Quantitative assessment of in vivo distribution of nanoplastics in bivalve Ruditapes philippinarum using reliable SERS tag-labeled nanoplastic models†
Hongyu
Du
ab,
Yunqing
Wang
*acd,
Panpan
Zhang
ab,
Rongchao
Mei
a,
Yunxia
Ji
a,
Xizhen
Zhao
a,
Zhiyang
Zhang
ac,
Jiping
Ma
e and
Lingxin
Chen
 *afcd
*afcd
aCAS Key Laboratory of Coastal Environmental Processes and Ecological Remediation, Shandong Key Laboratory of Coastal Environmental Processes, Yantai Institute of Coastal Zone Research, Chinese Academy of Sciences, Yantai 264003, China. E-mail: yqwang@yic.ac.cn
bUniversity of Chinese Academy of Sciences, Beijing 100049, China
cCenter for Ocean Mega-Science, Chinese Academy of Sciences, Qingdao 266071, China
dSchool of Pharmacy, Binzhou Medical University, Yantai 264003, China
eSchool of Environmental & Municipal Engineering, Qingdao University of Technology, Qingdao 266033, China
fLaboratory for Marine Biology and Biotechnology, Pilot National Laboratory for Marine Science and Technology, Qingdao 266237, China. E-mail: lxchen@yic.ac.cn
First published on 26th April 2022
Abstract
Nanoplastics (NPs) as emerging marine pollutants can be taken up by seafood organisms. It is crucial to quantitatively assess NP's distribution behavior in organisms to elucidate concentration dependent biological effects. Such a knowledge gap has remained due to the lack of reliable NP models and analytical methods. Herein, surface enhanced Raman scattering (SERS)-labeled NP models were developed and their bioavailability, distribution and accumulation in Ruditapes philippinarum, a typical marine bivalve, were quantitatively studied. Taking advantage of the sensitive and characteristic SERS signals of the NP models, distribution could be quickly and accurately obtained by the Raman imaging technique. Moreover, quantitative analysis of NPs could be performed by the detection of gold element contents via inductively coupled plasma mass spectroscopy (ICP-MS) detection. ICP-MS results revealed that after 3 days exposure of monodispersed NPs (100 nm, 0.2 mg L−1), the digestive gland accumulated 86.7% of whole-body NPs followed by gill (5.2%), mantle (5.1%), foot (1.3%), exhalant siphon (1.1%), and adductor (0.6%). Upon 11 days depuration, 98.7% of NPs in the digestive gland were excreted, whereas the clearance ratios in other organs were much lower. NP aggregates (around 1.5 μm) demonstrated similar distribution and clearance trends to the monodispersed ones. However, the accumulation amount in each organ was 15.2% to 77.6% lower. Surface adherence and passive ingestion routes resulted in NP accumulation, which contributed to the comparable NP abundance in these organs. Additionally, boiling treatment (mimicking a cooking process) did not decrease the NP amount in these organs. This work provided a dual-mode and quantitative analysis protocol for NPs for the first time, and suggested the risk of NP uptake by humans via bivalve seafood diets.
1. Introduction
Marine plastic pollution has become one of the critical global environmental problems.1 Once plastics are released into the environment, they go through physical and biological degradation processes, resulting in the formation of micro-nanoplastics.2 These particles as emerging contaminants have been detected in numerous marine ecosystems from the equator to the poles, from surface water to sediment, and from coast to open sea. Because of their small size and plentiful abundance, growing studies have evidenced that they could be readily ingested by over 690 marine organisms at different trophic levels.3Bivalves are proposed as a target group to research marine plastic pollution because of their global distribution, vital ecological niches, susceptibility to plastic uptake and close connection with marine predators and human health. In recent years, research on the distribution and effects of microplastics (MPs) on bivalves has intensified.4 For example, field-survey studies revealed that seven types of MPs existed in the digestive systems of four locally cultured bivalve species (scallop Chlamys farreri, mussel Mytilus galloprovincialis, oyster Crassostrea gigas, and clam Ruditapes philippinarum) in Qingdao city over four seasons.5 In a study in South Korea, MPs in bivalve seafood mainly sold in the market were in an average concentration of 0.15 ± 0.2 n g−1.6 Laboratory research indicated that 80 μm MPs could be ingested and accumulated on the surface of mussel gills and transferred to the digestive gland.7 Ingestion of MPs might cause histopathological damage to the digestive system, alteration in metabolic profiles, changes in energy metabolism and inflammatory responses, starvation, or malnutrition. For example, Urban-Malinga et al.8 investigated the responses of two common burrowing bivalves to PE MPs and found that bivalve behaviors changed after the addition of MPs to the sediment surface. Woods et al.9 exposed Mytilus edulis to PET microfibers shorter than 0.5 mm over 9 h and demonstrated that the filtration rate decreased under exposure to a concentration of 3 MPs mL−1. In a study by Detree and Gallardo-Escarate,10 after two periods of exposure to 1–50 μm of HDPE MPs, mussels were found to show significant changes between the transcriptomes of different tissues inducing immune system modulation and homeostasis alteration. Furthermore, the organs that are most affected by MP exposure are the digestive gland and gills. Brate et al.11 exposed Mytilus galloprovincialis to PE MPs (0.01 mg mL−1; 50–570 μm) and found that PE MP ingestion resulted in structural changes to the gills and digestive gland, as well as necrosis in other tissues such as the mantle. Comparatively, the understanding of the in vivo behaviors of nanoplastics (NPs, less than 1 μm), which probably have a higher binding affinity, biological barrier penetration ability and biological toxicity, has remained elusive.12,13
The main reason is the lack of an ideal in vivo analysis method for NPs. Raman technology is a popular method for MP detection. It is a non-contact technique with a highly specific fingerprint spectrum and with negligible interference from water.14 By collecting the unique Raman spectra of plastic polymers at each pixel, a Raman image can be generated to directly visualize MPs.15 However, it is challenging to analyze NPs due to the microscale spatial resolution.16 When the detected NP size was down to 100 nm, Raman mapping could only produce an image with a much larger scale.17 Dark-field hyperspectral microscopy has been used to identify MPs and NPs confined within nematodes in an intuitive and fast manner.18 This optical technique is suitable for the analysis of thin and transparent biological samples, and deep-tissue measurements for large organisms can hardly be realized. To date, many studies have used fluorescently-labeled plastic models to assess the fate of NPs in organisms with the aid of fluorescence imaging.19 However, the accuracy of this method was questioned because the fluorescent dye can leach out of the NPs and stain biological tissues, causing false-positive results.20,21 In addition, autofluorescence of certain organisms is strong and causes severe interference in the imaging results, preventing the general application of fluorescent models.22 Moreover, restricted by the sensitivity, the organs ready for detection were mostly limited to those with relatively high NP abundance;23 whole-body distribution investigations have been hardly carried out. To overcome these problems, some novel labeling and detection techniques have been proposed recently. For example, 14C-radiolabeled NPs were developed to study the NP accumulation in clam Pecten maximus.24 Metal-doped NPs were produced to investigate their fate and behavior in complex environmental systems by inductively coupled plasma mass spectroscopy (ICP-MS).25 These methods are sensitive and specific; however, they could not realize in situ and real-time imaging analysis. To date, the development of highly robust and sensitive NP model particles and rapid imaging methods has been in great demand for NP studies.
Surface enhanced Raman scattering (SERS) tags have attracted much attention for bioimaging analysis due to their high sensitivity, multiplex labeling ability, and satisfactory biocompatibility.26 SERS tags are composed of noble metal nanoparticle cores and organic Raman reporter molecules attached to the core surface. Upon laser irradiation, SERS tags emit strong characteristic SERS signals of Raman reporters, which can be used to track the tags in living cells or organisms. The SERS labeling protocol is superior to fluorescent dye labeling towards NPs because it prevents not only false-positive results but also biological background interference.27 Moreover, dual information, i.e., SERS signal and mapping results obtained by the Raman imaging technique, as well as gold element content obtained from ICP-MS detection, can be applied to indicate the distribution and accurate amount of the NPs. The two analysis results can complement and confirm each other.
Herein, SERS tag-labeled NP model particles with satisfactory sensitivity, anti-interference ability and reliable in vivo signal stability were developed. Based on this model, the uptake, in vivo distribution and excretion behaviors of NPs in a marine bivalve model, Ruditapes philippinarum, were quantitatively studied. The highlighted findings were as follows: (1) ingested NPs translocate from the digestive gland to peripheral organs such as muscle, exhalant siphon and adductor, and NPs in these organs are excreted at different rates in clean seawater; (2) monodispersed NPs (100 nm) accumulate more than NP aggregates (1.5 μm) in the organs with the same NP concentration; (3) the accumulation of NPs through adherence on the organ surface cannot be neglected; and (4) cooking processes cannot effectively remove the accumulated NPs.
2. Materials and methods
2.1 Materials
Chloroauric acid (HAuCl4), hydroxylamine hydrochloride (NH2OH·HCl), silver nitrate (AgNO3), L-ascorbic acid (AA), styrene (St), hydrochloric acid (HCl), and ethanol were purchased from Sinopharm Chemical Reagent Co. Ltd. Sodium citrate, sodium borohydride (NaBH4), and poly-(vinylpyrrolidone) (PVP) were obtained from Aladdin. Cetyltrimethylammonium bromide (CTAB) was purchased from Sigma-Aldrich. Divinylbenzene (DVB) and 2,2-azobis(2-methylpropyl) dihydrochloride (AIBA) were purchased from Macklin. Cyanine 7 chlorine (Cy7) was synthesized in our laboratory. Bovine serum albumin (BSA) was purchased from Beyotime Biotechnology. The water used in the experiment was ultrapure water obtained by secondary purification (18.2 MΩ cm−1).2.2 Characterization
The SERS spectra were recorded by using a DXR Raman microscope (Thermo Scientific, USA). A 780 nm laser (89 mW) was focused by a 10× microscope objective for NP sample solution and bivalve measurements. Dark-field images of NPs were captured by using an optical microscope (Eclipse Ti2, Nikon, Japan) equipped with a dark-field condenser (dry 0.95–0.80). Transmission electron microscopy (TEM) images were obtained on a JEM-1400 transmission electron microscope (JEOL, Japan). Scanning electron microscopy (SEM) images were acquired on an S-4800 field emission scanning electron microscope (Hitachi, Japan). Fourier transform infrared (FTIR) spectra were recorded on an FTIR-6300 infrared spectrometer (Jacso, Japan). 1 mL of SERS@PS and SERS@PS@BSA (500 mg L−1) was vacuum-dried for 12 hours and the powders were pressed into KBr tablets for analysis. The zeta potential and dynamic light scattering (DLS) measurements were performed on a Zetasizer NanoZS90 (Malvern Instruments, U.K.). Inductively coupled plasma mass spectrometry (ICP-MS) was carried out on a PerkinElmer SCIEX ELAN DRC II.2.3 Preparation of SERS labeled NPs
SERS@PS NPs were prepared according to our previous works.27,28 Gold nanostar (AuNS) based SERS tags were first synthesized (details in the ESI†), followed by a polystyrene (PS) shell coating process. Briefly, 19.5 mL of water and 82.5 mL of ethanol were added to a mixture of purified St (0.95 mL), DVB (0.05 mL), and PVP (300 mg) in a 250 mL two-necked flask equipped with a reflux condenser and a Teflon coated magnetic stirring bar. The reaction mixture was stirred at 70 °C for 1 h under a nitrogen atmosphere. Then, 3 mL of an aqueous AIBA solution (1.7 wt%) was added to the flask. After 8 min, 15 mL of a SERS tag solution was introduced into the reaction mixture. The reaction was continued with heating at 70 °C for another 12 h under a nitrogen atmosphere. Then SERS@PS NPs were obtained and washed 5 times by being centrifuged in water to remove the residual reactants and free PS NPs. To prepare NP models covered with BSA corona (SERS@PS@BSA), SERS@PS NPs were dispersed with BSA solution (1 mg mL−1) and incubated for 12 h, followed by centrifugation (6000 rpm, 2503g) to remove the residual BSA.2.4 Bivalve collection and experimental design
Healthy bivalve Ruditapes philippinarum samples (2.0 ± 0.5 cm shell length) were collected from Sishili Bay (Shandong, China). Before particle exposure, the bivalves were temporarily kept for one week in glass tanks equipped with mute air pump ventilation systems to acclimate to the laboratory conditions, and natural seawater was changed every day. Natural seawater (pH ∼ 8, temperature at 24.0 °C, salinity ∼33‰) was sand filtered (10 μm) followed by ultraviolet irradiation at a dose of 46 mJ cm−2. Bivalves were fed daily with a suspension of Chlorella vulgaris (0.1 g L−1) during acclimation or experimental assay.After the acclimation period, the bivalves were randomly divided into three tanks with 10 L capacity, each containing 50 individuals (one tank per concentration). Bivalves were exposed to separate treatments (each with three replicates) containing 0.2 mg L−1 (about 5.9 × 1011 particles per mL, see the calculation in the ESI†) SERS@PS NPs and SERS@PS@BSA NPs, and a control group without NPs. During the exposure period, the NPs and seawater were entirely changed every day. After 3 days exposure, bivalves were transferred to separate tanks with clean seawater for 11 days depuration. During the depuration period, seawater was changed every day. The total experiment time was 14 days. The in vivo distribution of NPs was determined by SERS measurement (at 1, 2, 3, 4, 8, 10, and 14 days, 3 bivalves each time) and via ICP-MS detection (at 1, 2 and 3 and 14 days, 3 bivalves each time).
2.5 SERS detection of NPs in bivalves
SERS spectra from the foot muscle, exhalant siphon, adductor, gill, mantle, and digestive gland of the bivalves were measured after dissection. The tissues were fixed on glass coverslips before recording the spectra. SERS mapping was carried out by the preparation of tissue slices with a scanning step size of 20 μm in both the x and y directions. The cross-section slices of muscular organs (adductor and foot) were prepared and fixed with paraffin. The false-color images were obtained from the intensity of the characteristic Raman peak of 1207 cm−1.2.6 Analysis of Au in bivalves by ICP-MS
The bivalves were dissected at 1, 3, and 14 days. The same organs (adductor, foot, exhalant siphon, mantle, gill, and digestive gland) of three bivalves were pooled together followed by acid digestion. 1.0 mL of HNO3 was added to each organ and heated for 12 h at 90 °C. Clear solutions could be obtained, the volumes of which were fixed to 10.0 mL for the ICP-MS analysis of Au. The main operating conditions were as follows: the auxiliary flow was 0.025 L min−1 and the plasma gas flow was 0.5 L min−1. The quantification was performed using an eight-point calibration curve between 1 and 500 ng mL−1, and 115In was chosen as an internal standard element. Sole SERS@PS NPs (100 nm, 5.9 × 1011 particles per mL) were used for NP recovery tests. At the end of the process, we were able to retrieve 85.5% of the NPs added. The recovery of NPs from the main organs was tested by a standard addition experiment and a satisfactory result of 86.2% was obtained (Table S1†).2.7 Statistical analysis
The experiments were conducted using a completely randomized design. Origin 8.0 (IBM) and Microsoft Excel 2019 (Microsoft) software were used to analyze the data, determine the variances, and prepare the diagrams. SPSS was used to examine the correlation of data obtained from ICP-MS and SERS methods, and the mode was set to “Spearman” and “two-tailed”. Values were expressed as means ± standard errors (SEs).3. Results
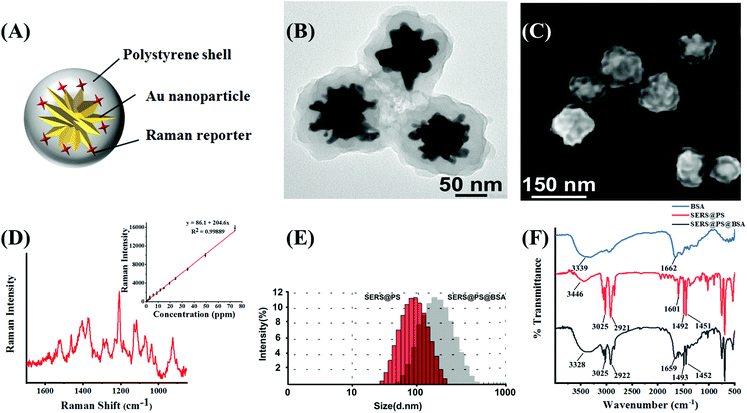
3.1 Characterization of NPs
Fig. 1A shows the schematic illustration of SERS@PS NPs, which were composed of three parts: (1) gold nanostars (AuNSs) as a SERS enhancement nanosubstrate; (2) Raman reporter molecules (Cy7 dye) attached to the surface of AuNSs, which generated a characteristic SERS signal, and this signal was distinguishable in biological matrices without interference and can indicate the presence of the NPs; and (3) a compact polystyrene (PS) shell over the bare SERS tag to simulate plastic materials. The TEM image (Fig. 1B) demonstrated the multi-branched structure of AuNS with a size around 60 nm, and the thickness of the PS shell is about 40 nm. SEM of the model NPs revealed the rough surface, which well mimics the irregular morphology of realistic NPs in the natural environment. The size distribution obtained from SEM images is uniform (Fig. S1†). Fig. 1D shows the Raman spectrum of SERS@PS. There was a good linear relationship between the Raman intensity at 1207 cm−1 and the concentration of SERS@PS.Apart from surface morphology, surface chemistry is also a key factor influencing the environmental behavior of NPs, which was also considered in this work. In natural water bodies, NPs are usually capped with numerous kinds of protein biosurfactants. The formation of a protein corona greatly influences the dispersive/aggregate and sediment behaviors of NPs, as well as the interaction with organisms and in vivo fate. It is challenging to identify and coat natural proteins in seawater on NPs, herein a commercially available bovine serum albumin (BSA) was selected as a capping agent for SERS@PS NPs to mimic the surface coating of the natural NPs. SERS@PS@BSA was prepared via hydrophobic and electrostatic interactions between BSA and the plastic surface29,30 and studied in parallel.
DLS results indicated the size increase of the NPs from 106.3 nm to 156.7 nm after BSA adsorption (Fig. 1E). The particle size distributions of SERS@PS and SERS@PS@BSA NPs were stable in seawater (Fig. S2†). The zeta potential changed from 47.0 mV to −18.9 mV (Fig. S3†). The polydispersity index values of SERS@PS and SERS@PS@BSA were 0.215 and 0.369, respectively, suggesting the uniform size distribution and good dispersity of both NPs in aqueous solutions. Fig. 1F shows the FTIR spectra of the NPs with and without the BSA corona. For the NP sample, the two bands at 2921 cm−1 and 3025 cm−1 were caused by the C–H deformation of the benzene ring, and the adsorption peaks at 1451 cm−1 and 1492 cm−1 were attributed to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretch of the benzene ring. After the attachment of BSA, the peaks at 1659 cm−1 and 3328 cm−1 greatly enhanced, which could be attributed to the amide and carboxyl groups of BSA, respectively.
C stretch of the benzene ring. After the attachment of BSA, the peaks at 1659 cm−1 and 3328 cm−1 greatly enhanced, which could be attributed to the amide and carboxyl groups of BSA, respectively.
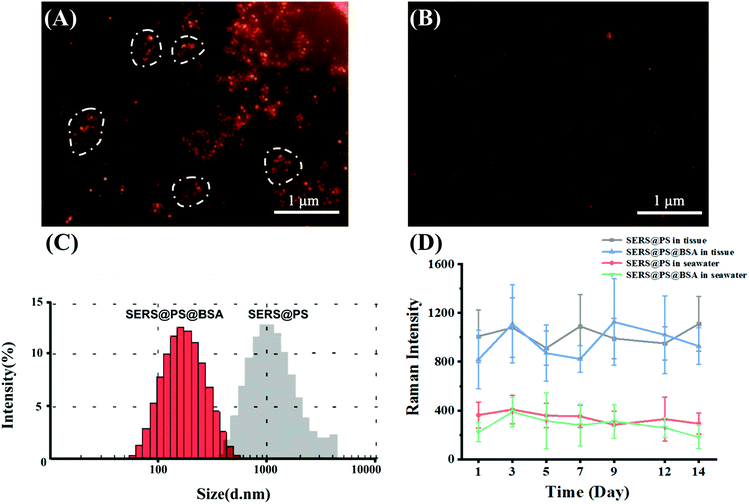
Before incubation with the marine bivalves, the dispersity and signal stability in seawater were studied. Taking advantage of the localized surface plasmon resonance (LSPR) properties of the AuNSs, the model NPs emit strong scattering light, and the image of a single NP can be captured under a dark-field microscope. This observation was performed in an aqueous solution and not affected by the high NaCl concentration, providing the facile and intuitive characterization of the NP state in seawater. Fig. 2A shows that after dilution of the SERS@PS NP solution with seawater, aggregation immediately occurred as suggested by the uneven NP clusters. By contrast, SERS@PS@BSA NPs homogeneously distributed as single dots in the observation area (Fig. 2B). DLS measurement showed similar results. The hydrodynamic diameters of SERS@PS@BSA slightly changed to 162.6 nm and kept stable for 14 days, whereas those of SERS@PS sharply increased to about 1.5 μm in seawater.
The aggregation status of PS NPs strongly affected their fate, transport, and ecological risks in aquatic environments.31 Sodium chloride (NaCl) is a dominant salt in natural seawater which accounts for 77.8% of the total inorganic salts in seawater. It has been reported that the NaCl concentration affected the aggregation of PS NPs. In pure water, NPs remained monodispersed due to the repulsive force from the positive surface charge. However, the increasing concentration of NaCl weakens the energy barrier. When the NaCl concentration reached its critical coagulation concentration value, which is the minimum electrolyte concentration to induce diffusion-limited aggregation, the Cl− ion neutralizes the positive charge on the NPs and consequently weakens their electrostatic repulsion, and the van der Waals attractive force dominated the particle interactions.32 By contrast, SERS@PS@BSA NPs are well dispersed as they took advantage of the steric hindrance and negative repulsion provided by the BSA corona.
Considering the long-term in vivo imaging study in the bivalves, the signal stability of both NPs in seawater and bivalve tissue homogenate was tested in 14 days. The intensities kept stable (Fig. 2D), laying a foundation for the following investigations.
3.2 Ingestion and accumulation of NPs in bivalves
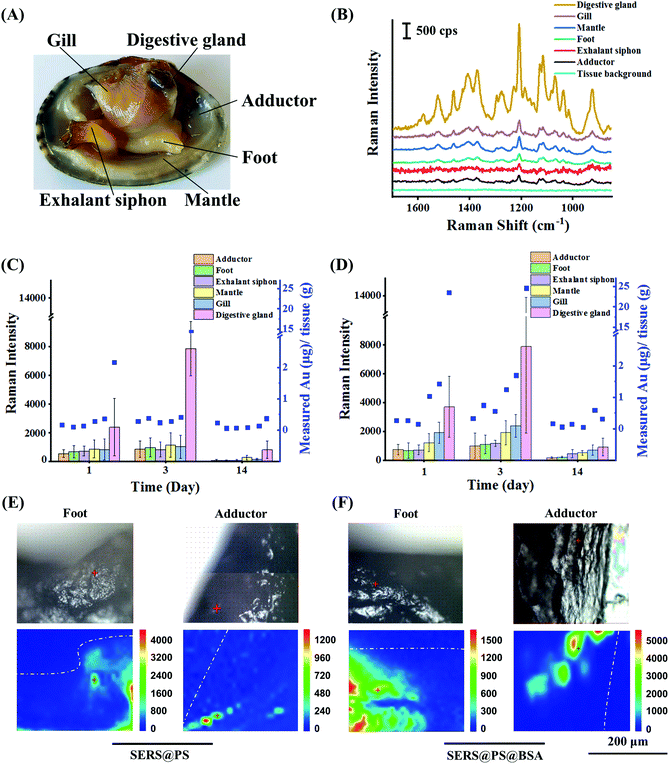
SERS@PS and SERS@PS@BSA NPs were added to the tanks containing bivalves, respectively, at a concentration of 0.2 mg L−1. This concentration was of realistic environmental meaning and simultaneously produced enough optical signals for the NP tracking. Despite no concentration data of NPs being available in real natural matrices, they were expected to be at least as abundant as MPs.33 Our exposure concentration is in the same magnitude of environmentally relevant concentrations as the maximum concentrations of MPs reported in multiple field investigations around 1.0 mg L−1 as surveyed in this literature.34After different exposure or depuration times, the bivalves were dissected and the number of NPs in six main organs was measured by SERS and ICP-MS (Fig. 3A). Fig. 3B shows that no background signal could be detected from all the tested organs of bivalves before exposure, whereas the characteristic SERS signal was recorded from all organs after 24 h exposure of SERS@PS NPs, suggesting that NPs widely distributed in the bivalves. The digestive gland showed the strongest signal, which was almost 6.4–12.0 times higher than those from the other organs, indicating that the digestive gland was a major NP accumulation organ.
Then the accumulation (0–3 day) and depuration (4–14 day) behaviors were systematically studied by measuring the SERS intensities (Fig. S4†) and gold element contents in these organs in a quantitative manner. Two-tailed Spearman analysis was performed to examine the correlation between the results of ICP-MS and SERS methods. The results showed that two methods had high correlation, and the characterization trends were consistent (SERS@PS: P < 0.01, r = 0.87 and SERS@PS@BSA: P < 0.01, r = 0.92). Fig. 3C shows the results from SERS@PS NPs. The apparent SERS signal and gold element could be detected from all the organs after 1 day exposure of SERS@PS NPs. The digestive gland accumulated the highest concentrations as revealed by the SERS intensity of 2380 cps and the gold concentration of 2.16 μg g−1 tissue. By contrast, the other five organs showed the SERS intensities in the range of 536–856 cps, and gold concentrations from 0.11 to 0.36 μg g−1 tissue. With prolonged exposure time to 3 days, the SERS signal from the digestive gland dramatically increased to 7881 cps. The gold content increased to 14.28 μg g−1 tissue, which was 6.6 times the value of the first day. By contrast, accumulation rates in the other organs were much lower. The SERS intensities slightly grew to around 1000 cps, and the gold concentration was still at the level of 0.28–0.42 μg g−1 tissue. The digestive gland accumulated the most NPs (94.0% of whole-body NPs) followed by the gill (1.8%), mantle (1.7%), exhalant siphon (0.9%), foot (0.9%) and adductor (0.8%).
After 11 days depuration, the NP concentration in the digestive glands significantly reduced to 0.37 μg g−1. The excretion ratio was 97.4%, indicating the strong clearance effect of the digestive glands. However, SERS signals in all organs were still detectable in the range of 50.6–794.4 cps, so it was the gold element in the adductor (0.23 μg g−1), gill (0.14 μg g−1), mantle (0.08 μg g−1), exhalant siphon (0.07 μg g−1), and foot (0.06 μg g−1). The excretion ratios of the exhalant siphon, foot, gill, and mantle were in the range of 66.7%–84.2%. For the adductor the value was much lower (17.9%).
The behaviors of SERS@PS@BSA NPs were also studied for comparison (Fig. 3D). The accumulation and depuration trends were similar to those of SERS@PS NPs. However, the accumulation rate and amount were higher. After 3 days exposure, the gold concentrations in the adductor (0.33 μg g−1), foot (0.75 μg g−1), exhalant siphon (0.56 μg g−1), mantle (1.25 μg g−1), gill (1.70 μg g−1), and digestive gland (24.59 μg g−1) were 1.18 to 4.46 times the values from SERS@PS in the corresponding organs. The digestive gland accumulated 86.7% of whole-body NPs followed by the gill (5.2%), mantle (5.1%), foot (1.3%), exhalant siphon (1.1%), and adductor (0.6%).
After depuration, the amounts remaining were 0.17 μg g−1 (adductor), 0.06 μg g−1 (foot), 0.15 μg g−1 (exhalant siphon), 0.05 μg g−1 (mantle), 0.60 μg g−1 (gill), and 0.31 μg g−1 (digestive gland). The excretion ratio in the digestive glands was 98.7%. SERS signals were in the range of 177.4–914.0 cps, indicating the remaining NPs in these organs. The elimination rate in the digestion gland was roughly calculated based on the elimination curve fitted by the elimination time and the corresponding gold concentration remaining in the digestion gland (Fig. S5†). The clearance of SERS@PS NPs in the digestive gland reached 90% after 5.7 days depuration.
SERS@PS@BSA was taken up more than SERS@PS, which was attributed to the different sizes and sediment behaviors in seawater. SERS@PS aggregated in seawater and the agglomerate (1.5 μm) tended to precipitate at the bottom of the culture tanks. In contrast, SERS@PS@BSA was stable and well dispersed in seawater. Considering the filter-feeding habit of bivalves, the NPs well dispersed in seawater gained a higher possibility to be inhaled into bivalves (i.e., higher bioavailability) compared to the NP aggregates deposited on the bottom of the tank.35 Moreover, particles with smaller sizes are more likely to come across the gut barrier and enter peripheral organs. This part of NPs was harder to be excreted and led to higher accumulation. These results implied that the dispersive behavior of NPs in the ocean was an important factor influencing the biological effects.36 Flocculation and sedimentation might weaken NP accumulation and adverse effects in bivalves.
The understanding of tissue distribution is the first step for future understanding of plastic debris’ toxicological effects and transfer within the food web.1 Despite MPs having been intensively studied, the knowledge on NPs was still lacking. Our study suggested that NPs presented in all main organs of bivalves, and particles in the digestive gland were much more numerous than those in the other organs. The results agreed with a previous study on bivalve C. fluminea, in which fluorescent NPs could accumulate in the mantle, visceral mass and gill.37 Another study reported that fluorescent NPs were concentrated in the siphons and remained there for longer than one month post-exposure.38 Upon the application of ultrasensitive 14C radiolabeled PS NPs, it was found that most organs of mollusk Pecten maximus were labeled (including the muscles, gonads, mantle, gills, intestine and kidneys), but the hepatopancreas had the highest 14C concentration.39 The wide tissue distribution indicated ingestion and accumulation in the digestive cavity and tubules and translocation into the circulatory system from which NPs reached the foot and adductor muscle.
To further confirm the translocation of NPs from the digestive gland to mussels, adductor muscle and foot cross-section tissue slices and the existence of NPs were examined by using the Raman imaging technique. The application of cross-section slices prevented possible interference from NPs adhered on the surface of the tissues. As shown in Fig. 3E and F, characteristic signals of the SERS tags were observed for both NPs in the two organs, providing solid evidence that the NPs had crossed intestinal barriers and transported to the muscular organs. The translocation of NPs might occur via two routes. On the one hand, small particles could pass through the biological barriers of animals and transfer from one tissue to another.40 This phenomenon was also observed in the gill membrane and the foregut of crabs.41 On the other hand, the immune cells which could uptake NPs might also play an important role.42 MPs were tracked in the hemolymph of the mussel Mytilus edulis, which translocated from the gut to the circulatory system within 3 days and persisted for over 48 days.31
3.3 Adherence of NPs to soft tissues
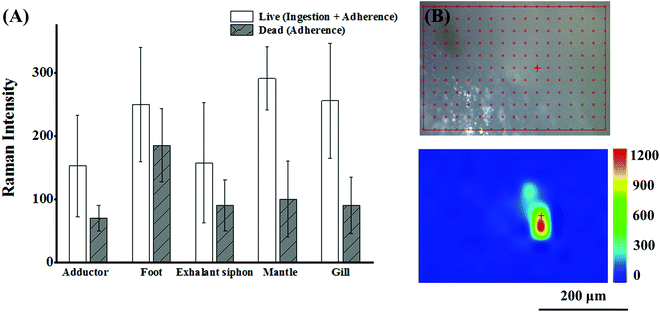
The surface of bivalves is rich in mucus that has a strong affinity to particulate matter. Thus adherence is also responsible for the accumulation of NPs in addition to the ingestion route.43 Shi's group reported the adherence of MPs to the soft tissue of mussels for the first time. They could draw this conclusion simply by the observation of MPs in the non-ingestion organs (foot and mantle) of mussels under the premise that the tested MPs larger than 100 μm could not enter the circulatory system and be transferred to the foot and mantle via an ingestion route. Adherence to the soft tissue is an exclusive way.44 In the case of NPs in our work, the situation was more complicated. Both ingestion and adherence were responsible for the NP abundance, and it is meaningful to clarify the contribution of the two routes. To make a rough estimation, SERS intensities were recorded from live and dead (sacrificed with a knife) bivalves after exposure to SERS@PS NPs for 12 h. The live and dead bivalves inferred the conditions of the sum of active ingestion and passive adherence, and sole adherence, respectively. As shown in Fig. 4(A), considerable SERS intensities were detected from various organs of dead bivalves, which were kept stable upon thorough rinsing with clean water. These results proved that the NPs firmly adhered to the tissue surface of various organs. The proportion of adhered NPs accounted for 34.4%–74.2% of the total NPs from the intensity ratios of the dead and live bivalves. Our results strongly suggested that both adherence and ingestion led to the accumulation of NPs in the muscular organs.To further explore whether NPs can penetrate organ surfaces, live clams were taken out of water and exposed to air. After a period, the clams extended their foot out of their shells. Then, we dripped SERS@PS NP solution (50 mg L−1) onto the foot surface. This air exposure made NPs adsorbed merely on the foot and avoided the possibility of passive NP uptake in water. After 4 hours, the clams were killed, and the surface of the foot was thoroughly washed until no Raman signal was detected on the foot surface. Then the foot was sliced to obtain a cross-section of the interior followed by Raman imaging. As shown in Fig. 4(B), characteristic SERS tag signals could be detected, which indicated that SERS@PS NPs could penetrate the foot surface.
3.4 Effect of cooking on the NP content
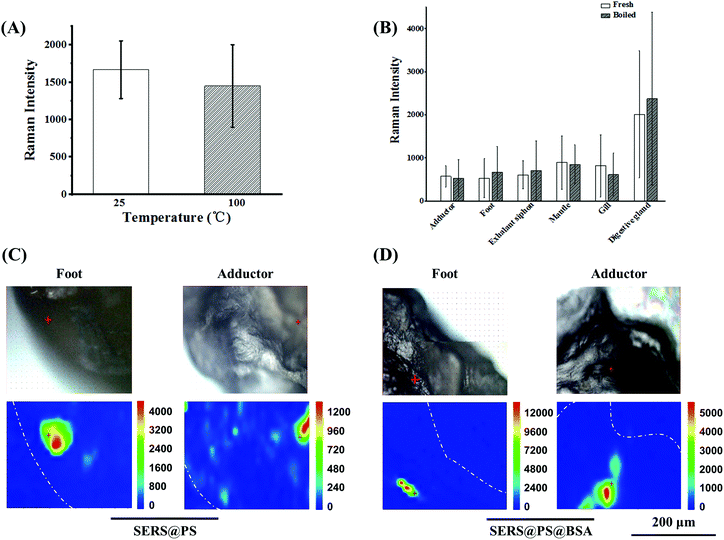
Shellfish is a popular kind of seafood and also a crucial source of plastic uptake by humans, posing a great threat to human health. A conventional cooking method was boiling. It is interesting to clarify whether the boiling treatment can reduce the content of NPs in the polluted bivalves, especially for the adherence proportion. To test this idea, the bivalves after 3 days exposure were boiled in water for 10 minutes, and the SERS intensities before and after boiling were compared. The signal stability of the model NPs upon boiling treatment was verified first. As shown in Fig. 5A, the signal intensity was almost maintained. On this basis, the intensities from the boiled bivalves were measured to indicate the NP amount. Fig. 5B shows that the SERS signals of each fresh and boiled tissue did not change obviously, suggesting that cooking did not expel NPs from bivalves’ tissues. Additionally, Raman mapping of the cross-section of the boiled adductor and foot (major edible organs) demonstrated intensive SERS signals for both NPs (Fig. 5C), further confirming that considerable NPs were entrapped in the cooked bivalve mussel.4. Conclusions
In this work, SERS-labeled NP models were developed to quantitatively assess the uptake and translocation of NPs in a typical marine bivalve Ruditapes philippinarum. The digestive gland was the major organ and could accumulate around 90% NPs of the whole body, whereas over 97% of NPs in the digestive gland could be eliminated after 11 days depuration. NPs were also detected from the tissue cross-section of the foot and adductor, implying that NPs could translocate from the digestive gland and accumulate in these organs. Boiling treatment did not work to remove the surface adhered or ingested NPs in the bivalves, suggesting the potential of NP uptake by humans via diet. These results implied that the removal of the digestive gland was suggested to alleviate the exposure risk to humans during bivalve seafood handling.To date, there has been an urgent demand for reliable analytical methods for NP studies in organisms. Single histological section observation is insufficient (or even impossible) to judge the absence of NPs in specific organs. Fluorescent labeled NPs and fluorescent imaging methods are criticized because they may produce false-positive results. This work provided a reliable NP model, and dual-mode SERS imaging and ICP-MS information with high sensitivity could be obtained. This protocol would facilitate researchers to gain a better understanding of the fate of NPs as well as ecological and human health impacts. In the future, the NPs still can be improved to overcome two limitations. One is that the metal core increases the density of plastic particles. Despite the model NPs uniformly dispersing in solution and not sinking, they are different from realistic NPs in this key physical parameter. Smaller gold cores should be used to alleviate this problem. The other is the NP model is prepared via an in situ polymerization way, which can only be realized for PS plastic types. A universal emulsion method may be helpful to extend labeling plastic types to PE, PP, PVC, etc.45
Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
Financial support from the National Natural Science Foundation of China (grant numbers 42076199, 21976209 and 21976099), the Youth Innovation Promotion Association CAS (grant number 2017256), the Instrument Developing Project of the Chinese Academy of Sciences (grant number YZ201662) and the Taishan Scholar Project Special Funding (grant number ts20190962) is gratefully acknowledged.References
- T. S. Galloway and C. N. Lewis, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 2331–2333 CrossRef CAS PubMed.
- D. Huang, J. Tao, M. Cheng, R. Deng, S. Chen, L. Yin and R. Li, J. Hazard. Mater., 2021, 407, 124399 CrossRef CAS PubMed.
- M. Carbery, W. O'Connor and T. Palanisami, Environ. Int., 2018, 115, 400–409 CrossRef PubMed.
- J. Li, Z. Wang, J. M. Rotchell, X. Shen, Q. Li and J. Zhu, Environ. Pollut., 2021, 286, 117543 CrossRef CAS PubMed.
- J. F. Ding, C. J. Sun, C. F. He, J. X. Li, P. Ju and F. M. Li, Sci. Total Environ., 2021, 782, 146830 CrossRef CAS PubMed.
- Y. Cho, W. J. Shim, M. Jang, G. M. Han and S. H. Hong, Environ. Pollut., 2019, 245, 1107–1116 CrossRef CAS PubMed.
- M. A. Browne, A. Dissanayake, T. S. Galloway, D. M. Lowe and R. C. Thompson, Environ. Sci. Technol., 2008, 42, 5026–5031 CrossRef CAS PubMed.
- B. Urban-Malinga, M. Jakubowska and M. Bialowas, Sci. Total Environ., 2021, 769, 144302 CrossRef CAS PubMed.
- M. N. Woods, M. E. Stack, D. M. Fields, S. D. Shaw and P. A. Matrai, Mar. Pollut. Bull., 2018, 137, 638–645 CrossRef CAS PubMed.
- C. Detree and C. Gallardo-Escarate, Fish Shellfish Immunol., 2018, 83, 52–60 CrossRef CAS PubMed.
- I. L. N. Brate, M. Blazquez, S. J. Brooks and K. V. Thomas, Sci. Total Environ., 2018, 626, 1310–1318 CrossRef PubMed.
- C. C. Gaylarde, J. A. B. Neto and E. M. da Fonseca, Environ. Pollut., 2021, 272, 115950 CrossRef CAS PubMed.
- S. Rist, A. Baun, R. Almeda and N. B. Hartmann, Mar. Pollut. Bull., 2019, 140, 423–430 CrossRef CAS PubMed.
- N. P. Ivleva, A. C. Wiesheu and R. Niessner, Angew. Chem., Int. Ed., 2017, 56, 1720–1739 CrossRef CAS PubMed.
- Z. Sobhani, M. Al Amin, R. Naidu, M. Megharaj and C. Fang, Anal. Chim. Acta, 2019, 1077, 191–199 CrossRef CAS PubMed.
- R. Ossikovski, Q. Nguyen, G. Picardi, J. Schreiber and P. Morin, J. Raman Spectrosc., 2008, 39, 661–672 CrossRef CAS.
- Z. Sobhani, X. Zhang, C. Gibson, R. Naidu, M. Megharaj and C. Fang, Water Res., 2020, 174, 115658 CrossRef CAS PubMed.
- L. Nigamatzyanova and R. Fakhrullin, Environ. Pollut., 2021, 271, 116337 CrossRef CAS PubMed.
- J. C. Prata, I. F. Sequeira, S. S. Monteiro, A. L. Patricio Silva, J. P. da Costa, P. Dias-Pereira, A. J. Silva Fernandes, F. M. da Costa, A. C. Duarte and T. Rocha-Santos, Sci. Total Environ., 2021, 783, 147065 CrossRef CAS PubMed.
- A. I. Catarino, A. Frutos and T. B. Henry, Sci. Total Environ., 2019, 670, 915–920 CrossRef CAS PubMed.
- C. Schuer, S. Rist, A. Baun, P. Mayer, N. B. Hartmann and M. Wagner, Environ. Toxicol. Chem., 2019, 38, 1495–1503 CrossRef CAS PubMed.
- Y. Liu, X. Qiu, X. Xu, Y. Takai, H. Ogawa, Y. Shimasaki and Y. Oshima, Ecotoxicol. Environ. Saf., 2021, 212, 112007 CrossRef CAS PubMed.
- M. Sendra, E. Sparaventi, B. Novoa and A. Figueras, Sci. Total Environ., 2021, 753, 142024 CrossRef CAS PubMed.
- M. Al-Sid-Cheikh, S. J. Rowland, K. Stevenson, C. Rouleau, T. B. Henry and R. C. Thompson, Environ. Sci. Technol., 2018, 52, 14480–14486 CrossRef CAS PubMed.
- D. M. Mitrano, A. Beltzung, S. Frehland, M. Schmiedgruber, A. Cingolani and F. Schmidt, Nat. Nanotechnol., 2019, 14, 362–368 CrossRef CAS PubMed.
- Y. Wang, B. Yan and L. Chen, Chem. Rev., 2013, 113, 1391–1428 CrossRef CAS PubMed.
- P. Zhang, Y. Wang, X. Zhao, Y. Ji, R. Mei, L. Fu, M. Man, J. Ma, X. Wang and L. Chen, J. Hazard. Mater., 2021, 425, 127959 CrossRef PubMed.
- Q. Yu, Y. Wang, R. Mei, Y. Yin, J. You and L. Chen, Anal. Chem., 2019, 91, 5270–5277 CrossRef CAS PubMed.
- P. M. Gopinath, V. Saranya, S. Vijayakumar, M. Mythili Meera, S. Ruprekha, R. Kunal, A. Pranay, J. Thomas, A. Mukherjee and N. Chandrasekaran, Sci. Rep., 2019, 9, 8860 CrossRef PubMed.
- Y. Ji, C. Wang, Y. Wang, L. Fu, M. Man and L. Chen, Environ. Sci. Nano, 2020, 7, 2313–2324 RSC.
- O. S. Alimi, J. F. Budarz, L. M. Hernandez and N. Tufenkji, Environ. Sci. Technol., 2018, 52, 1704–1724 CrossRef CAS PubMed.
- Y. Liu, Z. Huang, J. Zhou, J. Tang, C. Yang, C. Chen, W. Huang and Z. Dang, Water Res., 2020, 186, 116316 CrossRef CAS PubMed.
- P. E. Redondo-Hasselerharm, G. Gort, E. T. H. M. Peeters and A. A. Koelmans, Sci. Adv., 2020, 6, eaay4054 CrossRef CAS PubMed.
- T. Sun, J. Zhan, F. Li, C. Ji and H. Wu, J. Hazard. Mater., 2021, 414, 125581 CrossRef CAS PubMed.
- J. E. Ward and D. J. Kach, Mar. Environ. Res., 2009, 68, 137–142 CrossRef CAS PubMed.
- A. ter Halle and J. F. Ghiglione, Environ. Sci. Technol., 2021, 55, 14466–14469 CrossRef CAS PubMed.
- Z. Li, C. Feng, Y. Wu and X. Guo, J. Hazard. Mater., 2020, 392, 122418 CrossRef CAS PubMed.
- R. L. Merzel, L. Purser, T. L. Soucy, M. Olszewski, I. Colon-Bernal, M. Duhaime, A. K. Elgin and M. M. Banaszak Holl, Glob Chall., 2020, 4, 1800104 CrossRef PubMed.
- M. Al-Sid-Cheikh, S. J. Rowland, R. Kaegi, T. B. Henry, M.-A. Cormier and R. C. Thompson, Commun. Mater., 2020, 1, 97 CrossRef.
- C. Campanale, C. Massarelli, I. Savino, V. Locaputo and V. F. Uricchio, Int. J. Environ. Res. Public Health, 2020, 17, 1212 CrossRef CAS PubMed.
- A. J. R. Watts, C. Lewis, R. M. Goodhead, S. J. Beckett, J. Moger, C. R. Tyler and T. S. Galloway, Environ. Sci. Technol., 2014, 48, 8823–8830 CrossRef CAS PubMed.
- M. Sendra, A. Saco, M. P. Yeste, A. Romero, B. Novoa and A. Figueras, J. Hazard. Mater., 2020, 388, 121788 CrossRef CAS PubMed.
- L. Tian, Q. Chen, W. Jiang, L. Wang, H. Xie, N. Kalogerakis, Y. Ma and R. Ji, Environ. Sci. Nano, 2019, 6, 2907–2917 RSC.
- P. Kolandhasamy, L. Su, J. Li, X. Qu, K. Jabeen and H. Shi, Sci. Total Environ., 2018, 610–611, 635–640 CrossRef CAS PubMed.
- K. Tanaka, Y. Takahashi, H. Kuramochi, M. Osako, S. Tanaka and G. Suzuki, Small, 2021, 17, 2105781 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2nr00157h |
| This journal is © The Royal Society of Chemistry 2022 |