Electrospun hydrogels for dynamic culture systems: advantages, progress, and opportunities
M. Gregory
Grewal
 a and
Christopher B.
Highley
a and
Christopher B.
Highley
 *ab
*ab
aDepartment of Chemical Engineering, University of Virginia, VA 22903, USA. E-mail: highley@virginia.edu
bDepartment of Biomedical Engineering, University of Virginia, VA 22903, USA
First published on 25th January 2021
Abstract
The extracellular matrix (ECM) is a water-swollen, tissue-specific material environment in which biophysiochemical signals are organized and influence cell behaviors. Electrospun nanofibrous substrates have been pursued as platforms for tissue engineering and cell studies that recapitulate features of the native ECM, in particular its fibrous nature. In recent years, progress in the design of electrospun hydrogel systems has demonstrated that molecular design also enables unique studies of cellular behaviors. In comparison to the use of hydrophobic polymeric materials, electrospinning hydrophilic materials that crosslink to form hydrogels offer the potential to achieve the water-swollen, nanofibrous characteristics of endogenous ECM. Although electrospun hydrogels require an additional crosslinking step to stabilize the fibers (allowing fibers to swell with water instead of dissolving) in comparison to their hydrophobic counterparts, researchers have made significant advances in leveraging hydrogel chemistries to incorporate biochemical and dynamic functionalities within the fibers. Consequently, dynamic biophysical and biochemical properties can be engineered into hydrophilic nanofibers that would be difficult to engineer in hydrophobic systems without strategic and sometimes intensive post-processing techniques. This Review describes common methodologies to control biophysical and biochemical properties of both electrospun hydrophobic and hydrogel nanofibers, with an emphasis on highlighting recent progress using hydrogel nanofibers with engineered dynamic complexities to develop culture systems for the study of biological function, dysfunction, development, and regeneration.
1 Introduction
The extracellular matrix (ECM) is a complex, dynamic, and tissue-specific scaffolding system that presents a myriad of biophysical and biochemical cues that influence cellular behaviors.1–4 The ECM is typically comprised of varying compositions of fibrous proteins and proteoglycans, coupled with soluble components such as growth factors;5–7 however, the state of this structure is constantly in flux as it is simultaneously degraded and synthesized by the resident cellular population.4–8 As the biophysical and biochemical attributes of the ECM at two distinct junctures are never identical, recapitulating tissue-specific milieus in vitro is challenging.5–7 To better understand cellular behaviors and processes occurring in physiologically-relevant systems, in vitro culture systems must continue to advance to accurately model the ECM.4,6,9–11Progress in developing more sophisticated in vitro culture platforms has advanced with new insights into the composition and properties of the ECM coupled with new technical capabilities to recreate its features. The heterogeneous material environment of the ECM is water-rich and nanofibrous in nature,1,4,12 typically comprised of single-fiber diameters on the order of tens to hundreds of nanometers (10–500 nm).12–16 Electrospinning is an accessible technique for depositing fibrous substrates with diameters analogous to those comprising native ECM,5–7 and has been established as an effective way to produce nanofibrous materials across many fields of research,17–21 including tissue engineering.22,23 Within tissue engineering and regenerative medicine, electrospun nanofibers have been applied to wound healing24 and the engineering of diverse tissue types including models of cardiac,25 vascular,26 neural,27,28 and musculoskeletal29 environments. In research applications addressing fundamental biological and physiological questions, electrospun substrates have also been tactically engineered to tease out cellular responses to differing environmental cues and perturbations for in vitro studies.2,3,30–32 For more information, Xue et al.33 and Rahmati et al.34 have recently published expansive reviews of the electrospinning process and extensive applications of electrospun materials.
Turning the focus from the process and applications onto the materials themselves, electrospun fibers utilized in tissue engineering applications throughout the years have been primarily comprised of hydrophobic polymers that were solubilized in organic solvents prior to electrospinning (Fig. 1). These materials were prevalent in the early waves of electrospinning due to their favorable performance in the electrospinning process and their ability to form fibrous substrates for cell culture without further stabilization steps, such as interpolymer crosslinking.12,35 A disadvantage of utilizing many of these hydrophobic polymers is they may lack desired cell-instructive biofunctionality in their fibrous form, and consequently require strategic chemistries to increase the bioactivity prior to seeding cells for culture.36,37 Furthermore, since these materials are foreign to physiological systems, it may be necessary to engineer them further to mediate biological responses in vivo during transplantation and degradation. There are many established methods to modify the surfaces of these hydrophobic nanofibers;36,37 however, a current shift towards using crosslinked polymers to develop hydrogel networks offers potential to reduce the complexity of post-processing (refer to Fig. 1) by drawing on the diversity of hydrogel functionalities available for modifying and controlling microenvironmental features and establishing dynamic materials.38
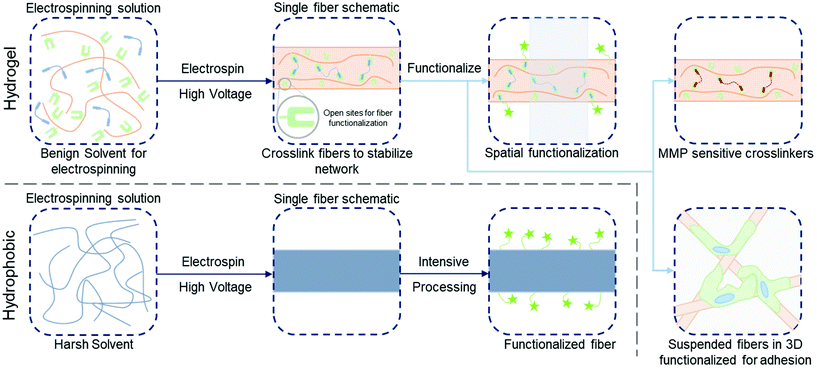 | ||
| Fig. 1 Functionalization of hydrogel versus hydrophobic nanofibers. (Top, left to right): electrospinning precursor solution containing a hydrophilic polymer with a crosslinker to stabilize hydrogel nanofibers; solution is electrospun and crosslinked (e.g. with UV irradiation) with leftover sites for further functionalization; three example pathways to functionalize the fibers – spatial control over bioactivity (green stars, shaded area indicates unfunctionalized region),86 fibers crosslinked with matrix metalloproteinase (MMP) sensitive crosslinkers for tunable degradation,8 suspended hydrogel fibers in a bulk gel for 3D models of the ECM.123 (Bottom, left to right): electrospinning precursor solution containing hydrophobic polymer (typically in a harsh solvent); solution is electrospun and fibers are ready for processing; intensive chemical processing is typically needed for fiber functionalization. | ||
Another advantage offered by electrospun hydrogel fibers compared to their hydrophobic material analogs is the water-swollen nature of native ECM and of natural fibers within ECM microenvironments.1,4,12 Furthermore, established chemistries used to modify polymeric backbones and engineer crosslinking in hydrogel fiber systems enables the facile development of functionality for controlling the biophysiochemical properties to recapitulate features of the endogenous ECM.1,39–41 Hydrogel systems for cell culture were originally introduced as advancements from tissue culture polystyrene,1 and as soon as they were developed for cell culture, researchers aimed to advance the technology towards dynamic culture systems.4,38 Electrospun fibers are mirroring this progression first through the development of hydrogel fibers, and now in trends towards dynamic fibrous environments that allow for modeling and probing of biological processes, while also affording control over the complexity of culture systems to reconstitute natural tissue as closely as possible. Significant progress in the engineering of fibrous culture substrates has been made, with the potential for further developments in materials design to continue to advance towards recapitulating endogenous tissue.42
This Review focuses on the methods developed to modify the biophysical and biochemical properties of electrospun polymers – both hydrophobic and hydrophilic – with an emphasis on the strengths provided by crosslinkable, hydrophilic polymers that form hydrogels. We further focus on the chemistries developed to modify hydrogel nanofibers to manipulate the complexity of biological systems in space and time, while additionally highlighting the advancements being made by researchers towards the development of dynamic scaffolding that effectively reconstitutes physiologically-relevant ECM. Furthermore, we also provide light commentary highlighting the advantages and associated challenges within these systems to ideally inform the next phase of advancements in nanofibrillar hydrogel design.
2 Hydrophobic polymer fibers for cell culture
The use of hydrophobic polymers has been central to the development of fibrous culture systems,43 and materials commonly used include polylactic acid (PLA),44–47 poly(lactic-co-glycolic acid) (PLGA),48 polycaprolactone (PCL),49 polyethylene terephthalate (PET),50 among many others.51,52 Since these materials are characteristically hydrophobic, they require nonpolar organic solvents to facilitate the electrospinning process.25,51,53,54 Therefore – in biomedical applications – water infiltration is limited to spaces between fibers, without substantially absorbing into the polymeric matrices of the fibers themselves.51 Despite this challenge, these materials are well-suited to the electrospinning process and have seen extensive use in the tissue engineering space. Part of the strength of these materials in electrospinning is that the morphological features of the resulting nanofibers can be readily tailored by simply controlling process parameters,12,54,55 yielding substrates with designed topographical characteristics that contribute to the biophysical properties that cells transduce. Similarly, post-electrospinning techniques have been employed to increase the bioactivity of the fibrous substrates. Since cells are heavily influenced by a combination of both biophysical and biochemical signals in their microenvironment,6,7 techniques have continuously progressed to introduce relevant signals to nanofibers based on these hydrophobic materials in order to influence the cells interacting with them.2.1 Hydrophobic nanofibers enabling control over physical properties
Work aiming to engineer and alter nanofibrous topographies is driven by cellular transduction of biophysical stimuli from their microenvironments to influence signaling pathways that direct downstream phenotypic fate decisions.56 Therefore, control over physical properties of culture systems is a critical consideration in biomedical applications including tissue engineering, regenerative medicine, and fundamental investigations into cellular processes and development. The diameters of electrospun fibers can be readily controlled through solution properties and variable parameters of the electrospinning process – in particular solution viscosity, polymer molecular weight, applied voltage, and solution flow rate.55,57,58 Even with this level of control, careful consideration is needed when developing fibers to match the tissue system of interest. For instance, Young's modulus of electrospun fibers exhibits an inverse relationship with fiber diameter;59 therefore, a balance is typically needed when engineering models that replicate tissue-specific systems in the body.60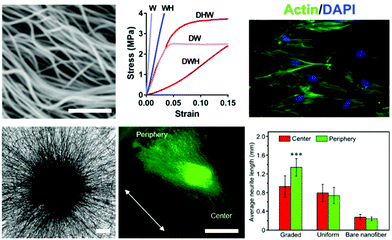 | ||
| Fig. 2 Cell culture on modified hydrophobic fibrous scaffolds. (Top, left to right): crimped PLLA fibers synthesized via heat treatment with sacrificial fibers by Szczesny et al.62 to develop a tendinous/ligament-like tissue structure; the crimped system (DWH) exhibited a traditional non-linear stress–strain curve similar to that of native tendon/ligament tissue, whereas controls (W, WH, DHW, DW) all were unable to replicate this behavior; actin/DAPI staining of cells seeded on these crimped systems demonstrated less alignment with the fibers and reoriented significantly upon mechanical strain. Scalebar = 1 μm. (Top) Reprinted and adapted with permission from Szczesny et al., copyright 2017 American Chemical Society.62 (Bottom, left to right): PCL fibers aligned radially due to a novel electrospinning collection setup, scalebar = 200 μm; Tuj-1 staining (green) of dorsal root ganglion cells shows significant neurite extension in the direction of fiber alignment (white arrow) and laminin gradient; quantification displaying average neurite length for the gradient experiments compared to controls of uniform laminin presentation and no laminin presentation. Scalebar = 1 mm, ***p < 0.001. (Bottom) Reprinted and adapted with permission from Wu et al., copyright 2018 American Chemical Society.76 | ||
2.2 Hydrophobic fibers enabling modulation of biochemical properties
3 Hydrogel nanofibers
The opportunities for increased control over the biophysical properties and spatiotemporal presentation of biochemical functionality has been a driving factor in the progression towards electrospun hydrogel fibers. Hydrogel fibers build on the strengths of hydrogel materials that can be chemically modified with functional moieties – for both crosslinking and introducing biomolecules.1,65 These strengths allow for the precise tailoring of mechanical and chemical properties to replicate the tissue system of choice.1,38 Thus, hydrogel nanofibers offer the potential for greater control over fiber properties compared to their hydrophobic analogs83 and the potential to provide a microenvironment that closely mirrors the water-swollen, fibrous characteristics of natural tissue.13–153.1 Fabrication of hydrogel nanofibers
Hydrogel nanofibers are produced via electrospinning similarly to other variants of polymeric nanofibers. Commonly, the solution consists of the hydrophilic polymer of choice (e.g. hyaluronic acid (HA), poly(ethylene glycol) (PEG), or dextran), a crosslinker (for systems that require a linker molecule), a photoinitiator (for photomediated reactions), and water as a solvent.2,84,85 For lower molecular weight polymers, like HA and PEG, a high molecular weight polymer, typically poly(ethylene oxide), is added to increase solution viscosity and induce chain entanglements.32,84–86 For higher molecular weight polymers, like dextran, this is not typically needed.2,31,87 This solution is then typically extruded though a needle at low flow rates, where an electric field is applied to the solution. This induces a competing interaction between polymer chain entanglements within the solution and electrostatic repulsion from the voltage – which due to solution extrusion, elongates into a Taylor cone. At the point of the Taylor cone, the solution vaporizes, which causes a polymeric fiber jet to form that whips and accelerates towards the grounded collection surface.12,58 Following the deposition of the fibers, they must then be stabilized through some variation of crosslinking (to be described in depth-below) in order to facilitate water absorption into the polymeric networks as opposed to fibers solubilizing upon hydration.2,84–86 Crosslinking also enables control over biophysical properties of hydrogel fibers, with degree of crosslinking directly affecting fiber parameters such as stiffness and diameter – which correlate with capacity for water swelling into the fibers.86,88 Once crosslinking is complete, functionalization of fibers is possible to introduce bioactivity into the fibrous hydrogel system.3.2 Introduction to hydrogel nanofiber crosslinking and stabilization
One group of hydrogel-forming materials are natural polymers with innate biocompatibility and presentation of relevant ligands.89,90 For example, collagen inherently presents bioactive sites for integrin-mediated cell adhesion.12 Other natural polymers may intrinsically interact with cells – such as hyaluronic acid (HA) (typically produced through fermentation processes1) with CD44.91–93 That being said, cells tend to exhibit low adhesion to some natural polymers, like HA, without chemical modifications to improve bioactivity.86 Therefore, HA, as well as other polysaccharide materials such as dextran,2 may need to be functionalized with bioactive molecules prior to being utilized for cell culture systems. Other hydrophilic polymers include synthetic polymers such as poly(ethylene glycol) (PEG).94 There are a variety of established chemistries to modify the backbones of these hydrophilic polymers with pendant functional moieties that can act as sites for crosslinking and biomolecule conjugation. Therefore, modification of these polymers thereby provides significant user control over biophysical and biochemical characteristics of the nanofibers.Unlike hydrophobic materials, as discussed previously, polymeric materials used in hydrogels are soluble in water and fibers generated by electrospinning can dissolve upon hydration without stabilization. Thus, hydrogel-based systems must generally be stabilized through some form of intermolecular crosslinking between the polymers that comprise the nanofibers. In many cases, regulation of crosslinking enables control over physical properties, as will be discussed at greater length in the next section. Naturally-derived polymers such as collagen95 and gelatin,96 for example, can be electrospun; however, though the native materials undergo physical crosslinking, the resultant nanofibers themselves typically are not robust enough for handling without further post-processing.95,96 To address this, crosslinking agents, like glutaraldehyde, have been utilized with collagen and gelatin to improve resultant mechanical properties.95–99 Furthermore, Kishan et al. developed a platform for electrospinning gelatin that crosslinks on-the-fly using a diisocyanate crosslinker to retain fiber mechanical properties.100 Another effective method to stabilize collagen/gelatin-based fibers leverages carbodiimide chemistry, such as EDC/NHS crosslinking, to introduce ‘zero-length’ crosslinks.101–103 Chemical crosslinking has also been used to stabilize nanofibers formed from synthetic hydrophilic materials,104 for example using glutaraldehyde to crosslink polyacrylamide (PA)105 and poly(vinyl alcohol) (PVA).106–108 Glutaraldehyde as a crosslinker readily reacts with pendant groups on PA and PVA to form linkages, and offers the potential to provide user-defined control over the stiffness and swelling of resultant electrospun fibers.105,106
3.3 Chemical modifications for covalent crosslinking of hydrophilic polymers
In many cases, the polymers forming the molecular backbones of these hydrogel materials are chemically modified using various strategies that enable their stabilization after electrospinning for use as fibrous hydrogel systems. Photoinitiated reactions represent a major platform for the stabilization of these hydrogel fibrous networks, and the common methodologies for photoinduced reactions utilize differing versions of the ene–ene scheme – for example through acrylate-based functional groups – and thiol–ene reactions. In the presence of light and a photoinitiator, ene–ene reactions undergo a chain-growth mechanism and form kinetic chains that crosslink the backbone polymers.109 In the case of the thiol–ene reaction, photoinitiation produces a thiyl radical, which opens and subsequently binds with an adjacent alkene enabling stoichiometric crosslinking.11,110–112 In addition to the crosslinking type, the degree of substitution on the polymeric backbone itself plays an important role in the regulation of downstream fiber mechanics113,114 – therefore, careful consideration is needed when designing the specific material system.Many hydrophilic polymers have been modified to present pendant alkenes (using methacrylates and vinyl sulfones, for example) for crosslinking post-electrospinning. Gelatin is commonly modified with methacrylate moieties to create a material (GelMA) that can be stabilized by photoinitiated crosslinking of electrospun fibers.115–117 Similar chemistry has been used to modify HA,30,118 silk fibroin,119,120 and PEG.32,94 Dextran, another polysaccharide, can also be modified with methacrylate2,3,31 or vinyl sulfone87 functional groups for crosslinking and subsequent reactions that aim at improving bioactivity. In most cases, alkene groups within nanofibers allow for anhydrous radical-induced polymerization within fibers to stabilize the polymeric networks prior to hydration.121 One of the strengths of photochemistries is the great potential for spatial control of reactions. Crosslinking, and therefore fiber stability (and ultimately mechanics), can be specified via selective irradiation of electrospun nanofibers through photomasks. Sundararaghavan et al. used this to introduce porosity within thick fibrous substrates that would aid in cell infiltration. By masking regions of fibers during anhydrous crosslinking of methacrylated HA nanofibers, leaving them unexposed to light, regions of fibers could be selectively dissolved during hydration122 (see Fig. 3A).
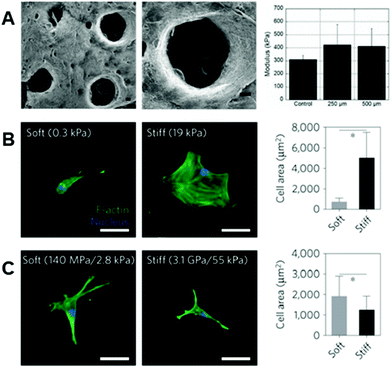 | ||
| Fig. 3 Importance of fiber physical properties for cell culture. (A, left to right): SEM micrographs of MeHA fibers with user-specified photopatterned pores, zoomed in micrograph of a photopatterned pore, and a column chart displaying modulus of scaffolds – with no significant difference between scaffolds with pores and scaffolds without pores. (A) Reprinted and adapted with permission from Sundararaghavan et al., copyright 2010 John Wiley and Sons;122 scalebars = 100 μm. (B, left to right): hMSCs show increased cell spreading on stiff hydrogels as opposed to soft hydrogels – quantified by the column chart illustrating cell area (*p < 0.05). (C, left to right): hMSCs demonstrate increased spreading on soft rather than stiff hydrogel fibers – quantified by the column chart showing cell area (*p < 0.05). These differing results emphasize the need for careful consideration when designing the biophysical properties of fibrous hydrogels for cell culture. (B) and (C) Reprinted and adapted with permission from Baker et al., copyright 2015 Springer Nature;2 scalebars = 50 μm. | ||
3.4 Disadvantages and considerations when electrospinning hydrogels
Although hydrogel materials have certain advantages over their non-hydrogel counterparts, there are some associated disadvantages that need to be considered when designing these material systems for electrospinning. For example, an important consideration when using some lower molecular weight polymers, like HA and PEG, is that a carrier polymer may be required during the electrospinning process to induce chain entanglements in the solution.85,118 High molecular weight polymers – like poly(ethylene oxide) – may be added to the electrospinning solution to facilitate fiber formation and subsequently be washed away when the scaffolds are hydrated.123 Furthermore, many biomaterials that form hydrogels are not ready for electrospinning ‘out-of-the-box’.1 Specifically, many of the materials require chemical functionalization to introduce reactive moieties such as methacrylates,2 vinyl sulfones,124 or norbornenes86 to the polymeric backbones. An additional consideration in using these functionalized materials is potential batch-to-batch variation in their synthesis, which may alter material properties.1 We refer to work reviewing hydrogels for cell culture1 for further information regarding synthesis and considerations of common hydrogel biomaterials. Finally, an inherent issue with these hydrophilic materials is the need to crosslink the fibers post-electrospinning, typically prior to any further functionalization.2,85,86 Once the material and crosslinking strategy are chosen, however, the resultant biophysical and biochemical properties can be easily modulated – as described in the following sections. Please refer to Table 1 for a representative list of hydrogel biomaterials that have been electrospun, along with a few established methods for crosslinking and modulating the resultant biophysiochemical properties.| Material | Example crosslinking method(s) | Modulation of biophysiochemical properties |
|---|---|---|
| Fully-synthetic materials | ||
| Polyacrylamide (PA) | Chemical: | Biochemical: |
| • Glutaraldehyde crosslinker105 | • Likely adsorption-based modifications | |
| Biophysical: | ||
| • Degree (extent) of crosslinking105 | ||
| Poly(vinyl alcohol) (PVA) | Chemical: | Biochemical: |
| • Glutaraldehyde crosslinker106 | • Likely adsorption-based modifications | |
| • PVA composites for crosslinking107 | ||
| Physical: | Biophysical: | |
| • Controlling hydrophobicity through PVA modifications108 | • Degree (extent) of crosslinking106 or PVA modification108 | |
| • Degree of hydrolysis (i.e. quantity of pendant reactive groups)107 | ||
| Poly(ethylene glycol) (PEG) | Chemical: | Biochemical: |
| • Pendant norbornenes (step-growth polymerization)85,136 | • Adsorption-based modifications32 | |
| • Pendant methacrylates (chain-growth polymerization)32 | • Pendant norbornenes provide sites for addition of biomolecules | |
| ∘ Light-mediated thiol-ene conjugation85 | ||
| Biophysical: | ||
| • Stiffness controlled via irradiation and crosslinker– for example: norbornenes136 and methacrylates32 | ||
| Naturally-derived materials | ||
| Collagen | Chemical: | Biochemical: |
| • Glutaraldehyde crosslinker95,97,99 | • Collagen provides natural bioactive sites for cell adhesion and interaction95 | |
| • Carbodiimide crosslinking (EDC/NHS)102 | Biophysical: | |
| • Degree (extent) of chemical crosslinking97 | ||
| Gelatin | Chemical: | Biochemical: |
| • Glutaraldehyde98 and diisocyanate crosslinkers100 | • Gelatin provides natural bioactive sites for cell adhesion and interaction96 | |
| • Carbodiimide crosslinking (EDC/NHS)101,103 | ||
| • Pendant methacrylates (chain-growth polymerization)115–117 | ||
| Physical: | Biophysical: | |
| • Dehydrothermal crosslinking (generally weaker fibers)96 | • Degree (extent) of chemical crosslinking96 | |
| • Degree of chain-growth polymerization (e.g. with methacrylates)115,116 | ||
| Hyaluronic acid (HA) | Chemical: | Biochemical: |
| • Pendant norbornenes (step-growth polymerization)86 | • Pendant molecules provide sites for addition of biomolecules | |
| • Pendant methacrylates (chain-growth polymerization)30,88,118,126,144 | ∘ Michael addition: thiolated biomolecules react with pendant alkenes in basic conditions8,118 | |
| • Pendant maleimides (chain-growth polymerization)8 | ∘ Light-mediated thiol-ene conjugation86 | |
| • Hydrazide/aldehyde proximity reactions to crosslink adjacent fibers145 | Biophysical: | |
| • Stiffness also controlled via irradiation time – for example: methacrylates88,122 | ||
| • Stiffness within norbornene modified systems can conceivably be controlled via crosslinker added, following from Gramlich et al.112 | ||
| Dextran | Chemical: | Biochemical: |
| • Pendant methacrylates (chain-growth polymerization)2,3,31 | • Pendant molecules provide sites for addition of biomolecules | |
| • Pendant vinyl sulfones (chain-growth polymerization)87,124,146 | ∘ Methacrylated heparin conjugated to free methacrylates within methacrylated-dextran fibers87 | |
| ∘ Michael addition: thiolated biomolecules react with pendant alkenes in basic conditions2,3,31,87,124,146 | ||
| Biophysical: | ||
| • Stiffness also controlled via irradiation time – for example: chain-growth polymerization2 | ||
3.5 Hydrogel nanofibers enabling control over physical properties
As noted, the physical properties of cellular microenvironments exert strong influences over cell behaviors and phenotypes.125,126 In nanofibrous systems, hydrogel-based materials offer possibilities for engineering these properties, such as the mechanical and viscoelastic environments with which cells interact, within a fiber-based environment to achieve certain outcomes or interrogate biological questions.Following the deposition and stabilization of hydrogel fibers, cell behaviors can be analyzed in in vitro tissue models that more closely mirror physiological features and enable experiments that assess cellular responses to perturbations of these environments. In ene–ene systems, control over mechanical properties, such as Young's modulus, has allowed cellular responses to environments of differing fiber stiffnesses to be assessed.2,3,30,31 For example, Baker et al. demonstrated, in a methacrylated-dextran system, that cell spreading behaviors on 2D stiff fibers (55 kPa, network stiffness) were inhibited in comparison to 2D soft fibers (2.8 kPa, network stiffness) – a phenomenon that is the inverse of what is seen on 2D hydrogels (Fig. 3B and C).2 Baker et al. propose that this is due to the cells’ superior ability to recruit fibers on soft substrates as opposed to stiff,2 a notion that is corroborated by a computational model presented by Cao et al. that suggests increased focal adhesion size when matrix fibers are recruited by cells.42 Highlighting the complexity of mechanoresponsive cellular behaviors that can be influenced and interrogated in these systems, modulating fiber stiffness allows for design of 3D environments with high cell infiltration, combating the poor infiltration typically seen through the small pores of electrospun scaffolds.127–129 Interestingly, Song et al. demonstrated that cellular infiltration can be improved by utilizing stiffer methacrylated-hyaluronic acid (MeHA) fibers,88 a concept that is seemingly contradictory to more cell spreading exhibited on soft fibers. This phenomenon can likely be attributed to the tendency of cells to recruit matrix fibers,88,130 which in turn decreases downstream pore size.88 In fact, Song et al. demonstrate that on short time scales, cells invade soft fibers quickly, but then are stagnant at longer time scales – whereas cells continually invade stiff fibers across these longer time scales.88 Furthering this, Heo et al. investigated the effect of nuclear stiffening as a response to matrix mechanics on cellular infiltration into these dense fibrous scaffolds.131 The result of this work demonstrated that momentary softening of the nucleus improves infiltration – suggesting that a combination of nuclear softening in conjunction with stiffer fibers can aid in cell migration into thick fibrous matrices.131
The ene–ene chain-growth polymerization is a common method for developing hydrogel fibers; however, in utilizing a chain-growth polymerization technique for crosslinking fibers and controlling mechanics, one must account for the continued growth and formation of kinetic chains in subsequent exposures to light. This additional exposure can result in increasingly stiff material environments and can cause heterogeneities leading to an inconsistent global network – an issue seen in aqueous chain-growth polymerization.132,133
To utilize thiol–ene chemistries to engineer the mechanical environment cells interacted with, Iglesias-Echevarria et al. designed a coaxial electrospinning method with PCL as the core polymer for structural stability, and PEG-norbornene (PEGNB) as the sheath for tunability.136 The PEGNB outer layer afforded control over resultant stiffness of the fibers, while also leaving behind residual norbornene groups for subsequent conjugation of thiolated RGD motifs for increased cell adhesion. The stiffness of the PEGNB sheath was modulated to investigate cellular response to differing environments. When bovine pulmonary artery endothelial cells were seeded on fibers of varying stiffnesses, higher cell infiltration and deposition of matrix materials (e.g. collagen, elastin) were seen on fibers with greater Young's moduli136 – a result in line with those mentioned above by Song et al. utilizing a MeHA fibrous system.88 Another interesting approach employed by Yang et al. involved electrospun poly((3-mercaptopropyl)methylsiloxane) (PMMS) with triallyl cyanurate (TAC) as the crosslinker.137 PMMS has pendant thiol groups that can react with any of the alkenes on TAC to form a crosslink that stabilizes the fibers, with residual thiols available for further modification. In addition to the flexibility in the crosslinking afforded by this system, Yang et al. leveraged the residual thiols on TAC to conjugate a maleimide-modified poly(N-isopropylacrylamide) (PNIPAAm) to the fibers – exploiting the thermal-responsiveness of PNIPAAm for user-control over resultant fiber hydrophobicity.137 In regard to physical properties, the thiol–ene reaction is a facile, powerful platform for the formation of hydrogel fibers for cell culture, providing high levels of control over the resultant fibrous scaffolds.
3.6 Hydrogel fibers enabling modulation of biochemical properties
Within hydrogel materials, modifications such as those described above allow for spatiotemporal modulation not just of the biophysical properties, as there has been considerable progress in utilizing the same chemistries in controlling biochemical properties. Hydrogels can be designed such that the functional groups used to bind crosslinking molecules might also bind biofunctional molecules, and careful control of the crosslinking process can leave unreacted sites within the hydrogel after crosslinking to couple molecules that increase bioactivity for cellular studies.86,87 The ene–ene and thiol–ene reaction pathways that have been described above are also commonly utilized to introduce these biochemical signals; however, there are alternative chemistries under development that achieve similar results. We aim to provide an overview of chemistries for incorporating biomolecules into nanofibrous scaffolds based on hydrogel materials, where, in comparison to hydrophobic polymers, aqueous media might be used for all reactions.36,37,86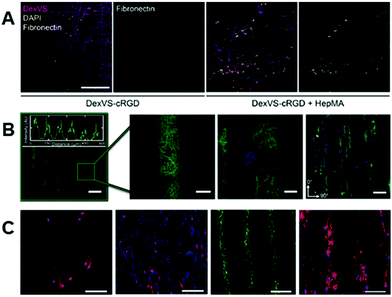 | ||
| Fig. 4 Introducing biochemical cues into fibrous hydrogels. (A, left to right): Dextran-vinyl sulfone (DexVS) fibers (magenta) were seeded with human lung fibroblasts (nuclei shown in yellow) in the presence of RGD or RGD + heparin. Conjugation of RGD + heparin to DexVS fibers increased the secretion and subsequent binding of fibronectin (white) onto the fibrous matrix. (A) Reprinted and adapted with permission from Davidson et al., copyright 2020 Elsevier;87 scalebar = 200 μm. (B, left to right): spatial patterning of thiolated fluorophores onto NorHA fibers via thiol–ene click chemistry. Zoomed in images show high pattern fidelity, and the ability to pattern multiple biomolecules on the same scaffold – indicated by the red, green, and blue fluorophores on the fibers. The ability to pattern adhesive regions, using an RGD motif, allows for preferential cellular localization in RGD + regions that elongate in the direction of fiber alignment. (B) Reprinted and adapted with permission from Wade et al., copyright 2015 John Wiley and Sons;86 scalebars (left to right) = 100 μm, 25 μm, 100 μm, and 100 μm. (C, left to right): patterning of bioactivity on synthetic fibers using UV irradiation. Rat Schwann cells exhibited a less elongated morphology on non-bioactive substrates (far left) when compared to substrates that were activated with UV light (middle left). The use of photomasks allowed for introduction of linear bioactive regions (middle right) which promoted cell attachment over non-bioactive regions (far right). (C) Reprinted and adapted with permission from Girão et al. 2019;147 scalebars (left to right) = 200 μm, 200 μm, 100 μm, and 100 μm. | ||
An important consideration in methods that functionalize fibers that were crosslinked via photoinitiated chain-growth polymerization through another photoinitiated reaction, is the effect of the subsequent reaction on kinetic chains formed during crosslinking. These kinetic chains can continue to propagate with the addition of radicals,87 and the Young's modulus of the fibers may increase with crosslinking. To surmount this challenge, researchers may leverage the Michael-type addition reaction, where thiolated molecules bind to double bonds at slightly elevated pH, in order to incorporate functional molecules onto the pendant alkenes within these systems, avoiding further polymerization.
4 Towards dynamic complexity and mimicking natural tissue
With technologies established to engineer nanofibrous substrates with specific biophysiochemical properties, it is possible to precisely control the spatial heterogeneity of biophysical and biochemical cues within the scaffolds. Because of this, there is exciting progress in the development of fibrous hydrogel systems that mimic natural tissue, with an emphasis on dynamic complexity – where properties of these systems might be designed to change or be controlled over time.4.1 Engineering degradability into hydrogel nanofibers
Advances in the engineering of bulk hydrogels, both in 2D and 3D, have demonstrated unique strengths in this area – for example in material designs using enzymatically degradable crosslinkers to allow for physiologically-mediated remodeling of the scaffolds149–151 – and it follows that nanofibers based on hydrogel systems would have similar potential. The potential to engineer materials technologies established in bulk hydrogels into hydrogel-based nanofibers is illustrated by the development of electrospun HA fibers crosslinked with a protease-sensitive crosslinker,8 establishing enzymatic degradability based on materials first used as bulk hydrogels.152 Wade and coworkers electrospun a maleimide-functionalized HA with a crosslinker peptide that was degradable enzymatically by rhMMP-2 and Type II collagenase8 (Fig. 5A). The addition of this degradability into fibrous hydrogels allows for dynamic restructuring of the fibrous ECM by resident cells via the secretion of enzymes and subsequent deposition of new matrix proteins. Wade et al. furthered this work by demonstrating degradation in vivo – highlighting aspects important to translation in a subcutaneous implantation model.8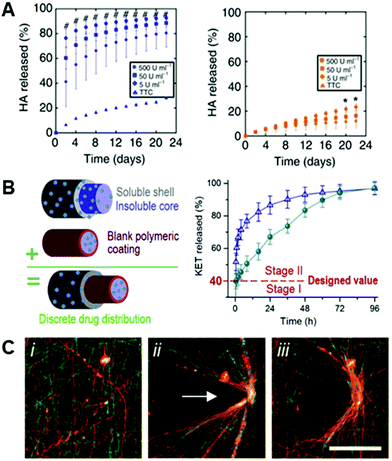 | ||
| Fig. 5 Dynamic complexity in electrospun fibers. (A): HA hydrogel fibers were crosslinked with a peptide crosslinker that was susceptible to degradation via matrix metalloproteinases (MMPs). (Left): degradation of MMP-sensitive HA fibers in the presence of differing concentrations of Type II collagenase (# p < 0.05, for all test groups versus control), and (right): degradation of HA fibers crosslinked with a peptide that is not sensitive to Type II collagenase (* p < 0.05, for 500 U mL−1 group versus control). There is a clear positive degradation effect when using an MMP-sensitive crosslinker. (A) Reprinted and adapted with permission from Wade et al., copyright 2015 Springer Nature.8 (B): Triaxial electrospun fibers for sustained drug release. (Left): schematic of the triaxial fibers that include a polymeric coating around the innermost fiber to slow drug release. (Right): Model drug release (KET) from core–shell fibers (blue triangles) and triaxial fibers (green circles). Core–shell and tri-layered fibers both exhibited quick release past stage I (40% of release), but tri-layered fibers slowed the release throughout stage II compared to core–shell fibers – due to the polymeric coating introduced around the core. (B) Reprinted and adapted with permission from Yang et al., copyright 2020 Elsevier.158 (C, left to right): Hydrazide and aldehyde-functionalized NorHA fibers (i) that react to form hydrazone bonds when in contact (ii) – allowing for permanent, covalent rearrangement of fibrous scaffolds (iii). (C) Reprinted and adapted with permission from Davidson et al., copyright 2019 John Wiley and Sons;145 scalebars = 100 μm. | ||
4.2 Dynamic fibers for selective molecule delivery
Dynamic properties in fibrous hydrogels are also embodied in applications that load the fibers with bioactive molecules to create temporal signaling. Temporal control over the release of chemokines or cytokines represent technologies with great potential for nanofibrous systems to influence cellular behavior and regeneration. Applications of controlled release from nanofibrous systems predominantly center on drug delivery applications, and there are several comprehensive reviews on this topic;55,82,153 we highlight systems here to illustrate technologies that might be applied in nanofibrous systems designed for tissue engineering and regenerative medicine.Non-hydrogel fibers have demonstrated effectiveness in the delivery of molecules by both coating fibers154,155 and incorporating bioactive molecules in the precursor solution.155 Ahire and coworkers adsorbed HA to the surface of poly(D,L, lactide) fibers and demonstrated a sustained, linear release of HA over time.154 Xia et al. also showed efficacy in the sustained delivery of adsorbed vascular endothelial growth factor (VEGF) to the surface of poly(L-lactic acid) fibers that included nerve growth factor (NGF) in the core.155 This two-step release allowed for sequential addition of biomolecules to the local environment and can, in theory, be applied to a multitude of growth/soluble factors.
Hydrogel fibers have also demonstrated promising results in the field of drug delivery. For example, Kishan and coworkers developed a platform that provides a sustained release of proteins to the local environment using different types of crosslinked gelatin fibers.156 Their methacrylated gelatin system relied on traditional mass transfer for the release of a model protein incorporated within the fibers. On the other hand, gelatin crosslinked using a diisocyanate molecule was loaded with a model protein that reacted with the gelatin backbone, and protein release in this scenario relied on gelatin degradation to free the protein from the fibers.156 These two gelatin systems can be employed together to provide a tunable, sustained release of desired proteins from hydrogel fibers to support tissue growth and regeneration.
Core–shell fibers have also proven to be advantageous in the release of bioactive molecules to the adjacent environment. In the spirit of hydrogel fibers, a core–shell fibrous system was developed for the thermally-responsive release of rhodamine B.157 The shell was comprised of poly-L-lactide-co-caprolactone (PLCL) and the core of poly(N-isopropylacrylamide-co-N-isopropylmethacrylamide) (P(NIPAAm-co-NIPMAAm)) – a thermally responsive polymer. The addition of the thermally-responsive P(NIPAAm-co-NIPMAAm) core allowed for a slower, more sustained release when compared to just a PLCL control.157 Extending this, Yang and coworkers developed triaxial nanofibers comprised of polyvinylpyrrolidone (PVP) and cellulose acetate (CA), using ketoprofen (KET) as a model drug.158 Yang et al. assert that the use of a tri-layered electrospun fiber yielded a more beneficial release profile initially, and the use of a CA blocking layer around the core provided a longer, more sustained release than a two layered system158 (Fig. 5B). While these are select examples of the extensive work in this area,55,82,153 they illustrate the potential to engineer nanofibers to control release profiles and deliver important bioactive molecules relevant in cellular systems. Continuing work in designing dynamic delivery systems has direct implications for engineering temporal complexity into electrospun fibers.
4.3 Improving cell infiltration
Incorporating dynamic properties into electrospun fibers is an important consideration in developing nanofibrous scaffolds that interface with cells and natural tissue, especially in translation of regenerative materials, as touched on above with respect to controlled release. Efforts to develop dynamic fibrous structures have sought to overcome a challenge faced by electrospun fibers in implantation: small pore sizes between fibers in larger, dense mats that are of clinically relevant dimensions prevent efficient cell infiltration into the scaffolds.127–129 One way to surmount this challenge, in addition to the aforementioned intrafiber modifications such as enzymatically degradable crosslinks, is to spin multiple fiber types into a single substrate, where a fiber type might confer dynamic features into the substrate, such as increasing its porosity upon implantation. Specifically, water-soluble poly(ethylene oxide) (PEO) sacrificial fibers that dissolve in water, but take up space during fiber deposition and contribute to the initial structure of a larger electrospun substrate, can be co-spun with a material that is stable and persists over longer timescales.159–161 This method has shown to improve infiltration, without hindering cellular transduction of microenvironmental cues.160 This technique has been extended to the development of an engineered intervertebral disc, where an annulus geometry was designed with PCL fibers as the outer shell and hydrogel as the inner core.162 The addition of PEO sacrificial fibers helped increase cell infiltration into this disc model which yielded superior matrix deposition when compared to the control that did not include sacrificial fibers.1624.4 Molecular-level dynamic complexity
Dynamic chemistries at the molecular level also offer the potential for engineering dynamic behaviors that emerge at the scales of individual fibers and fibrous systems. Chemical crosslinking approaches that allow for fibers to rearrange in response to outside perturbations—either during assembly of structures or through interactions with cells—have been demonstrated to enable the creation of complex fibrous constructs and to allow cells to modify the physical environment they experience over time. For example, dynamic supramolecular crosslinking, where non-covalent, reversible interactions occur between complementary molecules on different polymers, can be used to assemble nanofibrous substrates and create structures with biomimetic complexity. Hyaluronic acid functionalized with methacrylates for covalent stabilization of fibers and also β-cyclodextrin (CD) (CD-MeHA) can be used to create nanofibers that form reversible bonds at interfaces with materials similarly functionalized with adamantane through supramolecular host–guest interactions.84 CD is a cyclic host molecule with a hydrophobic core that hydrophobically interacts with guest molecules, such as adamantane (Ad) in noncovalent bonds that can be dynamically disrupted and restored.163–166 By designing nanofibers that present complementary functionalities on their surfaces, a nanofibrous substrate presenting CD could be adhered to another presenting Ad, offering capabilities to generate layers of aligned fibers that might be useful in cartilage or cardiac tissue engineering applications, where they might reproduce fibrous tissue structures.84Reversible bonds, like the Ad-CD guest–host system, have been demonstrated to introduce viscoelasticity into hydrogel tissue culture systems – allowing for cells to easily deform and remodel the local microenvironment.157,167,168 Nanofibrous systems with dynamic properties that enable cells to remodel their physical surroundings offer unique capabilities beyond bulk hydrogels, to observe, study, and perturb cellular behaviors through their interaction with fibrous materials. As discussed, these materials can be designed to offer ECM-like topographies as well as ECM-mimetic biophysical and biochemical features which offer cells more freedom of motion than might be achieved by encapsulating cells within a 3D hydrogel network. Towards establishing nanofibrous systems that allow dynamic, cell-responsive rearrangements of microenvironmental physical features, Davidson et al. used NorHA that was additionally modified with either hydrazide or aldehyde groups (NorHA-Hyd and NorHA-Ald, respectively) to dual-electrospin a fibrous blend of NorHA-Hyd and NorHA-Ald.145 At the fiber surfaces, hydrazide and aldehyde functional groups reacted to form hydrazone bonds when the two fiber types were in contact, i.e. an adhesive interaction145,152,169 (Fig. 5C). The interaction is proposed to allow cells to dynamically remodel the surrounding matrix by recruiting fibers with traction forces – with the recruited fibers subsequently reacting to preserve the structure.145 Xu et al. also employed this chemical functionality within poly(oligoethylene glycol methacrylate) (POEGMA) fibers. POEGMA was functionalized with hydrazide/aldehyde moieties, which allowed for immediate in situ crosslinking following double-barrel electrospinning.169 Xu et al. found that the hydrazide/aldehyde reaction allowed for the quick formation of crosslinks that were degradable both hydrolytically and enzymatically.169
4.5 Hydrogel fibers in the third dimension
Towards increasing the dimensionality of fibrous constructs or adding fibrous features to 3D tissue models, electrospun fibers have also been employed in 3D contexts – such as dispersion into bulk hydrogels124 and shape-shifting 3D scaffolds,170 as highlighted here. The addition of fibrous networks dispersed within amorphous bulk hydrogels allows for recapitulation of the fibrillar nature of endogenous ECM, in a physiologically relevant 3D environment.4 For example, Matera et al. demonstrated increased human dermal fibroblast spreading in hydrogels with dispersed dextran fibers, as well as cellular morphological changes in a fiber density-dependent manner124 (Fig. 6 Top). This example reinforces the influence of the biophysical signals that fibers provide within 3D cell culture systems as researchers progress towards perfecting models of ECM in vitro.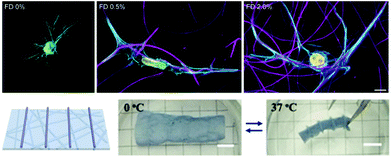 | ||
| Fig. 6 Fiber suspensions in 3D hydrogels. (Top): Dispersion of DexVS fibers in 3D GelMA hydrogels. Increasing concentrations of suspended fibers (from left to right) demonstrates stark influence of fiber density on cell morphology – 0% and 2% show high levels of spread, whereas 0.5% shows a uniaxial morphology. (Top) Reprinted and adapted with permission from Matera et al., copyright 2019 American Chemical Society;124 scalebar = 10 μm. (Bottom): P(NIPAAm-ABP) electrospun fibers with 3D printed supports. (from left to right): schematic of 3D printed supports atop of the nanofibrous P(NIPAAm-ABP) substrate; scaffold is suspended in water and adopts a relaxed conformation since the temperature is below the LCST (0° C); scaffold rolls and deforms when suspended in water with a temperature above the LCST (37° C) – thus acting as a shape-shifting hydrogel nanofiber system. (Bottom) Reprinted and adapted with permission from Chen et al., copyright 2018 John Wiley and Sons;170 scalebars = 5 mm. | ||
In an application combining electrospinning with 3D printing, Chen and coworkers demonstrated the ability to electrospin poly(N-isopropylacrylamide) (P(NIPAAm)) hydrogel nanofibrous scaffolds that were secondarily crosslinked via UV light with acryloylbenzophenone (P(NIPAAm-ABP)) to form thermo-responsive mats.170 Photocrosslinkable P(NIPAAm) solutions were also 3D printed onto these electrospun mats to provide rigid structure (i.e. trusses) to the mats. Due to P(NIPAAm)'s conformational changes above and below its lower critical solution temperature (LCST), the electrospun mats with supports exhibit shape changes upon temperature transition around the LCST due to the amount of water that is contained within the fibrous network. Below the LCST (0° C), P(NIPAAm-ABP) scaffolds demonstrated a relaxed structure; however, once the temperature was increased to above the LCST (37° C), the scaffolds rolled into shapes that were dictated by the structures 3D printed atop of the mats – hence shape-shifting nanofibrous hydrogel scaffolds (Fig. 6 Bottom).170 This system demonstrates efficacy in controlling the topography of nanofibrous hydrogel culture systems and can be extended to virtually any tissue system where 3D geometric structure is of interest.
4.6 Summary – dynamic complexity and mimicking natural tissue
Work in the field continues to advance dynamic features in fibrous cell culture systems that will be central to mimicking natural tissue systems, probing fundamental biological questions, and successfully designing systems for regenerative medicine. The inclusion of protease degradable crosslinkers, dynamic remodeling, sacrificial fibers for increased cellular infiltration, and the extension towards 3D scaffolds are key progressions in the development of fiber systems. However, the field of electrospun fibers can build on progress in 2D/3D bulk hydrogel systems, and there exists clear potential for hydrogel-based nanofibers to continue to be engineered to recapitulate native physiology and control cell behaviors.5 Next generation hydrogel fibers
As the field continues to progress towards fibrous hydrogel systems that recapture the salient features of a tissue system of interest, technology developed for engineering 2D/3D bulk hydrogels offers considerable opportunities for application in electrospun hydrogel systems. For example, expanding upon chemistries enabling dynamic degradation via the usage of a protease-sensitive crosslinker, chemical functionalities exist that allow directed degradation, such as photocleavable crosslinking through nitrobenzyl ether groups developed and demonstrated by the Anseth group.171,172 These have allowed for user-defined degradation at extremely short timescales relative to protease degradation.Technologies that allow reversible biochemical cues to be incorporated into bulk hydrogels offer the potential for dynamic spatiotemporal control over microenvironmental features. The presentation of relevant biomolecules within the ECM is in constant flux,4–7 and the ability to replicate this signaling complexity within an engineered microenvironment is critical to studying and replicating biological processes. Work that has reversibly, and repeatedly, introduced bioactive molecules into culture systems has utilized both covalent and supramolecular chemistries. Light-based approaches include nitrobenzyl ether techniques to photocleave the molecules from the scaffolds,10,173 and an allyl-sulfide chemistry has mediated multiple thiol–ene click reactions for incorporation and subsequent removal of desired molecules.11,111 These studies were conducted in PEG hydrogels, but can conceivably be applied to PEG electrospun fibers or other hydrogel fibers that are modified to support these chemistries.
Groups have also employed supramolecular chemistries to reversibly incorporate bioactive molecules in hydrogel materials. Guest–host interactions allow for self-assembly of molecules, but can be easily disrupted via the addition of a competing molecule.174 For example, Boekhoven et al. utilized β-cyclodextrin as a host molecule and took advantage of differing affinities of naphthyl and adamantane to reversibly incorporate biomolecules.174 To develop technology enabling greater control over these reversible interactions, oligonucleotides with toeholds have been employed for their ability to provide bioactive domains on hydrogel surfaces.175 Bioactivity was removed via the addition of complementary oligonucleotides that took advantage of the toehold region – providing a system with defined bioactivity by cyclical addition of these oligonucleotides.175 Both of these examples demonstrated the ability to control cell morphology and spreading based on the presentation of these bioactive ligands on alginate surfaces.174,175 Extending technologies such as these onto established hydrogel fibers would broaden opportunities to dynamically modulate complexity in water-swollen fibrous networks.
With continued progress and innovation in the materials design of fibrous hydrogel systems – and building upon exciting observations enabled by these platforms – we believe that it is inevitable that the technologies mentioned above will pave the way for platforms with increasing capabilities for recapitulating the endogenous ECM. With the growing understanding of the hydrated, fibrillar structure and function of the extracellular matrix, this progress is needed in probing fundamental physiological processes in vitro. As we progress, new capabilities to precisely define the biophysiochemical properties of an in vitro system offer opportunities for engineering biomimetic environments and controlling perturbations to homeostasis in order to understand fundamental physiological function, dysfunction, development, and regeneration. Moreover, in addition to exploring fundamental biological phenomena, technology that replicates natural tissue would enable strides towards engineered therapeutics for tissue regeneration. With applications ranging the full scale of tissue engineering – from fundamental studies to clinical translation – the development of dynamic, fibrillar hydrogels offers great potential as the field continues to develop.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the University of Virginia and the National Institutes of Health through UVA Biotechnology Training Program NIGMS 5T32 GM008715. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.References
- S. R. Caliari and J. A. Burdick, Nat. Methods, 2016, 13, 405–414 CrossRef CAS.
- B. M. Baker, B. Trappmann, W. Y. Wang, M. S. Sakar, I. L. Kim, V. B. Shenoy, J. A. Burdick and C. S. Chen, Nat. Mater., 2015, 14, 1262–1268 CrossRef CAS.
- C. D. Davidson, W. Y. Wang, I. Zaimi, D. K. P. Jayco and B. M. Baker, Sci. Rep., 2019, 9, 12 CrossRef.
- B. M. Baker and C. S. Chen, J. Cell Sci., 2012, 125, 3015–3024 CrossRef CAS.
- C. Frantz, K. M. Stewart and V. M. Weaver, J. Cell Sci., 2010, 123, 4195–4200 CrossRef CAS.
- R. J. Wade and J. A. Burdick, Mater. Today, 2012, 15, 454–459 CrossRef CAS.
- M. P. Lutolf and J. A. Hubbell, Nat. Biotechnol., 2005, 23, 47–55 CrossRef CAS.
- R. J. Wade, E. J. Bassin, C. B. Rodell and J. A. Burdick, Nat. Commun., 2015, 6, 6639 CrossRef CAS.
- A. Velasco-Hogan, J. Xu and M. A. Meyers, Adv. Mater., 2018, 30(52), 1800940 CrossRef.
- C. A. Deforest and K. S. Anseth, Angew. Chem., Int. Ed., 2012, 51, 1816–1819 CrossRef CAS.
- J. C. Grim, T. E. Brown, B. A. Aguado, D. A. Chapnick, A. L. Viert, X. Liu and K. S. Anseth, ACS Cent. Sci., 2018, 4, 909–916 CrossRef CAS.
- R. J. Wade and J. A. Burdick, Nano Today, 2014, 9, 722–742 CrossRef CAS.
- M. D. Shoulders and R. T. Raines, Annu. Rev. Biochem., 2009, 78, 929–958 CrossRef CAS.
- R. O. Hynes, Science, 2009, 326, 1216–1219 CrossRef CAS.
- L. G. Griffith and M. A. Swartz, Mol. Cell Biol., 2006, 7, 211–224 CAS.
- M. M. Stevens and J. H. George, Science, 2005, 310, 1135–1138 CrossRef CAS.
- C. Liu, P.-C. Hsu, H.-W. Lee, M. Ye, G. Zheng, N. Liu, W. Li and Y. Cui, Nat. Commun., 2015, 6, 6205 CrossRef CAS.
- Y. Wang, W. Li, Y. Xia, X. Jiao and D. Chen, J. Mater. Chem. A, 2014, 2, 15124–15131 RSC.
- T. E. Herricks, S.-H. Kim, J. Kim, D. Li, J. H. Kwak, J. W. Grate, S. H. Kim and Y. Xia, J. Mater. Chem., 2005, 15, 3241–3245 RSC.
- X. Ji, P. Wang, Z. Su, G. Ma and S. Zhang, J. Mater. Chem. B, 2014, 2, 181–190 RSC.
- M. S. Islam, B. C. Ang, A. Andriyana and A. M. Afifi, SN Appl. Sci., 2019, 1, 1248 CrossRef.
- H. Liu, X. Ding, G. Zhou, P. Li, X. Wei and Y. Fan, J. Nanomater., 2013, 495708 Search PubMed.
- A. P. Kishan and E. M. Cosgriff-Hernandez, J. Biomed. Mater. Res., Part A, 2017, 105, 2892–2905 CrossRef CAS.
- L. Sun, W. Gao, X. Fu, M. Shi, W. Xie, W. Zhang, F. Zhao and X. Chen, Biomater. Sci., 2018, 6, 340–349 RSC.
- B. Schoen, R. Avrahami, L. Baruch, Y. Efraim, I. Goldfracht, O. Elul, T. Davidov, L. Gepstein, E. Zussman and M. Machluf, Adv. Funct. Mater., 2017, 27(34), 1700427 CrossRef.
- S. De Valence, J. C. Tille, J. P. Giliberto, W. Mrowczynski, R. Gurny, B. H. Walpoth and M. Möller, Acta Biomater., 2012, 8, 3914–3920 CrossRef CAS.
- G. T. Christopherson, H. Song and H. Mao, Biomaterials, 2009, 30, 556–564 CrossRef CAS.
- S. H. Lim, X. Y. Liu, H. Song, K. J. Yarema and H. Mao, Biomaterials, 2010, 31, 9031–9039 CrossRef CAS.
- V. N. Chamundeswari, L. Y. Siang, Y. J. Chuah, J. S. Tan, D. A. Wang and S. C. J. Loo, Biomed. Mater., 2017, 13(1), 015019 CrossRef.
- M. D. Davidson, K. H. Song, M. H. Lee, J. Llewellyn, Y. Du, B. M. Baker, R. G. Wells and J. A. Burdick, ACS Biomater. Sci. Eng., 2019, 5, 3899–3908 CrossRef CAS.
- W. Y. Wang, C. D. Davidson, D. Lin and B. M. Baker, Nat. Commun., 2019, 10, 1186 CrossRef.
- K. Wingate, W. Bonani, Y. Tan, S. J. Bryant and W. Tan, Acta Biomater., 2012, 8, 1440–1449 CrossRef CAS.
- J. Xue, T. Wu, Y. Dai and Y. Xia, Chem. Rev., 2019, 119, 5298–5415 CrossRef CAS.
- M. Rahmati, D. K. Mills, A. M. Urbanska, M. R. Saeb, J. R. Venugopal, S. Ramakrishna and M. Mozafari, Prog. Mater. Sci. DOI:10.1016/j.pmatsci.2020.100721.
- J. D. Schiffman and C. L. Schauer, Polym. Rev., 2008, 48, 317–352 CrossRef CAS.
- A. M. Jordan, V. Viswanath, S. Kim, J. K. Pokorski and L. T. J. Korley, J. Mater. Chem. B, 2016, 4, 5958–5974 RSC.
- O. I. Kalaoglu-Altan, R. Sanyal and A. Sanyal, Polym. Chem., 2015, 6, 3372–3381 RSC.
- J. A. Burdick and W. L. Murphy, Nat. Commun., 2012, 3, 1269 CrossRef.
- M. W. Tibbitt and K. S. Anseth, Biotechnol. Bioeng., 2009, 103, 655–663 CrossRef CAS.
- C. B. Highley, G. D. Prestwich and J. A. Burdick, Curr. Opin. Biotechnol., 2016, 40, 35–40 CrossRef CAS.
- J. A. Burdick and G. D. Prestwich, Adv. Healthcare Mater., 2011, 23, H41–H56 CrossRef CAS.
- X. Cao, E. Ban, B. M. Baker, Y. Lin, J. A. Burdick, C. S. Chen and V. B. Shenoy, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E4549–E4555 CrossRef CAS.
- W.-J. Li, C. T. Laurencin, E. J. Caterson, R. S. Tuan and F. K. Ko, J. Biomed. Mater. Res., 2002, 60, 613–621 CrossRef CAS.
- B. L.-P. Lee, H. Jeon, A. Wang, Z. Yan, J. Yu, C. Grigoropoulos and S. Li, Acta Biomater., 2012, 8, 2648–2658 CrossRef CAS.
- J. F. Piai, M. A. da Silva, A. Martins, A. B. Torres, S. Faria, R. L. Reis, E. C. Muniz and N. M. Neves, Appl. Surf. Sci., 2017, 403, 112–125 CrossRef CAS.
- I. Shabani, V. Haddadi-Asl, E. Seyedjafari and M. Soleimani, Biochem. Biophys. Res. Commun., 2012, 423, 50–54 CrossRef CAS.
- L. He, S. Tang, M. P. Prabhakaran, S. Liao, L. Tian, Y. Zhang, W. Xue and S. Ramakrishna, Macromol. Biosci., 2013, 13, 1601–1609 CrossRef CAS.
- Y.-G. Ko and O. H. Kwon, J. Ind. Eng. Chem., 2020, 89, 147–155 CrossRef CAS.
- S. Soliman, S. Sant, J. W. Nichol, M. Khabiry, E. Traversa and A. Khademhosseini, J. Biomed. Mater. Res., Part A, 2011, 96, 566–574 CrossRef.
- H. Savoji, A. Hadjizadeh, M. Maire, A. Ajji, M. R. Wertheimer and S. Lerouge, Macromol. Biosci., 2014, 14, 1084–1095 CrossRef CAS.
- X. Hu, S. Liu, G. Zhou, Y. Huang, Z. Xie and X. Jing, J. Controlled Release, 2014, 185, 12–21 CrossRef CAS.
- C. A. Bashur, R. D. Shaffer, L. A. Dahlgren, S. A. Guelcher and A. S. Goldstein, Tissue Eng. Part A, 2009, 15, 2435–2445 CrossRef CAS.
- A. Haider, S. Haider and I. K. Kang, Arabian J. Chem., 2018, 11, 1165–1188 CrossRef CAS.
- S. Agarwal, J. H. Wendorff and A. Greiner, Polymer, 2008, 49, 5603–5621 CrossRef CAS.
- S. Chakraborty, I.-C. Liao, A. Adler and K. W. Leong, Adv. Drug Delivery Rev., 2009, 61, 1043–1054 CrossRef CAS.
- A. E. Miller, P. Hu and T. H. Barker, Adv. Healthcare Mater., 2020, 9, 1901445 CrossRef CAS.
- T. Peijs, in Comprehensive Composite Materials II, 2018, pp. 162–200 Search PubMed.
- Q. P. Pham, U. Sharma and A. G. Mikos, Tissue Eng., 2006, 12, 1197–1211 CrossRef CAS.
- S. S. Ojha, in Electospun Nanofibers, 2017, pp. 239–253 Search PubMed.
- R. M. Nezarati, M. B. Eifert, D. K. Dempsey and E. Cosgriff-Hernandez, J. Biomed. Mater. Res., Part B, 2015, 103, 313–323 CrossRef.
- S. M. Park, S. Eom, D. Choi, S. J. Han, S. J. Park and D. S. Kim, Chem. Eng. J., 2018, 335, 712–719 CrossRef CAS.
- S. E. Szczesny, T. P. Driscoll, H.-Y. Tseng, P.-C. Liu, S.-J. Heo, R. L. Mauck and P.-H. G. Chao, ACS Biomater. Sci. Eng., 2017, 3, 2869–2876 CrossRef CAS.
- H. Chen, D. F. Baptista, G. Criscenti, J. Crispim, H. Fernandes, C. van Blitterswijk, R. Truckenmüller and L. Moroni, Nanoscale, 2019, 11, 14312–14321 RSC.
- D. Seliktar, Science, 2012, 336, 1124–1129 CrossRef CAS.
- W. L. Murphy, T. C. McDevitt and A. J. Engler, Nat. Mater., 2014, 13, 547–557 CrossRef CAS.
- L. G. Griffith and G. Naughton, Science, 2002, 295, 1009–1014 CrossRef CAS.
- K. E. Kador, H. S. Alsehli, A. N. Zindell, L. W. Lau, F. M. Andreopoulos, B. D. Watson and J. L. Goldberg, Acta Biomater., 2014, 10, 4939–4946 CrossRef.
- H. Lee, J. Rho and P. B. Messersmith, Adv. Mater., 2009, 21, 431–434 CrossRef CAS.
- H. Cho, S. K. Madhurakkat Perikamana, J. Lee, J. Lee, K.-M. Lee, C. S. Shin and H. Shin, ACS Appl. Mater. Interfaces, 2014, 6, 11225–11235 CrossRef CAS.
- N. Nazeri, R. Karimi and H. Ghanbari, J. Biomed. Mater. Res., Part A, 2021, 109(2), 159–169 CrossRef CAS.
- Y. M. Shin, I. Jun, Y.-M. Lim, T. Rhim and H. Shin, Macromol. Mater. Eng., 2013, 298, 555–564 CrossRef CAS.
- M. E. Lynge, R. van der Westen, A. Postma and B. Städler, Nanoscale, 2011, 3, 4916–4928 RSC.
- J. H. Ryu, P. B. Messersmith and H. Lee, ACS Appl. Mater. Interfaces, 2018, 10, 7523–7540 CrossRef CAS.
- X. Qiu, B. L.-P. Lee, X. Ning, N. Murthy, N. Dong and S. Li, Acta Biomater., 2017, 51, 138–147 CrossRef CAS.
- M. L. Tanes, J. Xue and Y. Xia, J. Mater. Chem. B, 2017, 5, 5580–5587 RSC.
- T. Wu, J. Xue, H. Li, C. Zhu, X. Mo and Y. Xia, ACS Appl. Mater. Interfaces, 2018, 10, 8536–8545 CrossRef CAS.
- E. Haldón, M. C. Nicasio and P. J. Pérez, Org. Biomol. Chem., 2015, 13, 9528–9550 RSC.
- N. E. Mbua, J. Guo, M. A. Wolfert, R. Steet and G.-J. Boons, ChemBioChem, 2011, 12, 1912–1921 CrossRef CAS.
- Q. Shi, X. Chen, T. Lu and X. Jing, Biomaterials, 2008, 29, 1118–1126 CrossRef CAS.
- L. A. Smith Callahan, S. Xie, I. A. Barker, J. Zheng, D. H. Reneker, A. P. Dove and M. L. Becker, Biomaterials, 2013, 34, 9089–9095 CrossRef CAS.
- J. Zheng, S. Xie, F. Lin, G. Hua, T. Yu, D. H. Reneker and M. L. Becker, Polym. Chem., 2013, 4, 2215–2218 RSC.
- V. Pillay, C. Dott, Y. E. Choonara, C. Tyagi, L. Tomar, P. Kumar, L. C. du Toit and V. M. K. Ndesendo, J. Nanomater., 2013, 789289 Search PubMed.
- G. G. de Lima, S. Lyons, D. M. Devine and M. J. D. Nugent, in Hydrogels, Springer, Singapore, 2018, pp. 219–258 Search PubMed.
- C. B. Highley, C. B. Rodell, I. L. Kim, R. J. Wade and J. A. Burdick, J. Mater. Chem. B, 2015, 2, 8110–8115 RSC.
- S. Sharma, M. Floren, Y. Ding, K. R. Stenmark, W. Tan and S. J. Bryant, Biomaterials, 2017, 143, 17–28 CrossRef CAS.
- R. J. Wade, E. J. Bassin, W. M. Gramlich and J. A. Burdick, Adv. Mater., 2015, 27, 1356–1362 CrossRef CAS.
- C. D. Davidson, D. K. P. Jayco, D. L. Matera, S. J. DePalma, H. L. Hiraki, W. Y. Wang and B. M. Baker, Acta Biomater., 2020, 105, 78–86 CrossRef CAS.
- K. H. Song, S.-J. Heo, A. P. Peredo, M. D. Davidson, R. L. Mauck and J. A. Burdick, Adv. Healthcare Mater., 2019, 9, 1901228 CrossRef.
- L. Moroni, J. A. Burdick, C. Highley, S. J. Lee, Y. Morimoto, S. Takeuchi and J. J. Yoo, Nat. Rev. Mater., 2018, 3, 21–37 CrossRef CAS.
- M. Li, M. J. Mondrinos, M. R. Gandhi, F. K. Ko, A. S. Weiss and P. I. Lelkes, Biomaterials, 2005, 26, 5999–6008 CrossRef CAS.
- M. Y. Kwon, C. Wang, J. H. Galarraga, E. Puré, L. Han and J. A. Burdick, Biomaterials, 2019, 222, 119451 CrossRef CAS.
- S. Misra, V. C. Hascall, R. R. Markwald and S. Ghatak, Front. Immunol., 2015, 6, 201 Search PubMed.
- C. Chung, M. Beecham, R. L. Mauck and J. A. Burdick, Biomaterials, 2009, 30, 4287–4296 CrossRef CAS.
- J. J. Roberts and S. J. Bryant, Biomaterials, 2013, 34, 9969–9979 CrossRef CAS.
- D. A. Castilla-Casadiego, H. V. Ramos-Avilez, S. Herrera-Posada, B. Calcagno, L. Loyo, J. Shipmon, A. Acevedo, A. Quintana and J. Almodovar, Macromol. Mater. Eng., 2016, 301, 1064–1075 CrossRef CAS.
- C. E. Campiglio, N. C. Negrini, S. Farè and L. Draghi, Materials, 2019, 12, 2476 CrossRef CAS.
- B. S. Jha, C. E. Ayres, J. R. Bowman, T. A. Telemeco, S. A. Sell, G. L. Bowlin and D. G. Simpson, J. Nanomater., 2011, 348268 Search PubMed.
- H. Aoki, H. Miyoshi and Y. Yamagata, Polym. J., 2015, 47, 267–277 CrossRef CAS.
- X. Zhang, K. Tang and X. Zheng, J. Bionic Eng., 2016, 13, 143–149 CrossRef.
- A. P. Kishan, R. M. Nezarati, C. M. Radzicki, A. L. Renfro, J. L. Robinson, M. E. Whitely and E. M. Cosgriff-Hernandez, J. Mater. Chem. B, 2015, 3, 7930–7938 RSC.
- L. Liu, K. Kamei, M. Yoshioka, M. Nakajima, J. Li, N. Fujimoto, S. Terada, Y. Tokunaga, Y. Koyama, H. Sato, K. Hasegawa, N. Nakatsuji and Y. Chen, Biomaterials, 2017, 124, 47–54 CrossRef CAS.
- R. L. Fischer, M. G. McCoy and S. A. Grant, J. Mater. Sci. Mater. Med., 2012, 23, 1645–1654 CrossRef CAS.
- Z. Ghassemi and G. Slaughter, Conf. Proc. IEEE Eng. Med. Biol. Soc., 2018, pp. 6088–6091.
- R. E. Young, J. Graf, I. Miserocchi, R. M. Van Horn, M. B. Gordon, C. R. Anderson and L. S. Sefcik, PLoS One, 2019, 14, 1–15 Search PubMed.
- P. Lu and Y.-L. Hsieh, Polymer, 2009, 50, 3670–3679 CrossRef CAS.
- A. G. Destaye, C. K. Lin and C. K. Lee, ACS Appl. Mater. Interfaces, 2013, 5, 4745–4752 CrossRef CAS.
- J. C. Park, T. Ito, K. O. Kim, K. W. Kim, B. S. Kim, M. S. Khil, H. Y. Kim and I. S. Kim, Polym. J., 2010, 42, 273–276 CrossRef CAS.
- C. K. Kim, B. S. Kim, F. A. Sheikh, U. S. Lee, M. S. Khil and H. Y. Kim, Macromolecules, 2007, 40, 4823–4828 CrossRef CAS.
- E. Hui, K. I. Gimeno, G. Guan and S. R. Caliari, Biomacromolecules, 2019, 20, 4126–4134 CrossRef CAS.
- C. E. Hoyle and C. N. Bowman, Angew. Chem., Int. Ed., 2010, 49, 1540–1573 CrossRef CAS.
- J. C. Grim, I. A. Marozas and K. S. Anseth, J. Controlled Release, 2015, 219, 95–106 CrossRef CAS.
- W. M. Gramlich, I. L. Kim and J. A. Burdick, Biomaterials, 2013, 34, 9803–9811 CrossRef CAS.
- J. L. Ifkovits and J. A. Burdick, Tissue Eng., 2007, 13, 2369–2385 CrossRef CAS.
- A. I. Van Den Bulcke, B. Bogdanov, N. De Rooze, E. H. Schacht, M. Cornelissen and H. Berghmans, Biomacromolecules, 2000, 1, 31–38 CrossRef CAS.
- A. A. Aldana, L. Malatto, M. A. U. Rehman, A. R. Boccaccini and G. A. Abraham, Nanomaterials, 2019, 9, 120 CrossRef.
- X. Zhao, X. Sun, L. Yildirimer, Q. Lang, Z. Y. Lin, R. Zheng, Y. Zhang, W. Cui, N. Annabi and A. Khademhosseini, Acta Biomater., 2017, 49, 66–77 CrossRef CAS.
- A. O. Lobo, S. Afewerki, M. M. M. de Paula, P. Ghannadian, F. R. Marciano, Y. S. Zhang, T. J. Webster and A. Khademhosseini, Int. J. Nanomed., 2018, 13, 7891–7903 CrossRef CAS.
- I. L. Kim, S. Khetan, B. M. Baker, C. S. Chen and J. A. Burdick, Biomaterials, 2013, 34, 5571–5580 CrossRef CAS.
- I. V. Bessonov, Y. A. Rochev, A. Y. Arkhipova, M. N. Kopitsyna, D. V. Bagrov, E. A. Karpushkin, T. N. Bibikova, A. M. Moysenovich, A. S. Soldatenko, I. I. Nikishin, M. S. Kotliarova, V. G. Bogush, K. V. Shaitan and M. M. Moisenovich, Biomed. Mater., 2019, 14, 034102 CrossRef CAS.
- S. Bin Bae, M. H. Kim and W. H. Park, Polym. Degrad. Stab., 2020, 179, 109304 CrossRef.
- A. R. Tan, J. L. Ifkovits, B. M. Baker, D. M. Brey, R. L. Mauck and J. A. Burdick, J. Biomed. Mater. Res., Part A, 2008, 87, 1034–1043 CrossRef.
- H. G. Sundararaghavan, R. B. Metter and J. A. Burdick, Macromol. Biosci., 2010, 10, 265–270 CrossRef CAS.
- B. Dong, O. Arnoult, M. E. Smith and G. E. Wnek, Macromol. Rapid Commun., 2009, 30, 539–542 CrossRef CAS.
- D. L. Matera, W. Y. Wang, M. R. Smith, A. Shikanov and B. M. Baker, ACS Biomater. Sci. Eng., 2019, 5, 2965–2975 CrossRef CAS.
- M. S. Hall, F. Alisafaei, E. Ban, X. Feng, C. Y. Hui, V. B. Shenoy and M. Wu, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 14043–14048 CrossRef CAS.
- H. G. Sundararaghavan, R. L. Saunders, D. A. Hammer and J. A. Burdick, Biotechnol. Bioeng., 2013, 110, 1249–1254 CrossRef CAS.
- B. M. Baker, A. S. Nathan, G. R. Huffman and R. L. Mauck, Osteoarthr. Cartil., 2009, 17, 336–345 CrossRef CAS.
- F. Qu, F. Guilak and R. L. Mauck, Nat. Rev. Rheumatol., 2019, 15, 167–179 CrossRef.
- B. M. Baker and R. L. Mauck, Biomaterials, 2007, 28, 1967–1977 CrossRef CAS.
- A. S. Abhilash, B. M. Baker, B. Trappmann, C. S. Chen and V. B. Shenoy, Biophys. J., 2014, 107, 1829–1840 CrossRef CAS.
- S. J. Heo, K. H. Song, S. Thakur, L. M. Miller, X. Cao, A. P. Peredo, B. N. Seiber, F. Qu, T. P. Driscoll, V. B. Shenoy, M. Lakadamyali, J. A. Burdick and R. L. Mauck, Sci. Adv., 2020, 6, 1–13 Search PubMed.
- A. M. Kloxin, C. J. Kloxin, C. N. Bowman and K. S. Anseth, Adv. Mater., 2010, 22, 3484–3494 CrossRef CAS.
- S. Lin-Gibson, R. L. Jones, N. R. Washburn and F. Horkay, Macromolecules, 2005, 38, 2897–2902 CrossRef CAS.
- K. Shanmuganathan, R. K. Sankhagowit, P. Iyer and C. J. Ellison, Chem. Mater., 2011, 23, 4726–4732 CrossRef CAS.
- O. I. Kalaoglu-Altan, B. Verbraeken, K. Lava, T. N. Gevrek, R. Sanyal, T. Dargaville, K. De Clerck, R. Hoogenboom and A. Sanyal, ACS Macro Lett., 2016, 5, 676–681 CrossRef CAS.
- M. Iglesias-Echevarria, L. Durante, R. Johnson, M. Rafuse, Y. Ding, W. Bonani, D. Maniglio and W. Tan, Biomater. Sci., 2019, 7, 3640–3651 RSC.
- H. Yang, Q. Zhang, B. Lin, G. Fu, X. Zhang and L. Guo, J. Polym. Sci., Part A: Polym. Chem., 2012, 50, 4182–4190 CrossRef CAS.
- E. A. Mays, S. S. Kallakuri and H. G. Sundararaghavan, J. Biomed. Mater. Res., Part A, 2020, 1–9 Search PubMed.
- D. P. Nair, M. Podgórski, S. Chatani, T. Gong, W. Xi, C. R. Fenoli and C. N. Bowman, Chem. Mater., 2014, 26, 724–744 CrossRef CAS.
- C. L. Petrou, T. J. D’Ovidio, D. A. Bolukbas, S. Tas, R. D. Brown, A. Allawzi, S. Lindstedt, E. Nozik-Grayck, K. R. Stenmark, D. E. Wagner and C. M. Magin, J. Mater. Chem. B, 2020, 8, 6814–6826 RSC.
- B. Trappmann, B. M. Baker, W. J. Polacheck, C. K. Choi, J. A. Burdick and C. S. Chen, Nat. Commun., 2017, 8, 371 CrossRef.
- S. P. Zustiak, H. Boukari and J. B. Leach, Soft Matter, 2010, 6, 3609–3618 RSC.
- K. Han, W.-N. Yin, J.-X. Fan, F.-Y. Cao and X.-Z. Zhang, ACS Appl. Mater. Interfaces, 2015, 7, 23679–23684 CrossRef CAS.
- H. G. Sundararaghavan and J. A. Burdick, Biomacromolecules, 2011, 12, 2344–2350 CrossRef CAS.
- M. D. Davidson, E. Ban, A. C. M. Schoonen, M.-H. Lee, M. D’Este, V. B. Shenoy and J. A. Burdick, Adv. Mater., 2020, 32(8), 1905719 CrossRef CAS.
- C. D. Davidson, D. K. P. Jayco, W. Y. Wang, A. Shikanov and B. M. Baker, J. Biomech. Eng., 2020, 142, 1–9 CrossRef.
- A. F. Girão, P. Wieringa, S. C. Pinto, P. A. A. P. Marques, S. Micera, R. van Wezel, M. Ahmed, R. Truckenmueller and L. Moroni, Front. Bioeng. Biotechnol., 2019, 7, 159 CrossRef.
- K. Saha, J. F. Pollock, D. V. Schaffer and K. E. Healy, Curr. Opin. Chem. Biol., 2007, 11, 381–387 CrossRef CAS.
- L. R. Nih, P. Moshayedi, I. L. Llorente, A. R. Berg, J. Cinkornpumin, W. E. Lowry, T. Segura and S. T. Carmichael, Data in Brief, 2017, 10, 202–209 CrossRef.
- L. R. Nih, E. Sideris, S. T. Carmichael and T. Segura, Adv. Mater., 2017, 29(32), 1606471 CrossRef.
- J. L. Holloway, H. Ma, R. Rai and J. A. Burdick, J. Controlled Release, 2014, 191, 63–70 CrossRef CAS.
- B. P. Purcell, D. Lobb, M. B. Charati, S. M. Dorsey, R. J. Wade, K. N. Zellars, H. Doviak, S. Pettaway, C. B. Logdon, J. A. Shuman, P. D. Freels, J. H. Gorman III, R. C. Gorman, F. G. Spinale and J. A. Burdick, Nat. Mater., 2014, 13, 653–661 CrossRef CAS.
- R. S. Bhattarai, R. D. Bachu, S. H. S. Boddu and S. Bhaduri, Pharmaceutics, 2019, 11(1) DOI:10.3390/pharmaceutics11010005.
- J. J. Ahire, D. Robertson, D. P. Neveling, A. J. Van Reenen and L. M. T. Dicks, RSC Adv., 2016, 6, 34791–34796 RSC.
- B. Xia and Y. Lv, Mater. Sci. Eng. C, 2018, 82, 253–264 CrossRef CAS.
- A. Kishan, T. Walker, N. Sears, T. Wilems and E. Cosgriff-Hernandez, J. Biomed. Mater. Res., Part A, 2018, 106, 1155–1164 CrossRef CAS.
- S. Pawłowska, C. Rinoldi, P. Nakielski, Y. Ziai, O. Urbanek, X. Li, T. A. Kowalewski, B. Ding and F. Pierini, Adv. Mater. Interfaces, 2020, 7(12), 2000247 CrossRef.
- Y. Yang, S. Chang, Y. Bai, Y. Du and D.-G. Yu, Carbohydr. Polym., 2020, 243, 116477 CrossRef CAS.
- M. C. Phipps, W. C. Clem, J. M. Grunda, G. A. Clines and S. L. Bellis, Biomaterials, 2012, 33, 524–534 CrossRef CAS.
- B. M. Baker, R. P. Shah, A. M. Silverstein, J. L. Esterhai, J. A. Burdick and R. L. Mauck, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 14176–14181 CrossRef CAS.
- B. M. Baker, A. O. Gee, R. B. Metter, A. S. Nathan, R. A. Marklein, J. A. Burdick and R. L. Mauck, Biomaterials, 2008, 29, 2348–2358 CrossRef CAS.
- B. G. Ashinsky, S. E. Gullbrand, E. D. Bonnevie, C. Wang, D. H. Kim, L. Han, R. L. Mauck and H. E. Smith, Acta Biomater., 2020, 111, 232–241 CrossRef CAS.
- M. E. Gomes, R. M. A. Domingues and R. L. Reis, Tissue Eng., Part B, 2017, 23, 211–224 CrossRef.
- C. B. Highley, C. B. Rodell and J. A. Burdick, Adv. Mater., 2015, 27, 5075–5079 CrossRef CAS.
- G. Sinawang, M. Osaki, Y. Takashima, H. Yamaguchi and A. Harada, Polym. J., 2020, 52, 839–859 CrossRef CAS.
- M. Tanaka, M. Nakahata, P. Linke and S. Kaufmann, Polym. J., 2020, 52, 861–870 CrossRef CAS.
- C. Loebel, R. L. Mauck and J. A. Burdick, Nat. Mater., 2019, 18, 883–891 CrossRef CAS.
- K. H. Vining, A. Stafford and D. J. Mooney, Biomaterials, 2019, 188, 187–197 CrossRef CAS.
- F. Xu, H. Sheardown and T. Hoare, Chem. Commun., 2016, 52, 1451–1454 RSC.
- T. Chen, H. Bakhshi, L. Liu, J. Ji and S. Agarwal, Adv. Funct. Mater., 2018, 28, 3–9 Search PubMed.
- A. M. Kloxin, A. M. Kasko, C. N. Salinas and K. S. Anseth, Science, 2009, 324, 59–63 CrossRef CAS.
- C. A. DeForest and K. S. Anseth, Nat. Chem., 2011, 3, 925–931 CrossRef CAS.
- C. A. Deforest and D. A. Tirrell, Nat. Mater., 2015, 14, 523–531 CrossRef CAS.
- J. Boekhoven, C. M. Rubert Pérez, S. Sur, A. Worthy and S. I. Stupp, Angew. Chem., Int. Ed., 2013, 52, 12077–12080 CrossRef CAS.
- R. Freeman, N. Stephanopoulos, Z. Álvarez, J. A. Lewis, S. Sur, C. M. Serrano, J. Boekhoven, S. S. Lee and S. I. Stupp, Nat. Commun., 2017, 8, 15982 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2021 |


