Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Unusual topological RNA G-quadruplex formed by an RNA duplex: implications for the dimerization of SARS-CoV-2 RNA†
Shiyu
Wang‡
a,
Yi
Song‡
a,
Zhiyong
He
a,
Hisao
Saneyoshi
a,
Rie
Iwakiri
a,
Pengyu
Xu
b,
Chuanqi
Zhao
 c,
Xiaogang
Qu
c,
Xiaogang
Qu
 cd and
Yan
Xu
cd and
Yan
Xu
 *a
*a
aDivision of Chemistry, Department of Medical Sciences, Faculty of Medicine, University of Miyazaki, 5200 Kihara, Kiyotake, Miyazaki 889-1692, Japan. E-mail: xuyan@med.miyazaki-u.ac.jp
bShonan Laboratory, Corporate R&D Headquarters, Otsuka Chemical Co., Ltd., 2Chome26-1, Muraokahigashi, Fujisawa, Kanagawa 251-8555, Japan
cLaboratory of Chemical Biology and State Key Laboratory of Rare Earth Resource Utilization, Changchun Institute of Applied Chemistry, Chinese Academy of Science, Changchun, Jilin 130022, P. R. China
dSchool of Applied Chemistry and Engineering, University of Science and Technology of China, Hefei, Anhui 230026, P. R. China
First published on 11th October 2023
Abstract
The infectious disease coronavirus 2019 (SARS-CoV-2) is caused by a virus that has RNA as its genetic material. To understand the detailed structural features of SARS-COV-2 RNA, we probed the RNA structure by NMR. Two RNA sequences form a duplex and self-associate to form a dimeric G-quadruplex. The FrG nucleoside was employed as a 19F sensor to confirm the RNA structure in cells by 19F NMR. A FRET assay further demonstrated that the dimeric G-quadruplex resulted in RNA dimerization in cells. These results provide the basis for the elucidation of SARS-COV-2 RNA function, which provides new insights into developing novel antiviral drugs against SARS-COV-2.
The infectious disease coronavirus 2019 (SARS-COV-2) is caused by a positive-sense, single-stranded RNA virus belonging to the family of enveloped coronaviruses (CoVs).1 The genomic RNA acts as messenger RNA (mRNA) and can be directly translated into viral proteins, including spike (S), envelope (E), membrane (M) and nucleocapsid (N) proteins.
Guanine-rich sequences in RNA form G-quadruplex structures, which were recently shown to exist in human cells.2 RNA G-quadruplexes are widely used as essential biological regulators, especially in transcriptional control, translation processes and telomere homeostasis.3 Therefore, G-quadruplexes have been identified as promising therapeutic targets.4 For example, we and other groups have demonstrated that telomeric RNA G-quadruplexes are formed in human cells and play an important role in providing a protective structure for telomere ends.5 It is also suggested that RNA G-quadruplexes exist in some viruses and regulate the life cycle of viruses.6 Bioinformatics analysis of the SARS-COV-2 RNA sequence suggests that there are several potential sequences that could form an RNA G-quadruplex.7 Defining the structural features of SARS-COV-2 RNA will be essential to understanding the molecular details of this virus and further identifying candidate drugs and treatments.
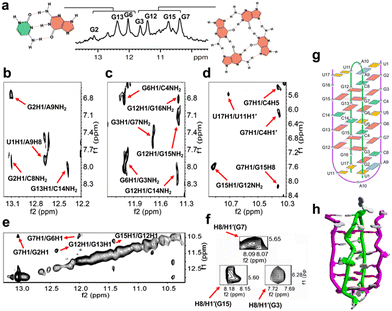
To achieve this goal, we revealed the detailed structure of the human SARS-COV-2 RNA sequence 5′-UGGCUGGCAAUGGCGGU-3′ (ORN-1) located in the open reading frame region of the nucleocapsid gene by 1H-NMR investigation (Fig. 1 and 2). The imino region of the 1H NMR spectrum of ORN-1 in the presence of K+ is shown in Fig. 2a. One set of imino proton resonances suggests one main conformation. Imino protons resonating in the range of 12.0–13.5 ppm are typical of Watson–Crick base pairs, and those at approximately 11.0 ppm are characteristic of Hoogsteen base pairs, such as G:G, indicating that the structure formed by ORN-1 is far more complex.
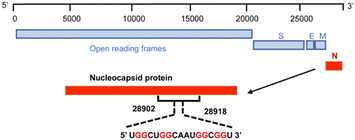 | ||
| Fig. 1 Scheme of 17-mer RNA located on the region of the nucleocapsid gene from 28902 to 28918 in the SARS-COV-2 RNA genome. | ||
The 2D NOESY spectrum was further used in the assignment of protons to determine the structural characteristics of SARS-COV-2 RNA (Fig. 2b–f). A continuous set of sequential H6/H8-H1′ NOE connectivities could be traced, as shown in Fig. S1, ESI.† The NOE peaks of G2H8/C8H5, G2H1/C8NH2, G6H1/C4NH2, and G13H1/C14NH2 were observed and allowed unambiguous identification of G2:C8, G6:C4 and G13:C14 Watson–Crick base pairs. Correspondingly, U1:A9 also exists in the conformation through analysis of the relative cross peaks of U1H1/A9H8. Relative NOE peaks of mismatch U17:U11 were observed, as indicated by U17H1/U11H1′ (Fig. 2b–d and g and Fig. S1, ESI†). A G-tetrad consisting of G3:G7:G15:G12 was observed and proven by the NOE peaks of G3H1/G7NH2, G7H1/G15H8, G12H1/G15NH2 and G15H1/G12NH2 (Fig. 2c and d), consistent with the imino NOE connection G12H1/G15H1 (Fig. 2e), which revealed the formation of a G-tetrad. The syn-glycosidic conformations of G3, G7 and G15 are supported by the observation of the strong presence of intraresidue H8/H1′ NOEs (Fig. 2f). Interaction signals of adjacent nucleosides near the G-tetrad framework were also discovered. For example, the NOE cross peaks of G2H1/A9NH2, G2H1/G7H1, G6H1/G3NH2, G7H1/C4H5, and G7H1/C4H1′ were observed and obviously indicate G3:G7 of the G-quartet in the middle of G2:C8 and G6:C4 (Fig. 2b–e). On the other side of the G-tetrad, G12H8/U17H1′, G12H1/G16NH2 and G12H1/C14NH2 were observed and demonstrate that G15:G12 was caught between G13:C14 and U11:U17 (Fig. 2c and Fig. S1, ESI†). U5 is bulged out in the intercalation region, adopting a looped-out conformation (Fig. S3, ESI†). All results demonstrated significant evidence for the formation of a unique architecture constructed by dimeric RNA (Fig. 2g, h and Fig. S2, ESI†). Next, a RNase T1 footprinting assay (Fig. S4, ESI†) was studied.8 RNase T1 catalyzes cleavage at rGs in single-stranded RNA but not rGs that are involved in secondary structure. The rGs were protected by a higher-order structure against scission by RNase T1 in the presence of 100 mM Na+ and K+ compared with the condition without ions. We performed the ESI-MS experiment to further analyze ORN-1. As shown in Fig. S5, ESI,† ions near m/z [1875.0580] were interpreted as [2M + 6Cl− + 3NH4+ − 3H+]6−. These results indicate the presence of a dimeric structure of ORN-1 with a mass of MW × 2 and consistently support the formation of dimeric G-quadruplexes.
CD experiments were performed on ORN-1 in the presence of K+ or Li+ ions (Fig. S6, ESI†). The CD spectrum of ORN-1 in the presence of K+ ions displayed the characteristic CD features of dimeric G-quadruplexes, including a positive peak at 270 nm and a negative peak at 240 nm, identical to those reported for dimeric G-quadruplexes.9 On the other hand, the CD spectrum of ORN-1 in the presence of Li+ ions exhibited a single positive peak at 265 nm, suggesting the presence of a non-G-quadruplex structure for ORN-1. CD melting study revealed a Tm value of 51 °C for the folded RNA architecture of ORN-1 in a 100 mM KCl solution, indicating its stability. However, no discernible Tm value was observed for ORN-1 in the presence of Li+ ions. Moreover, a comparison with the NMR spectra of ORN-1 in K+ or Li+ indicated that ORN-1 is essentially unstructured in Li+ (Fig. S7, ESI†). Similarly, the gel electrophoresis assay yielded consistent results, showing a band with high mobility in the presence of Li+ ions (Fig. S8, ESI†). The CD spectra exhibited nearly identical structural features for 5, 10, and 100 μM ORN-1. With increasing RNA concentrations, a slight increment in the Tm value was observed (Fig. S9a and S9b, ESI†). No hysteresis is observed for ORN-1 (Fig. S9c, ESI†).
We next investigated whether G-quadruplex ligands can stabilize the dimeric structure. PDP, as a pyridostatin derivative, is an ideal alternative whereas certain pyridostatin derivatives have been reported to exhibit capacity to bind dimeric G-quadruplexes.10 Here, a significantly increased thermal stability of the dimeric G-quadruplex with PDP at physiological ionic strength was found, demonstrating the binding of PDP to dimeric RNA (Fig. S10, ESI†).
To provide additional evidence supporting the formation of dimeric G-quadruplex structures, we synthesized a mutated RNA (mutant-RNA1) in which we replaced G16 with adenosine. The CD spectrum of mutant-RNA1 exhibited structural characteristics consistent with ORN-1 in the presence of K+ ions, while an unfolded structural profile was observed in the presence of Li+ ions. Furthermore, the CD melting curve indicated the presence of a stable structure with Tm of approximately 51 °C in K+. In contrast, mutant-RNA1 displayed a fragile structural state in the presence of Li+ ions (Fig. S11, ESI†). Native gel electrophoresis was employed, and the corresponding results revealed the presence of a slower-migrating band of mutant-RNA1, similar to that of ORN-1, in the presence of K+ ions. Conversely, a band with higher mobility was observed in the presence of Li+ ions for mutant-RNA1 (Fig. S12, ESI†). Moreover, the imino proton NMR spectrum demonstrated distinct resonances in mutant-RNA1, which closely resembled the spectrum of ORN-1 (Fig. S13, ESI†). Conversely, no discernible proton signals were evident in the presence of Li+ ions. This observation strongly indicates the formation of a G-quadruplex structure by mutant-RNA1. This is consistent with the substitution of G16 with adenosine disrupting the formation of the intramolecular monomeric G-quadruplex while having no effect on the intermolecular dimeric G-quadruplex.
As a complementary approach, we synthesized mutant-RNA2 by replacing crucial guanines involved in G-quartet formation and G:C base pairing, excluding G16, with adenosines (Fig. S14 and S15, ESI†). Our observations revealed that mutant-RNA2 did not exhibit characteristic signals in the CD spectrum, nor did it produce any discernible resonances in the imino-proton spectra when exposed to either K+ or Li+ ions. Furthermore, in a native gel electrophoresis experiment comparing ORN-1 with mutant-RNA2, ORN-1 displayed a lower mobility (Fig. S12b and S16, ESI†). These consistent findings provide further confirmation that dimeric G-quadruplex formation is primarily driven by intermolecular G-quartets and G:C base pairs, with G16 not playing a role in this process.
Next, we synthesized a longer RNA (ORN-3) which extends from 3′- and 5′-termini of ORN-1 and investigated its structural feature (Fig. S17, ESI†). The CD spectrum of ORN-3 in K+ shows a positive peak at 270 nm and a negative peak around 240 nm, demonstrating dimeric G-quadruplex formation. In contrast, ORN-3 is found to be a single strand similar to ORN-1 in the presence of Li+ ions. The CD melting assay reveals the presence of a stabilized structure in ORN-3, as evidenced by a Tm value of 43 °C. Furthermore, the imino proton spectrum obtained from NMR analysis shows almost identical resonances as those observed in ORN-1, whereas no imino proton signals were observed from ORN-3 in Li+ (Fig. S18, ESI†). Furthermore, the native gel electrophoresis assay indicates a lower mobility from ORN-3 in K+ compared with Li+ (Fig. S19, ESI†). Similar results were also observed from ORN-3 in a mixture of K+ and Li+ (Fig. S20, ESI†). These results consistently indicate that ORN-3 possesses a dimeric G-quadruplex structure in the presence of K+ ions.
Furthermore, a competing assay was employed to investigate the structural stability and interaction of ORN-1 and ORN-3 with other RNAs. We performed a native gel electrophoresis assay by adding telomere RNA r(UAGGGU)2 into the ORN-1 solution (Fig. S21, ESI†). The results showed distinct bands representing ORN-1 and r(UAGGGU)2, and no additional band appeared when the two were mixed together at different ratios. Similarly, mixing ORN-3 with telomere RNA r(UAGGGU)2 or r(UAGGGU)4 showed distinct, individually integral bands representing ORN-3, r(UAGGGU)2, and r(UAGGGU)4 (Fig. S22 and S23, ESI†). These results indicate a lack of significant complex formation between ORN-1, ORN-3 and telomere RNAs.
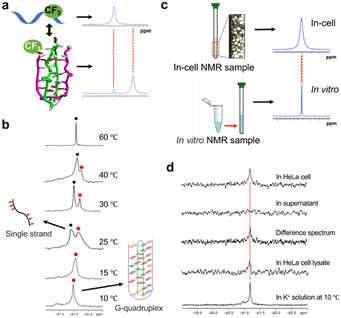
To investigate whether a dimeric G-quadruplex exists in cells, we developed an 8-trifluoromethyl-guanosine (FrG) nucleoside as a 19F sensor and employed it to study the RNA structure in cells by 19F NMR (Fig. 3 and Fig. S24, S27–S43, ESI†).
FrG is site-specific and incorporated at the G16 position of ORN-1, as a 19F sensor provides 19F NMR signals. Thus, it is possible to distinguish different RNA structures by the different chemical shifts in 19F NMR spectra (Fig. 3a). A 19F NMR experiment was performed to investigate the ORN-2 containing FrG in K+ solution (Fig. 3b). One sharp peak at −61.18 ppm was observed at 10 °C, which is consistent with the 1H NMR result showing that the ORN-2 sequence could form an unusual topological RNA G-quadruplex in K+ solution. As the temperature increased, the intensity of the signal decreased; upon heating to 25 °C, a new peak corresponding to the unfolded single strand appeared, and at temperatures over 60 °C, only this peak remained with a strong intensity. 19F NMR experiments further provide explicit evidence that the RNA forms an unusual topological RNA G-quadruplex.
Fig. 3c shows the in-cell 19F NMR spectroscopy strategy, in which comparing the in-cell 19F-NMR spectrum to the in vitro result as a reference enables reliable determination of the intracellular G-quadruplex. ORN-2 was transfected into HeLa cells using an SLO treatment approach with high cell viability and efficient transfection of exogenous biological molecules.11Fig. 3d shows a comparison of the in vitro and in-cell NMR spectra. The in-cell 19F NMR results showed a major peak at approximately −61.18 ppm, which was almost identical to those observed in the K+ solution. Thus, the in-cell 19F NMR spectrum demonstrates that 19F-labeled RNA can be derived from RNA G-quadruplexes in living human cells. We also collected and examined the cell suspension medium by 19F NMR spectroscopy. No signal was observed in this supernatant (Fig. 3d), indicating that all the NMR signals originated from the 19F-labeled RNA within the HeLa cells. A difference spectrum between the HeLa cells and the suspension was produced to eliminate the signal from the supernatant (Fig. 3d). The clear signal in the difference spectrum supports the observation of the RNA G-quadruplex structure in the cells.
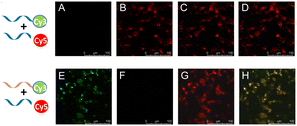
To further confirm that the RNA dimerization resulted from dimeric G-quadruplexes, Förster resonance energy transfer (FRET) was employed to monitor dimer formation (Fig. S25, ESI†). Cy3-RNA and Cy5-RNA sequences containing respectively a Cy3 and a Cy5 fluorophore at the 5′ terminus were prepared for FRET studies, in addition to a control strand Cy3-RNA1 containing Cy3 at the 5′ end. The formation of dimeric G-quadruplexes brings the Cy3 and Cy5 fluorophores into close proximity to display FRET (Fig. S25-a, ESI†). The addition of Cy5-RNA to Cy3-RNA indicated that the Cy3 signal at 570 nm markedly decreased, while the signal for Cy5 at 670 nm increased (Fig. S25-b, ESI†). The control experiment indicated no emission at 670 nm when Cy5-RNA was added to Cy3-RNA1 under the same conditions. This FRET assay enabled visual detection of RNA dimeric G-quadruplex formation. A clear red color was observed by the naked eye when the mixture of Cy5-RNA and Cy3-RNA was excited at the Cy3 absorption wavelength of 540 nm, and Cy5 emission at 670 nm was observed, but not in assays including Cy3-RNA alone or a mixture of Cy5-RNA and control Cy3-RNA1 (Fig. S25-c, ESI†). We next performed an electrophoretic mobility shift assay using Cy3- and Cy5-labeled RNA to confirm the RNA G-quadruplex structure (Fig. S25-d, ESI†). Lines 1, 2 and 3 show the control Cy3-RNA1, Cy3-RNA and complex of Cy3-RNA and Cy5-RNA, respectively. Line 1 was visualized as a green band with a high mobility compared to Cy3-RNA (Line 2) and a complex of Cy3-RNA and Cy5-RNA (Line 3) in Cy3-mode (excitation wavelength = 550 nm, emission wavelength = 570 nm), suggesting that Cy3-RNA forms a G-quadruplex structure. Line 3 showed two bands, which indicated a dimeric G-quadruplex formed by a complex of Cy3-RNA and Cy5-RNA. The dimeric G-quadruplex is visualized as a red band in Cy5 mode (excitation wavelength = 640 nm, emission wavelength = 670 nm), consistent with a yellow band visualized in the overlay of the two modes.
To document direct evidence of RNA dimerization in living cells, RNA probes were applied to living cells (Fig. 4). The key result was the observation of a red signal of Cy3-RNA and Cy5-RNA in image B of Fig. 4 upon excitation at the Cy3 absorption wavelength of 540 nm and the Cy5 emission wavelength of 670 nm, with a control study using Cy3-RNA1 and Cy5-RNA eliciting no emission (image F, Fig. 4) under these conditions. When the Cy3 chromophore was excited using a 540 nm laser and observed at Cy3 emission at 570 nm, no Cy3 green signal was observed (image A, Fig. 4), whereas image E of Fig. 4 clearly shows a significant Cy3 green signal in control experiments using Cy3-RNA1 and Cy5-RNA. In images C and G, a red color was observed in Cy5 mode (excitation wavelength = 640 nm, emission wavelength = 670 nm). Furthermore, RNA dimerization was examined at various concentrations. Overall Cy3-RNA (1–10 μM)/Cy5-RNA (10–100 μM) showed the FRET phenomenon, but control Cy3-RNA1 (1–10 μM) was not observed when mixed with Cy5-RNA in living cells (Fig. S26, ESI†).
In this study, we found a unique RNA architecture formed by SARS-COV-2 RNA, in which two RNA sequences form a duplex and self-associate to form a dimeric G-quadruplex. Thus, such structures may be valuable targets for antiviral agents.
This work was supported by Japan Society for the Promotion of Science KAKENHI [21H02081] and Otsuka Chemical Co., Ltd.
Conflicts of interest
There are no conflicts to declare.References
- P. V'Kovski, A. Kratzel, S. Steiner, H. Stalder and V. Thiel, Nat. Rev. Microbiol., 2021, 19, 155–170 CrossRef PubMed.
- G. Biffi, M. Di Antonio, D. Tannahill and S. Balasubramanian, Nat. Chem., 2014, 6, 75–80 CrossRef CAS PubMed.
- (a) Y. Xu, Chem. Soc. Rev., 2011, 40, 2719–2740 RSC; (b) K. Kawauchi, W. Sugimoto, T. Yasui, K. Murata, K. Itoh, K. Takagi, T. Tsuruoka, K. Akamatsu, H. Tateishi-Karimata, N. Sugimoto and D. Miyoshi, Nat. Commun., 2018, 9, 1–12 CrossRef CAS PubMed.
- (a) J. Abraham Punnoose, Y. Ma, Y. Li, M. Sakuma, S. Mandal, K. Nagasawa and H. Mao, J. Am. Chem. Soc., 2017, 139, 7476–7484 CrossRef CAS PubMed; (b) K.-B. Wang, J. Dickerhoff and D. Yang, J. Am. Chem. Soc., 2021, 143, 16549–16555 CrossRef CAS PubMed.
- (a) Y. Xu, K. Kaminaga and M. Komiyama, J. Am. Chem. Soc., 2008, 130, 11179–11184 CrossRef CAS PubMed; (b) H. Martadinata and A. Phan, J. Am. Chem. Soc., 2009, 131, 2570–2578 CrossRef CAS PubMed.
- (a) E. Ruggiero and S. N. Richter, Nucleic Acids Res., 2018, 46, 3270–3283 CrossRef CAS PubMed; (b) E. Butovskaya, E. Heddi, B. Bakalar, S. N. Richter and A. T. Phan, J. Am. Chem. Soc., 2018, 140, 13654–13662 CrossRef CAS PubMed; (c) E. Ruggiero and S. N. Richter, Bioorg. Med. Chem. Lett., 2023, 79, 129085 CrossRef CAS PubMed.
- C. Zhao, G. Qin, J. Niu, Z. Wang, C. Wang, J. Ren and X. Qu, Angew. Chem., Int. Ed., 2021, 60, 432–438 CrossRef CAS PubMed.
- C. D. Xiao, T. Shibata, Y. Yamamoto and Y. Xu, Chem. Commun., 2018, 54, 3944–3946 RSC.
- G. D. Balkwill, T. P. Garner, H. E. Williams and M. S. Searle, J. Mol. Biol., 2009, 385, 1600–1615 CrossRef CAS PubMed.
- (a) R. Rocca, F. Moraca, G. Costa, M. Nadai, M. Scalabrin, C. Talarico, S. Distinto, E. Maccioni, F. Ortuso and A. Artese, Biochim. Biophys. Acta, Gen. Subj., 2017, 1861, 1329–1340 CrossRef CAS PubMed; (b) M. Arévalo-Ruiz, S. Amrane, F. Rosu, E. Belmonte-Reche, P. Peñalver, J.-L. Mergny and J. C. Morales, Bioorg. Chem., 2020, 99, 103786 CrossRef PubMed; (c) N. H. Campbel, M. Patel, A. B. Tofa, R. Ghosh, G. N. Parkinson and S. Neidle, Biochemistry, 2009, 48, 1675–1680 CrossRef PubMed.
- (a) H. L. Bao, T. Masuzawa, T. Oyoshi and Y. Xu, Nucleic Acids Res., 2020, 48, 7041–7051 CAS; (b) H. L. Bao, T. Ishizuka, T. Sakamoto, K. Fujimoto, T. Uechi, N. Kenmochi and Y. Xu, Nucleic Acids Res., 2017, 45, 5501–5511 CrossRef CAS PubMed; (c) H. L. Bao and Y. Xu, Nat. Protoc., 2018, 13, 652–665 CrossRef CAS PubMed; (d) H.-L. Bao and Y. Xu, Chem. Commun., 2020, 56, 6547–6550 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3cc03192f |
| ‡ The two authors contributed equally: Yi Song, Shiyu Wang. |
| This journal is © The Royal Society of Chemistry 2023 |