Peptide modified polycations with pH triggered lytic activity for efficient gene delivery†
Xiaojing
Chen
a,
Kai
Xu
a,
Jing
Yu
a,
Xiaodan
Zhao
b,
Qiang
Zhang
a,
Yanfeng
Zhang
 a and
Yilong
Cheng
a and
Yilong
Cheng
 *a
*a
aSchool of Chemistry, Xi'an Jiaotong University, Xi'an 710049, China. E-mail: yilongcheng@mail.xjtu.edu.cn
bKey Laboratory of Shaanxi Province for Craniofacial Precision Medicine Research, College of Stomatology, Xi'an Jiaotong University, Xi'an 710049, China
First published on 15th September 2020
Abstract
Endo/lysosome entrapment is the key barrier for gene delivery using synthetic polycations. Although the introduction of a membrane-lytic peptide into polycations could facilitate efficient endo/lysosome release and improve gene delivery efficiency, it is always accompanied by serious safety concerns. In this work, the widely used polycations, poly(2-dimethylaminoethyl methacrylate (PDMAEMA), poly(L-lysine) (PLL) and polyethylenimine (PEI), are modified with a pH-sensitive peptide (C6M3) with selective lytic activity to produce three functional polycations to address the issue of endo/lysosome entrapment and facilitate efficient gene transfer. Hemolysis study shows that the functionalized polycations show good biocompatibility toward red blood cells at neutral pH, and exhibit potent membrane lysis activity under acidic conditions, which are both on-demand for the ideal gene carriers. In vitro transfection studies demonstrate that the peptide modified polycations mediate promising gene delivery efficiency with the luciferase plasmid and the green fluorescence protein plasmid in HeLa cells compared to the parent polycations. Owing to the facile preparation and selective lysis activity of the C6M3 modified polycations, these smart gene vectors may be good candidates for the transfer of various nucleic acids and further clinical gene therapy.
1. Introduction
Delivery of therapeutic nucleic acids into target cells is regarded as an efficient way to tackle a myriad of acquired or congenital diseases.1–6 However, the poor stability under physiological conditions and the impermeability of cell membranes to nucleic acids significantly compromise the gene therapy efficiency. Hence, the assistance of a carrier to successfully transfer nucleic acids into the intracellular parts is of importance for gene delivery.7–12 Compared to viral carriers, non-viral carriers, especially polycations, feature various advantages, such as affordable facile preparation and functionalization, low immunogenicity and inflammation, which have been widely employed for the delivery of plasmid DNA (pDNA),13–17 small interfering RNA (siRNA),18–24 messenger RNA (mRNA) and micro RNA (miRNA).25–31 However, due to the limited capability to overcome various physiological barriers, the gene delivery efficiency is usually orders of magnitude lower than those of their viral counterparts.32–35Endo/lysosome entrapment is the key obstacle for polycation-based intracellular gene delivery.36 If the formed complexes of nucleic acids and polycations are trapped in the endosome and cannot escape in time, it will be automatically routed into lysosome degradation, leading to failed gene transfer.37–39 Tremendous efforts have been made to address this issue.40–45 The proton sponge effect is the widely used method for polycation mediated endosomal release.46,47 However, it cannot be translated into in vivo applications due to the requirement of a large amount of polymer accumulation in the endo/lysosome, which may raise safety concerns. As an alternative way, peptides with membrane-lytic activity, such as melittin and sHGP, have been introduced into polycations to facilitate endo/lysosomal escape.48–50 The peptide-conjugated polymers can significantly improve the gene delivery efficiency compared to the parent polymers. But due to off-site lysis, serious cytotoxicity was also detected. We recently designed a pH-sensitive block copolymer, called VIPER (virus-inspired polymer for endosomal release), aiming to minimize the side effect.28,42 VIPER can assemble into nanoparticles with a lytic peptide shielded in the core; that responds to endosomal acidification by revealing a membrane-lytic peptide that is conjugated to a reversibly hydrophobic polymer block, followed by promising gene transfer efficiency in various cell lines in vitro and different tissues in vivo. However, the cleavage of the disulfide linkage between melittin and the polymer may raise undesired safety concerns because of the native lytic activity of melittin under physiological conditions.51
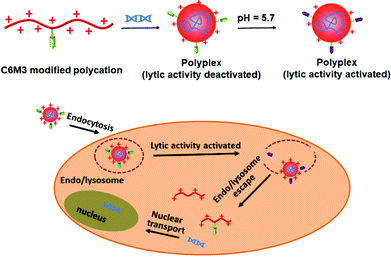
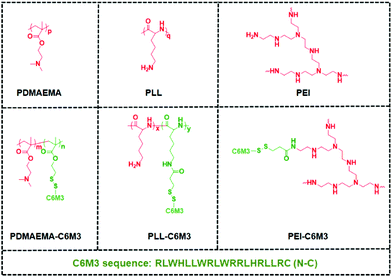
Previous studies showed that the C6M3 peptide (a peptide derived from C6) features pH-triggered lytic activity, and promising membrane lysis was observed under endo/lysosomal acidic conditions rather than at a normal pH value.51,52 We hypothesize that the incorporation of polycations with C6M3 could not only enhance the gene delivery efficiency through the promotion of endo/lysosomal escape but also avoid off-site toxicity. As shown in Scheme 1, the C6M3 modified polycations condense DNA into nanoparticles, which can be endocytosed by cells through the endosome pathway. Afterward, C6M3 can be activated to mediate endo/lysosomal escape of the polyplexes. In this work, three widely used polycations, poly(2-dimethylaminoethyl methacrylate (PDMAEMA), poly(L-lysine) (PLL) and branched polyethylenimine (PEI), are modified with the C6M3 peptide (Fig. 1), and the pH selective hemolysis was evaluated with mouse red blood cells to verify the desired membrane-lytic activity. The functionalized polycations are also well assessed. To explore the potential for gene therapy, the luciferase plasmid and the green fluorescence protein (GFP) plasmid as reporter genes are employed to evaluate the gene delivery efficiency of the polycations in vitro.
2. Experimental section
2.1. Materials
The chemical and biological reagents used in this work are listed in the ESI.†2.2. Characterization
The 1H nuclear magnetic resonance (1H NMR) spectra were recorded on a Bruker Avance 400 spectrometer (400 MHz). The size and surface charge of the polyplexes were tested on a Malvern ZEN 3690 system. The morphology of all the polyplexes in the dried state was imaged with a JEOL JEM-2100 electron microscope using an acceleration voltage of 120 kV. UV–vis absorption spectra were measured with a Shimadzu UV-2550 spectrometer.2.3. Polymer synthesis
2.4. Gel retardation
For polyplex preparation, the parent and peptide modified polymer solutions with different concentrations were added to the luciferase plasmid solution (v/v = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), and the formed polyplex solutions were vortexed for 15 seconds and incubated at room temperature for 10 min. The polyplex solution (12 μL) with different N/P ratios was subjected to electrophoresis on agarose gel (1%) at 100 V for 45 min to test the DNA condensing ability.
1), and the formed polyplex solutions were vortexed for 15 seconds and incubated at room temperature for 10 min. The polyplex solution (12 μL) with different N/P ratios was subjected to electrophoresis on agarose gel (1%) at 100 V for 45 min to test the DNA condensing ability.
2.5. Hemolysis of polycations
Hemolysis assay was used to evaluate the acid-triggered membrane-lytic activity of the synthetic materials at pH 7.4 and 5.7. The details are presented in the ESI.†2.6. In vitro transfection
Luciferase and GFP plasmids were used to evaluate the gene delivery efficiency of the polycations. HeLa cells were seeded with a density of 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well (24 well plate) in MEM medium supplemented with 10% FBS and 1% antibiotic/antimicrobial. Cells were firstly incubated at 37 °C under 5% CO2 for 24 h. Polyplexes were prepared at different N/P ratios using 1 μg of pGL3 in 20 μL total volume. Each sample was diluted with 180 μL OptiMEM medium. For transfection study, the cells were washed with PBS, followed by the addition of the polyplex solution. After incubation for 4 h, the cells were rinsed with PBS twice and complete cell culture medium (500 μL) was added. After an additional 44 h of incubation, luciferase activity was quantified with a luciferase assay kit (Promega Corp, Fitchburg, WI). The total protein content in each well was measured using a BCA Protein Assay Kit. The GFP plasmid delivery study was the same as that with the luciferase plasmid. Flow cytometry was used to analyze the percentage of the transfected cells.42 All experiments were conducted in triplicate.
000 cells per well (24 well plate) in MEM medium supplemented with 10% FBS and 1% antibiotic/antimicrobial. Cells were firstly incubated at 37 °C under 5% CO2 for 24 h. Polyplexes were prepared at different N/P ratios using 1 μg of pGL3 in 20 μL total volume. Each sample was diluted with 180 μL OptiMEM medium. For transfection study, the cells were washed with PBS, followed by the addition of the polyplex solution. After incubation for 4 h, the cells were rinsed with PBS twice and complete cell culture medium (500 μL) was added. After an additional 44 h of incubation, luciferase activity was quantified with a luciferase assay kit (Promega Corp, Fitchburg, WI). The total protein content in each well was measured using a BCA Protein Assay Kit. The GFP plasmid delivery study was the same as that with the luciferase plasmid. Flow cytometry was used to analyze the percentage of the transfected cells.42 All experiments were conducted in triplicate.
2.7. Statistical analysis
All statistical analyses were performed using a two-tailed Student's t-test with unequal variance.3. Results and discussion
3.1. Polymer synthesis and characterization
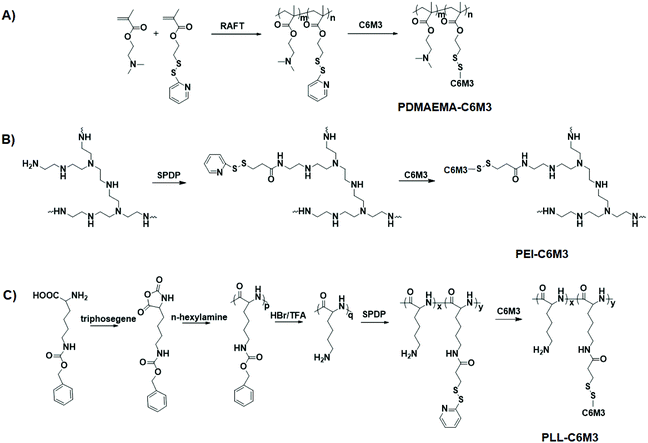
The synthetic routes of the polycations are shown in Scheme 2A–C. To synthesize C6M3 modified PDMAEMA, a functional monomer, PDSEMA, was first copolymerized with DMAEMA to afford P(DMAEMA-co-PDSEMA). The 1H NMR spectra are shown in Fig. S1,† and all the signals were assigned. The composition of the polymer was determined to be P(DMAEMA98-co-PDSEMA2) based on the conversion (∼100%) of the monomers. The GPC results showed that the molecular weight of the copolymer was 218![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, and the polydispersity index was 1.19. PZLL was synthesized by the ring-opening polymerization of 3-benzyloxycarbonyl-L-lysine N-carboxyanhydride (ZLL NCA) in the presence of hexylamine. The degree of polymerization of PZLL was determined to be 44 based on the 1H NMR spectrum (Fig. S2†), and the molecular weight of PZLL was 166
000, and the polydispersity index was 1.19. PZLL was synthesized by the ring-opening polymerization of 3-benzyloxycarbonyl-L-lysine N-carboxyanhydride (ZLL NCA) in the presence of hexylamine. The degree of polymerization of PZLL was determined to be 44 based on the 1H NMR spectrum (Fig. S2†), and the molecular weight of PZLL was 166![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 with a polydispersity index of 1.24. After deprotection in the presence of trifluoroacetic acid, the obtained PLL was used to react with SPDP by EDC/NHS chemistry. Based on the 1H NMR spectra shown in Fig. S2,† there were 1.2 SPDP linked to the side chain of PLL. The SPDP modified PEI was prepared using the same protocol, and the signals at 8.3, 7.8 and 7.2 ppm demonstrated the successful incorporation of SPDP (Fig. S3†). We used the UV spectrum to quantify the amount of the functional group (Fig. S4†), and the results showed that there was 0.9 PDSPA per PEI. Next, cysteine-C6M3 was introduced into the polycations through a disulfide exchange reaction, and the reactions were monitored by UV spectroscopy (Fig. S5†). After 24 h, the absorption was saturated, indicating the completion of the modification. Based on the above results, three C6M3 modified polycations were successfully obtained, and there were 2, 1.2 and 0.9 C6M3 for PDMAEMA, PLL and PEI, respectively.
000 with a polydispersity index of 1.24. After deprotection in the presence of trifluoroacetic acid, the obtained PLL was used to react with SPDP by EDC/NHS chemistry. Based on the 1H NMR spectra shown in Fig. S2,† there were 1.2 SPDP linked to the side chain of PLL. The SPDP modified PEI was prepared using the same protocol, and the signals at 8.3, 7.8 and 7.2 ppm demonstrated the successful incorporation of SPDP (Fig. S3†). We used the UV spectrum to quantify the amount of the functional group (Fig. S4†), and the results showed that there was 0.9 PDSPA per PEI. Next, cysteine-C6M3 was introduced into the polycations through a disulfide exchange reaction, and the reactions were monitored by UV spectroscopy (Fig. S5†). After 24 h, the absorption was saturated, indicating the completion of the modification. Based on the above results, three C6M3 modified polycations were successfully obtained, and there were 2, 1.2 and 0.9 C6M3 for PDMAEMA, PLL and PEI, respectively.
3.2. Hemolysis testing of the polycations
It is reported that C6M3 exhibits pH dependence for membrane lysis. To verify the pH selective lytic activity, we incubated the peptide modified polycations (10–100 μg mL−1) with mouse red blood cells at different pH values for 1 h at 37 °C. The absorption at 541 nm attributed to the characteristic peak of hemoglobin in the UV spectrum was then used to quantify the hemolytic activity. As shown in Fig. 2, negligible hemolysis was detected when the concentration of the polymers was lower than 80 μg mL−1 at pH 7.4. However, obvious hemolysis was detected when the pH of the culture media was 5.7, and the lytic activity showed concentration dependence. Previous work demonstrated that the EC50 (the concentration of free peptide for 50% hemolysis) value of C6M3 was ∼16 μg mL−1,52 which was much lower than those in our work (29.7, 27.0 and 33.8 μg mL−1 for PDMAEMA-C6M3, PEI-C6M3 and PLL-C6M3, respectively). This may be attributed to the fact that the conjugation of C6M3 to polycations compromises the lytic activity. Meanwhile, no obvious hemolysis was observed for PDMAEMA, PEI and PLL at pH 7.4 and 5.7 in the tested concentration range, indicating that membrane lysis is caused by the pH triggered activation of C6M3 rather than the polymer backbone.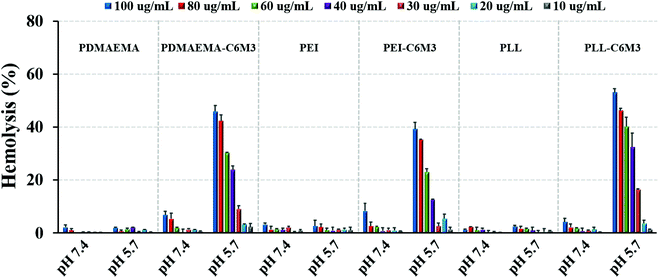 | ||
| Fig. 2 Hemolysis activity of polycations and C6M3 modified polycations at various concentrations and pH values. Data are shown as mean ± SD (n = 3). | ||
3.3. Polyplex characterization
We performed agarose gel electrophoresis assay to evaluate the DNA binding ability of the parent polycations and the C6M3 functionalized polycations. As shown in Fig. S6,† it was observed that all the polymers can completely retard the mobility of DNA when the N/P ratio (amine to phosphate) was higher than 2. Moreover, we found that the introduction of the C6M3 peptide had no obvious effect on the DNA condensing capability of the polycations. DLS was then employed to test the size of the polyplexes with different N/P ratios. It was found that all the polycations could condense DNA into nanoparticles with the hydrodynamic diameter in the range of 70–200 nm (Fig. 3A), which is beneficial for cellular endocytosis. The increase in the N/P ratio led to a smaller size owing to the enhanced condensing capability. Besides, the incorporation of C6M3 into polycations slightly increased the size of the polyplexes compared to the parent polymers, which may be attributed to the relatively hydrophobic nature of the peptide at neutral pH. Furthermore, the morphology of the polyplexes was imaged by TEM at an N/P ratio of 5 (Fig. 3B). We can find that all the polyplexes featured compact structures with a relatively uniform spherical shape, and the diameter was around 100 nm. The size based on DLS testing was larger than that obtained by TEM, which may be attributed to the shrinkage of the polyplexes during sample preparation in TEM testing.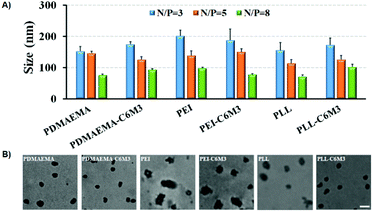 | ||
| Fig. 3 (A) Average hydrodynamic diameters of different formulations. (B) TEM images of polymer/DNA complexes formed at N/P = 5 (scale bar: 100 nm). | ||
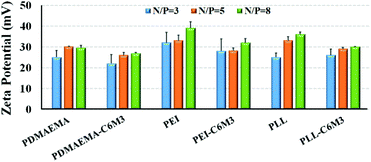
The positively charged surface of the nanoparticles is favorable to cellular endocytosis since the cell membrane is negatively charged. The zeta potentials of all the polyplexes were above 20 mV (Fig. 4), which is beneficial for cell membrane binding. And the increase of the N/P ratio resulted in the increase of the zeta potential. Because of the partial shielding of the peptide, the surface charge of the polyplexes by C6M3 modified polyplexes was slightly lower than that by the parent polycations.
3.4. In vitro transfection
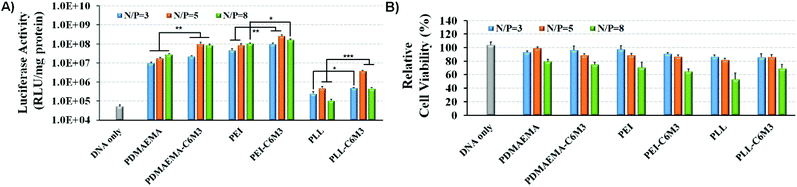
C6M3 modified polycations not only showed good biocompatibility to cells at neutral pH (negligible hemolysis to red blood cells), but also exhibited promising membrane lysis under acidic conditions, both of which are desirable for an ideal non-viral gene vector. Then we performed the gene transfection study in vitro using the luciferase plasmid and the GFP plasmid as reporter genes in HeLa cells to evaluate the delivery efficiency using the six polycations. As shown in Fig. 5A, for PDMAEMA, the transfection efficiency increased with the N/P ratio from 3 to 8, and the same trend was also observed for PEI. However, the cell viability at the N/P ratio of 8 was lower than 80%, indicating potential cytotoxicity due to the excess polycations. PLL also showed enhanced delivery efficiency when the N/P ratio increased from 3 to 5, but further improvement of the N/P ratio led to compromised luciferase transgene expression, which may be attributed to the hard unpackaging of the payloads in the cytoplasm. When the pH-sensitive peptide was introduced into the polycations, the transfection efficiency was significantly enhanced. The luciferase expression mediated by PDMAEMA-C6M3, PEI-C6M3 and PLL-C6M3 at the N/P ratio of 5 was around 5.7, 3.1 and 7.8 times higher than that by PDMAEMA, PEI and PLL, respectively. The relative cell viability results in Fig. 5B reveal that the polycations showed acceptable cell biocompatibility at N/P ratios of 3 and 5 (>80%).Furthermore, based on the promising luciferase plasmid delivery efficiency and acceptable cell compatibility mediated by the N/P ratio of 5, we selected the corresponding formulations to deliver the GFP plasmid to quantify the percentage of the transfected cells using a flow cytometer. As shown in Fig. 6, we found that around 22% and 29% cells were transfected by PDMAEMA-C6M3 and PEI-C6M3, which were much higher than those mediated by the parent PDMAEMA and PEI (7.0% and 16.3%). Although the percentage of transfected cells by PLL and PLL-C6M3 was much lower, the obvious improvement also can be observed after the introduction of C6M3 into PLL. Fig. S7† shows the representative results of the flow cytometry analysis. Furthermore, the GFP positive cells in the transfection study were also observed by fluorescence microscopy (Fig. S8†). We can find that cells transfected by the C6M3 modified polycations showed enhanced fluorescence intensity, suggesting that more cells were transfected.
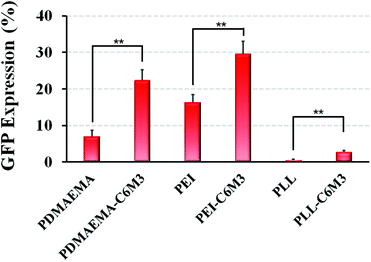 | ||
| Fig. 6 In vitro GFP plasmid delivery efficiency with HeLa cells using different polyplexes (N/P = 5). Data are shown as mean ± SD (n = 3; Student's t-test, **p < 0.01). | ||
It is reported that melittin modified polycations could improve the gene delivery efficiency owing to the membrane lytic nature, but serious cytotoxicity was observed when the N/P ratio was higher than 3, which was attributed to off-site lysis.48 In order to address the off-site toxicity, the Wagner group developed functional polycations with anhydride masked melittin to selectively shield the undesirable lysis for gene delivery. The resulting polycations can deactivate the peptide at neutral pH, and mediated successful endosomal escape of the polyplexes through the acidic pH triggered hydrolysis of the anhydride, further leading to promising siRNA delivery efficiency.43 Although acceptable biocompatibility and excellent delivery efficiency were achieved, the anhydride protecting group is reversible and susceptible to hydrolysis even at neutral pH,54 which may raise safety issues during long-term storage. Since C6M3 features pH selective lytic activity, the modified polycations could avoid the toxicity toward cells extracellularly; in the acidic endo/lysosomal environment, the activated lysis capability induces the disruption of the membrane structure and facilitates efficient escape from endo/lysosome entrapment. Furthermore, the peptide can be de-activated again after release, and recover the safe nature. Hence, our C6M3 modified polycations could realize successful nucleic acid delivery while avoiding off-site cytotoxicity, which may endow the polycations with outstanding advantages for clinical applications.
4. Conclusions
In summary, three widely used polycations (PDMAEMA, PLL and PEI) functionalized with a pH-sensitive peptide C6M3 were employed as gene vectors to mediate efficient endo/lysosome release and further successful gene delivery. PDMAEMA-C6M3, PLL-C6M3 and PEI-C6M3 showed selective membrane lysis behavior under endo/lysosomal acidic conditions, and did not cause off-site safety concerns as the parent polycations. Furthermore, in vitro transfection results demonstrated that the C6M3 modified polycations exhibited higher gene transfer capability using luciferase and GFP plasmids in HeLa cells than that by the parent polycations with good biocompatibility. Our work provides a facile, safe and efficient way to address the issue of endo/lysosome entrapment for polycation-based gene therapy, and may promote the translation of non-viral vectors for clinical applications.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge the financial support from the National Natural Science Foundation of China (NSFC 51803165), the Natural Science Basic Research Plan in Shaanxi Province of China (2019JQ-167), the “Young Talent Support Plan” of Xi'an Jiaotong University, and the Fundamental Research Funds for the Central Universities (xjj2018050). We also acknowledge the support of the Opening Project of Key Laboratory of Shaanxi Province for Craniofacial Precision Medicine Research, College of Stomatology, Xi'an Jiaotong University (2019LHM-KFKT007).Notes and references
- C. E. Dunbar, K. A. High, J. K. Joung, D. B. Kohn, K. Ozawa and M. Sadelain, Science, 2018, 359, eaan4672 CrossRef.
- Y. Liu, C. F. Xu, S. Iqbal, X. Z. Yang and J. Wang, Adv. Drug Delivery Rev., 2017, 115, 98–114 CrossRef CAS.
- L. J. Chang and A. S. Lewin, Curr. Gene Ther., 2018, 18, 1–1 CrossRef CAS.
- S. L. Ginn, A. K. Amaya, I. E. Alexander, M. Edelstein and M. R. Abedi, J. Gene Med., 2018, 20, e3015 CrossRef.
- L. Naldini, Nature, 2015, 526, 351–360 CrossRef CAS.
- Y. Yang, B. Xue, K. Shi, Y. Jia, Y. Hao, Y. Xiao and Z. Qian, J. Biomed. Nanotechnol., 2019, 15, 431–442 CrossRef CAS.
- M. F. Naso, B. Tomkowicz, W. L. Perry and W. R. Strohl, BioDrugs, 2017, 31, 317–334 CrossRef CAS.
- L. Peng and E. Wagner, Biomacromolecules, 2019, 20, 3613–3626 CrossRef CAS.
- D. Pepin, A. Sosulski, K. Hendren, L. H. Zhang, F. Nicolaou, D. Wang, G. P. Gao and P. K. Donahoe, Clin. Cancer Res., 2015, 21 DOI:10.1158/1557-3265.OVCASYMP14-AS25.
- H. Wei, L. R. Volpatti, D. L. Sellers, D. O. Maris, I. W. Andrews, A. S. Hemphill, L. W. Chan, D. S. H. Chu, P. J. Horner and S. H. Pun, Angew. Chem., Int. Ed., 2013, 52, 5377–5381 CrossRef CAS.
- F. J. Xu and W. T. Yang, Prog. Polym. Sci., 2011, 36, 1099–1131 CrossRef CAS.
- H. Yin, R. L. Kanasty, A. A. Eltoukhy, A. J. Vegas, J. R. Dorkin and D. G. Anderson, Nat. Rev. Genet., 2014, 15, 541–555 CrossRef CAS.
- B. R. Olden, E. Cheng, Y. L. Cheng and S. H. Pun, Biomater. Sci., 2019, 7, 789–797 RSC.
- H. P. Fang, Z. P. Guo, L. Lin, J. Chen, P. J. Sun, J. Y. Wu, C. N. Xu, H. Y. Tian and X. S. Chen, J. Am. Chem. Soc., 2018, 140, 11992–12000 CrossRef CAS.
- X. Liu, J. J. Xiang, D. C. Zhu, L. M. Jiang, Z. X. Zhou, J. B. Tang, X. R. Liu, Y. Z. Huang and Y. Q. Shen, Adv. Mater., 2016, 28, 1743–1752 CrossRef CAS.
- L. H. Wang, D. C. Wu, H. X. Xu and Y. Z. You, Angew. Chem., Int. Ed., 2016, 55, 755–759 CrossRef CAS.
- L. Zou, S. Y. Lee, Q. Wu, H. Zhang, A. Bastian, C. Orji, G. Payne, A. Galvez, T. Thomas, Z. Zhang and H. Dou, J. Biomed. Nanotechnol., 2018, 14, 1785–1795 CrossRef CAS.
- H. He, N. Zheng, Z. Y. Song, K. H. Kim, C. Yao, R. J. Zhang, C. L. Zhang, Y. H. Huang, F. M. Uckun, J. J. Cheng, Y. F. Zhang and L. C. Yin, ACS Nano, 2016, 10, 1859–1870 CrossRef CAS.
- D. P. Feldmann, Y. L. Cheng, R. Kandil, Y. R. Xie, M. Mohammadi, H. Harz, A. Sharma, D. J. Peeler, A. Moszczynska, H. Leonhardt, S. H. Pun and O. M. Merkel, J. Controlled Release, 2018, 276, 50–58 CrossRef CAS.
- M. E. Davis, J. E. Zuckerman, C. H. J. Choi, D. Seligson, A. Tolcher, C. A. Alabi, Y. Yen, J. D. Heidel and A. Ribas, Nature, 2010, 464, U1067–U1140 CrossRef.
- W. W. Shen, Q. W. Wang, Y. Shen, X. Gao, L. Li, Y. Yan, H. Wang and Y. Y. Cheng, ACS Cent. Sci., 2018, 4, 1326–1333 CrossRef CAS.
- J. E. Dahlman, C. Barnes, O. F. Khan, A. Thiriot, S. Jhunjunwala, T. E. Shaw, Y. P. Xing, H. B. Sager, G. Sahay, L. Speciner, A. Bader, R. L. Bogorad, H. Yin, T. Racie, Y. Z. Dong, S. Jiang, D. Seedorf, A. Dave, K. S. Sandhu, M. J. Webber, T. Novobrantseva, V. M. Ruda, A. K. R. Lytton-Jean, C. G. Levins, B. Kalish, D. K. Mudge, M. Perez, L. Abezgauz, P. Dutta, L. Smith, K. Charisse, M. W. Kieran, K. Fitzgerald, M. Nahrendorf, D. Danino, R. M. Tuder, U. H. von Andrian, A. Akinc, D. Panigrahy, A. Schroeder, V. Koteliansky, R. Langer and D. G. Anderson, Nat. Nanotechnol., 2014, 9, 648–655 CrossRef CAS.
- E. Wagner, Acc. Chem. Res., 2012, 45, 1005–1013 CrossRef CAS.
- L. Du, C. Wang, L. Meng, Q. Cheng, J. Zhou, X. Wang, D. Zhao, J. Zhang, L. Deng, Z. Liang, A. Dong and H. Cao, Biomaterials, 2018, 176, 84–93 CrossRef CAS.
- C. Xu, Y. Z. Z. Zhang, K. Xu, J. J. Nie, B. R. Yu, S. J. Li, G. Cheng, Y. L. Li, J. Du and F. J. Xu, Nat. Commun., 2019, 10, 3184 CrossRef.
- H. Lopez-Bertoni, K. L. Kozielski, Y. Rui, B. Lal, H. Vaughan, D. R. Wilson, N. Mihelson, C. G. Eberhart, J. Laterra and J. J. Green, Nano Lett., 2018, 18, 4086–4094 CrossRef CAS.
- Y. H. Jiang, Q. Lu, Y. H. Wang, E. Xu, A. Ho, P. Singh, Y. F. Wang, Z. Z. Jiang, F. Yang, G. T. Tietjen, P. Cresswell and W. M. Saltzman, Nano Lett., 2020, 20, 1117–1123 CrossRef CAS.
- A. Yen, Y. L. Cheng, M. Sylvestre, H. H. Gustafson, S. Puri and S. H. Pun, Mol. Pharmaceutics, 2018, 15, 2268–2276 CrossRef CAS.
- S. W. L. Lee, C. Paoletti, M. Campisi, T. Osaki, G. Adriani, R. D. Kamm, C. Mattu and V. Chiono, J. Controlled Release, 2019, 313, 80–95 CrossRef CAS.
- M. A. Islam, E. K. G. Reesor, Y. J. Xu, H. R. Zope, B. R. Zetter and J. J. Shi, Biomater. Sci., 2015, 3, 1519–1533 RSC.
- Y. Weng, H. Xiao, J. Zhang, X.-J. Liang and Y. Huang, Biotechnol. Adv., 2019, 37, 801–825 CrossRef CAS.
- Y. Zhang, A. Satterlee and L. Huang, Mol. Ther., 2012, 20, 1298–1304 CrossRef CAS.
- J. Chen, K. Wang, J. Y. Wu, H. Y. Tian and X. S. Chen, Bioconjugate Chem., 2019, 30, 338–349 CrossRef CAS.
- H. C. Kang, K. M. Huh and Y. H. Bae, J. Controlled Release, 2012, 164, 256–264 CrossRef CAS.
- I. Lostale-Seijo and J. Montenegro, Nat. Rev. Chem., 2018, 2, 258–277 CrossRef.
- D. H. Pei and M. Buyanova, Bioconjugate Chem., 2019, 30, 273–283 CrossRef CAS.
- E. L. Eskelinen and P. Saftig, Biochim. Biophys. Acta, Mol. Cell Res., 2009, 1793, 664–673 CrossRef CAS.
- H. Haisma, Hum. Gene Ther., 2011, 22, A14–A14 Search PubMed.
- W. Stoorvogel, G. J. Strous, H. J. Geuze, V. Oorschot and A. L. Schwartz, Cell, 1991, 65, 417–427 CrossRef CAS.
- A. J. Convertine, D. S. W. Benoit, C. L. Duvall, A. S. Hoffman and P. S. Stayton, J. Controlled Release, 2009, 133, 221–229 CrossRef CAS.
- M. K. G. Jayakumar, A. Bansal, K. Huang, R. Yao, B. N. Li and Y. Zhang, ACS Nano, 2014, 8, 4848–4858 CrossRef CAS.
- Y. L. Cheng, R. C. Yumul and S. H. Pun, Angew. Chem., Int. Ed., 2016, 55, 12013–12017 CrossRef CAS.
- M. Meyer, A. Philipp, R. Oskuee, C. Schmidt and E. Wagner, J. Am. Chem. Soc., 2008, 130, 3272–3273 CrossRef CAS.
- M. M. Wang, H. M. Liu, L. Li and Y. Y. Cheng, Nat. Commun., 2014, 5, 3053 CrossRef.
- Y. Y. Yuan, C. J. Zhang and B. Liu, Angew. Chem., Int. Ed., 2015, 54, 11419–11423 CrossRef CAS.
- J.-P. Behr, Chimia, 1997, 51, 34–36 CAS.
- A. E. Nel, L. Madler, D. Velegol, T. Xia, E. M. V. Hoek, P. Somasundaran, F. Klaessig, V. Castranova and M. Thompson, Nat. Mater., 2009, 8, 543–557 CrossRef CAS.
- J. G. Schellinger, J. A. Pahang, R. N. Johnson, D. S. H. Chu, D. L. Sellers, D. O. Maris, A. J. Convertine, P. S. Stayton, P. J. Horner and S. H. Pun, Biomaterials, 2013, 34, 2318–2326 CrossRef CAS.
- E. J. Kwon, S. Liong and S. H. Pun, Mol. Pharmaceutics, 2010, 7, 1260–1265 CrossRef CAS.
- J. G. Schellinger, J. A. Pahang, J. L. Shi and S. H. Pun, ACS Macro Lett., 2013, 2, 725–730 CrossRef CAS.
- D. J. Peeler, S. N. Thai, Y. L. Cheng, P. J. Horner, D. L. Sellers and S. H. Pun, Biomaterials, 2019, 192, 235–244 CrossRef CAS.
- B. L. Chen, K. Yoo, W. Xu, R. Pan, X. X. Han and P. Chen, Drug Delivery Transl. Res., 2017, 7, 507–515 CrossRef CAS.
- J. J. Chen, J. X. Ding, Y. C. Wang, J. J. Cheng, S. X. Ji, X. L. Zhuang and X. S. Chen, Adv. Mater., 2017, 29, 1701170 CrossRef.
- J. Z. Du, T. M. Sun, W. J. Song, J. Wu and J. Wang, Angew. Chem., Int. Ed., 2010, 49, 3621–3626 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0bm01231a |
| This journal is © The Royal Society of Chemistry 2020 |