Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Correction: Triplex-forming peptide nucleic acid modified with 2-aminopyridine as a new tool for detection of A-to-I editing
Chiara
Annoni
a,
Tamaki
Endoh
a,
Dziyana
Hnedzko
b,
Eriks
Rozners
b and
Naoki
Sugimoto
*ac
aFrontier Institute for Biomolecular Engineering Research (FIBER), Konan University, 7-1-20 Minatojima-minamimachi, Chuo-ku, Kobe 650-0047, Japan. E-mail: sugimoto@konan-u.ac.jp
bDepartment of Chemistry, Binghamton University, State University of New York, Binghamton, New York 13902, USA
cFaculty of Frontiers of Innovative Research in Science and Technology (FIRST), Konan University, 7-1-20 Minatojima-minamimachi, Chuo-ku, Kobe 650-0047, Japan
First published on 28th October 2016
Abstract
Correction for ‘Triplex-forming peptide nucleic acid modified with 2-aminopyridine as a new tool for detection of A-to-I editing’ by Chiara Annoni et al., Chem. Commun., 2016, 52, 7935–7938.
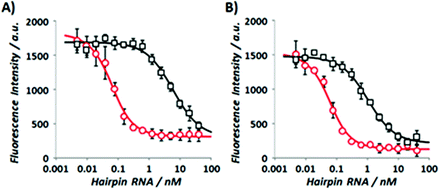
The authors regret that minor deviations in buffer pH due to a faulty pH meter have been identified in the experiments that provided the data presented in the original article, all measured pH values have since been found to be slightly higher than the true pH values. As the affinities of the triplex-forming peptide nucleic acids for both the wild-type and the edited RNA hairpins are sensitive to buffer pH, the experiments furnishing the binding constants and other relevant data, reported in Fig. 1, Table 1 and Fig. 2 of the original article, as well as Fig. S2, S3, S6 and S7 and Tables S2–S5 of the original ESI, have been re-run. Revised versions of Fig. 1, Table 1 and Fig. 2 are included herein. The ESI attached to the original article has also been updated accordingly.
| Hairpin RNA | K A25 (×109 M−1) | |||||
|---|---|---|---|---|---|---|
| pH 6.4 | pH 6.6 | pH 6.8 | pH 7.0 | pH 7.2 | pH 7.4 | |
| a The reported values are means ± standard deviations from at least three independent measurements. b The value could not be determined due to low affinity. | ||||||
| P5AC | 8.66 ± 1.8 | 2.44 ± 0.44 | 0.522 ± 0.02 | 0.173 ± 0.02 | 0.051 ± 0.02 | n.d.b |
| P5IC | 125 ± 17 | 97.2 ± 11 | 140 ± 42 | 27.2 ± 7.5 | 5.16 ± 1.1 | 0.750 ± 0.13 |
| P5AU | 63.8 ± 11 | 27.0 ± 2.4 | 7.82 ± 0.62 | 0.971 ± 0.05 | 0.157 ± 0.04 | n.d.b |
| P5IU | 101 ± 17 | 106 ± 16 | 82.1 ± 12 | 29.5 ± 1.4 | 5.28 ± 0.56 | 1.18 ± 0.12 |
As a result of the identified errors, the originally quoted limit of detection (LOD) values for the inosine-containing hairpin RNAs, P3IC, P4IC, P5IC and P5IU, are slightly increased to 17.7, 23.7, 13.0, and 17.3 pM, respectively (Table S4), but still remain 1000 times lower than the reported experimental conditions adopted by fluorometric methodology.
The Royal Society of Chemistry apologises for these errors and any consequent inconvenience to authors and readers.
| This journal is © The Royal Society of Chemistry 2016 |