Fujiyoshi, A new method to measure bilayer thickness: cryo-electron microscopy of frozen hydrated liposomes and image simulation, Micron, 1994, 25(2), 141–149 CrossRef CAS PubMed.
- K. D. Sharma, F. A. Heberle and M. N. Waxham, Visualizing lipid membrane structure with cryo-EM: past, present, and future, Emerging Top. Life Sci., 2023, 7(1), 55–65 CrossRef CAS PubMed.
- F. A. Heberle and M. N. Waxham, Phase separation in model lipid membranes investigated with cryogenic electron microscopy, Methods Enzymol., 2024, 700, 189–216 Search PubMed.
- E. Lafarge, C. Marques, M. Schmutz, P. Muller and A. Schroder, Thickness determination of hydroperoxidized lipid bilayers from medium-resolution cryo-TEM images, Biophysical Approaches for the Study of Membrane Structure-Part A: Experimental, 2024, vol. 700, pp. 329–348 Search PubMed.
- L. Wang, P. S. Bose and F. J. Sigworth, Using cryo-EM to measure the dipole potential of a lipid membrane, Proc. Natl. Acad. Sci. U. S. A., 2006, 103(49), 18528–18533 CrossRef CAS PubMed.
- F. A. Heberle, M. Doktorova, H. L. Scott, A. D. Skinkle, M. N. Waxham and I. Levental, Direct label-free imaging of nanodomains in biomimetic and biological membranes by cryogenic electron microscopy, Proc. Natl. Acad. Sci. U.S.A., 2020, 117(33), 19943–19952 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2025 |

