A novel application of porphyrin nanoparticles as an effective fluorescent assay platform for nucleic acid detection†
Junfeng
Zhai‡
a,
Hailong
Li‡
ab and
Xuping
Sun
*a
aState Key Lab of Electroanalytical Chemistry, Changchun Institute of Applied Chemistry, Chinese Academy of Sciences, Changchun, 130022 Jilin, China. E-mail: sunxp@ciac.jl.cn; Fax: (+86) 431-85262065; Tel: (+86) 431-85262065
bGraduate School of the Chinese Academy of Sciences, Beijing 100039, China
First published on 21st July 2011
Abstract
The present communication demonstrates the preparation of 2,3,7,8,12,13,17,18-octaethyl-21H,23H-porphine iron(III) chloride nanoparticles (FePNPs) by reprecipitation method. In a further effort, the potential application of FePNPs was exploited as an effective fluorescent assay platform for nucleic acid detection. This assay system exhibits high sensitivity and selectivity down to a single-base mismatch.
Developing rapid, cost-effective and sensitive methods for sequence-specific detection of nucleic acid is of great importance in medical research, clinical disease diagnostics, and environmental monitoring.1 The increasing availability of nanostructures has created widespread interest in their use in biotechnological systems for diagnostic applications,2 and indeed, the use of a variety of nanostructures for this purpose has been well developed.3 Recently, nanostructures have also proven for use as “nanoquenchers” in fluorescence-enhanced nucleic acid detection.4–6 Because the same nanostructure is capable of quenching dyes with different emission frequencies, the selection issue of a fluorophore-quencher pair is eliminated from the nanostructure-based system. Since the pioneering discovery of the excellent fluorescence quenching ability of gold nanoparticles (AuNPs),5 considerable efforts have been made to develop fluorescent methods for DNA detection based on AuNPs.6 Recently, both single-walled carbon nanotubes7 and graphene oxide8 have found their significant application for fluorescence-enhanced DNA detection. Until now, many other structures have been used in this assay, including silver nanoparticles,9amulti-walled carbon nanotubes,9bcarbon nanoparticles,9ccarbon nanospheres,9dnano-C60,9e mesoporous carbon microparticles,9f poly(p-phenylenediamine) nanobelts,9gpolyaniline nanofibres,9h poly(o-phenylenediamine) colloids,9i poly(2,3-diaminonaphthalene) microspheres,9j coordination polymer colloids and nanobelts,9k–mAg@poly(m-phenylenediamine) core–shell nanoparticles,9ntetracyanoquinodimethane nanoparticles,9o and supramolecular microparticles.9p
Porphyrin and related compounds possess many desirable structural and functional properties and they have been widely studied due to their importance in biological process.10 Although the past years have witnessed their application in the development of artificial photosynthesis, catalysis, and sensor systems,11 their use for nucleic acid detection has not been explored so far. In this communication, we report the first use of 2,3,7,8,12,13,17,18-octaethyl-21H,23H-porphine iron(III) chloride (FeP) nanoparticles (FePNPs) obtained by the reprecipitation method12 as a very effective assay platform for fluorescence-enhanced nucleic acid detection with high sensitivity and selectivity down to single-base mismatch. The detection is accomplished by two steps: (1) FePNP absorbs and quenches the fluorescence of dye-labeled single-stranded DNA (ssDNA) serving as the probe; (2) the subsequent hybridization of the probe with its target produces a double-stranded DNA (dsDNA) which detaches from the FePNP, leading to fluorescence recovery.
Fig. 1a shows a typical SEM image of the products thus formed (see ESI† for preparation details), indicating the formation of a large amount of nanoparticles. The TEM image (Fig. 1b) further reveals that they are nearly spherical in shape and in the range of 50–200 nm. The morphology of one single FePNP is shown in the inset. The chemical composition of the nanoparticles was determined by the energy-dispersed spectrum (EDS), as shown in Fig. S1 (ESI†). The peaks of C, N, Fe, and Cl elements are observed, indicating that the nanoparticles are formed from FeP. All these observations indicate that FePNPs can be prepared by the reprecipitation method.
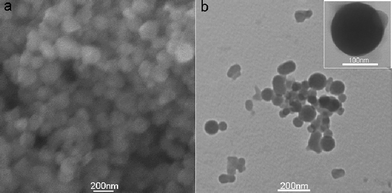 | ||
| Fig. 1 (a) SEM and (b) TEM images of the products thus formed. Inset: a single FePNP. | ||
The feasibility of using FePNPs as a fluorescent assay platform for nucleic acid detection was firstly evaluated. Fig. 2 shows the fluorescence emission spectra of the ssDNA probe, PHIV, at different conditions. The sequence of PHIV is associated with human immunodeficiency virus (HIV) and all oligonucleotide sequences are available in the ESI.† In the absence of FePNPs, PHIV displayed strong fluorescence emission due to the fluorescein-based dye (curve a) in Tris–HCl buffer. However, the presence of FePNPs leads to 85% quenching of the dye fluorescence (curve c), indicating that FePNPs can strongly adsorb ssDNA and quench the fluorescent dye very effectively. On the other hand, the PHIV–FePNP complex had significant fluorescence enhancement upon its incubation with complementary target T1 over a period of 30 min, leading to an 83% fluorescence recovery (curve d). It should be noted that the fluorescence of the free PHIV was, however, scarcely influenced by the addition of T1 in the absence of FePNPs (curve b). The fluorescence intensity changes (F/F0–1) of PHIV–FePNP complex were further determined upon addition of different concentrations of T1 ranging from 5 to 300 nM, where F0 and F correspond to the fluorescence intensities at 522 nm in the absence and the presence of the target, respectively. A notable increase in the fluorescence intensity was observed as the T1 concentration increased, indicating that the FePNP/DNA assembly approach is effective in probing biomolecular interactions (inset). Although such FePNPs tend to aggregate, they can be well-dispersed by shaking and kept stable during our measurements.
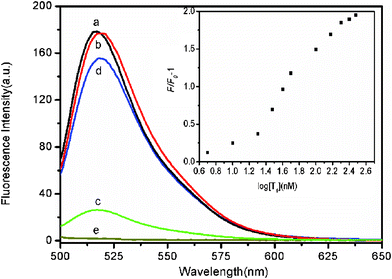 | ||
| Fig. 2 Fluorescence emission spectra of PHIV (50 nM) at different conditions: (a) PHIV; (b) PHIV + 300 nM T1; (c) PHIV + FePNPs; (d) PHIV + FePNPs +300 nM T1. Curve e is the emission spectra of FePNPs. Inset: fluorescence intensity changes (F/F0–1) of PHIV–FePNP complex plotted against the logarithm of the concentration of T1 (where F0 and F are the fluorescence intensities without and with T1, respectively). Excitation was at 480 nm, and the emission was monitored at 522 nm. All measurements were done in Tris-HCl buffer in the presence of 5 mM Mg2+ (pH: 7.4). | ||
FePNP is a π-rich structure and thus there should be strong π–π stacking interactions between unpaired ssDNA bases and FePNP,13 leading to strong adsorption of ssDNA on PePNP. The zeta potential of the FePNP was measured to be about 0.70 mV, indicating that the FePNP has a very low positively charged surface. So, the slight electrostatic attractive interactions between FePNP and negatively charged backbone of ssDNA only contribute little to the adsorption of ssDNA on FePNP. In contrast, FePNP should have weak or no binding with dsDNA due to the absence of unpaired DNA bases and the rigid conformation of dsDNA. Scheme 1 shows a schematic to illustrate the nucleic acid detection mechanism involved. The DNA detection is accomplished by two steps: (1) FePNP binds FAM-ssDNA via both electrostatic attraction and π–π stacking interactions between unpaired DNA bases and FePNP, leading to substantial fluorescence quenching of the dye due to their close proximity; (2) the hybridization of FAM-ssDNA with its target produces a dsDNA which detaches from FePNP, leading to fluorescence recovery.
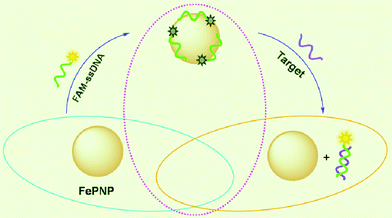 | ||
| Scheme 1 A schematic (not to scale) illustrating the fluorescence-enhanced nucleic acid detection using FePNP as an assay platform. | ||
The kinetic behaviors of PHIV and FePNPs, as well as of the PHIV–FePNP complex with T1, were monitored by collecting the time-dependent fluorescence spectra. Fig. 3 shows the fluorescence quenching of PHIV by FePNPs (square) and fluorescence recovery of PHIV–FePNP by T1 in Tris-HCl buffer (round) as a function of incubation time. In the absence of the target, the fluorescence intensity of PHIV exhibits a rapid decrease in the first 5 min and gradually reaches equilibrium in the following 25 min, indicating that the adsorption of ssDNA on FePNP is a fast process. In the presence of a complementary target, the fluorescence intensity of PHIV–FePNP complex displays a rapid increase in the first 5 min, followed by a slow enhancement over a period of 25 min. The best fluorescence response was obtained after 30 min incubation.
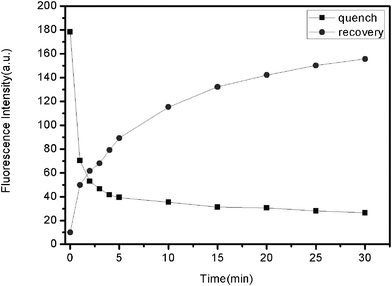 | ||
| Fig. 3 Fluorescence quenching of PHIV (50 nM) by FePNP (square) and fluorescence recovery of PHIV–FePNP complex by T1 (300 nM) (round) as a function of incubation time. Excitation wavelength was 480 nm, and the emission was monitored at 522 nm. All measurements were done in Tris–HCl buffer in the presence of 5 mM Mg2+ (pH: 7.4). | ||
It should point out that this assay system is capable of distinguishing complementary and single-base mismatched sequences. Fig. 4 shows the fluorescence responses of PHIV–FePNP complex toward complementary target T1 and the single-base mismatched target T2. It is found that the F/F0 value obtained upon addition of 300 nM T2 is about 66% of that obtained upon addition of 300 nM T1 into the corresponding PHIV–FePNP complex. However, only a very small fluorescence intensity change was observed for the PHIV–FePNP complex upon addition of 300 nM noncomplementary target T3, indicating that the observed fluorescence enhancement in our present system is indeed due to the base pairing between probe and its target. The inset in Fig. 4 is the corresponding fluorescence intensity histograms with error bars. All the above observations clearly indicate the present nucleic acid detection system has a high selectivity down to single-base mismatch and the results exhibit good reproducibility.
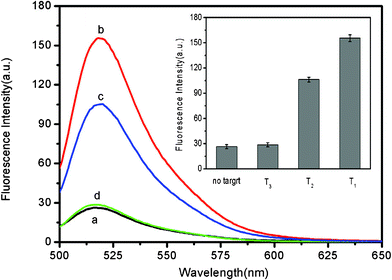 | ||
| Fig. 4 Fluorescence emission spectra of PHIV (50 nM) at different conditions: (a) PHIV–FePNP complex; (b) PHIV–FePNP complex + 300 nM T1; (c) PHIV–FePNP complex + 300 nM T2; (d) PHIV–FePNP complex + 300 nM T3. Inset: fluorescence intensity histograms with error bars. Excitation was at 480 nm, and the emission was monitored at 522 nm. All measurements were done in Tris-HCl buffer in the presence of 5 mM Mg2+ (pH: 7.4). | ||
We also studied the influence of the amount of FePNPs used on the fluorescence quenching and the subsequent fluorescence recovery of seven samples with the use of different amounts of FePNPs. Fig. 5 shows the corresponding results. It suggests that the quenching efficiency increases but the recovery efficiency decreases with the volume of FePNPs increasing from 0 to 35 μL. Such an observation can be reasonably explained as follows: FePNP has a strong affinity for ssDNA. It is obvious that an increase of FePNP in amount leads to more efficient adsorption of ssDNA on it and thus a higher quenching efficiency. However, during the subsequent recovery process, the adsorption of target on excessive FePNPs, which will compete with the hybridization of target with ssDNA adsorbed on FePNP, leads to decreased hybridization and recovery efficiency.
![Fluorescence intensity histograms of PHIV + FePNPs and PHIV + FePNPs + T1 with the use of 0, 10, 15, 20, 25, 30, and 35 μL of FePNPs respectively ([PHIV]=50 nM; [T1]=300 nM). Excitation was at 480 nm, and the emission was monitored at 522 nm. All measurements were done in Tris-HCl buffer in the presence of 5 mM Mg2+ (pH: 7.4).](/image/article/2011/RA/c1ra00026h/c1ra00026h-f5.gif) | ||
| Fig. 5 Fluorescence intensity histograms of PHIV + FePNPs and PHIV + FePNPs + T1 with the use of 0, 10, 15, 20, 25, 30, and 35 μL of FePNPs respectively ([PHIV]=50 nM; [T1]=300 nM). Excitation was at 480 nm, and the emission was monitored at 522 nm. All measurements were done in Tris-HCl buffer in the presence of 5 mM Mg2+ (pH: 7.4). | ||
In summary, FePNPs have been proven to be a very effective fluorescent assay platform for nucleic acid detection with high sensitivity and selectivity down to single-base mismatch. Our present observations are significant for the following two reasons: (1) it extends the application of porphyrin nanoparticles to nucleic acid detection; (2) this assay platform is promising for universal and effective fluorescence-enhanced detection sensitive and selective to the target molecule studied.
Acknowledgements
This work was supported by National Basic Research Program of China (No. 2011CB935800).References
- D. Gresham, D. M. Ruderfer, S. C. Pratt, J. Schacherer, M. J. Dunham, D. Botstein and L. Kruglyak, Science, 2006, 311, 1932 CrossRef CAS.
- R. Brayner, Nano Today, 2008, 3, 48 CrossRef.
- N. L. Rosi and C. A. Mirkin, Chem. Rev., 2005, 105, 1547 CrossRef CAS.
- P. C. Ray, G. K. Darbha, A. Ray, J. Walker and W. Hardy, Plasmonics, 2007, 2, 173 CrossRef CAS.
- B. Dubertret, M. Calame and A. J. Libchaber, Nat. Biotechnol., 2001, 19, 365 CrossRef CAS.
- (a) D. J. Maxwell, J. R. Taylor and S. Nie, J. Am. Chem. Soc., 2002, 124, 9606 CrossRef CAS; (b) H. Li and L. J. Rothberg, Anal. Chem., 2004, 76, 5414 CrossRef CAS; (c) S. Song, Z. Liang, J. Zhang, L. Wang, G. Li and C. Fan, Angew. Chem. Int. Ed., 2009, 48, 8670 CrossRef CAS; (d) D. Li, S. Song and C. Fan, Acc. Chem. Res., 2010, 43, 631 CrossRef CAS.
- (a) R. Yang, Z. Tang, J. Yan, H. Kang, Y. Kim, Z. Zhu and W. Tan, Anal. Chem., 2008, 80, 7408 CrossRef CAS; (b) R. Yang, J. Jin, Y. Chen, N. Shao, H. Kang, Z. Xiao, Z. Tang, Y. Wu, Z. Zhu and W. Tan, J. Am. Chem. Soc., 2008, 130, 8351 CrossRef CAS.
- (a) C. Lu, H. Yang, C. Zhu, X. Chen and G. Chen, Angew. Chem. Int. Ed., 2009, 48, 4785 CrossRef CAS; (b) S. He, B. Song, D. Li, C. Zhu, W. Qi, W. Wen, L. Wang, S. Song, H. Fang and C. Fan, Adv. Funct. Mater., 2010, 20, 453 CrossRef.
- (a) Y. Zhang, H. Li and X. Sun, Chinese J. Anal. Chem., 2011, 39, 998 Search PubMed; (b) H. Li, J. Tian, L. Wang, Y. Zhang and X. Sun, J. Mater. Chem., 2011, 21, 824 RSC; (c) H. Li, Y. Zhang, L. Wang, J. Tian and X. Sun, Chem. Communu., 2011, 47, 961 Search PubMed; (d) H. Li, Y. Zhang, T. Wu, S. Liu, L. Wang and X. Sun, J. Mater. Chem., 2011, 21, 4663 RSC; (e) H. Li, Y. Zhang, Y. Luo and X. Sun, Small, 2011, 7, 1562 CrossRef CAS; (f) S. Liu, H. Li, L. Wang, J. Tian and X. Sun, J. Mater. Chem., 2011, 21, 339 RSC; (g) L. Wang, Y. Zhang, J. Tian, H. Li and X. Sun, Nucleic Acids Res., 2011, 39, e37 CrossRef CAS; (h) S. Liu, L. Wang, Y. Luo, J. Tian, H. Li and X. Sun, Nanoscale, 2011, 3, 967 RSC; (i) J. Tian, H. Li, Y. Luo, L. Wang, Y. Zhang and X. Sun, Langmuir, 2011, 27, 874 CrossRef CAS; (j) J. Tian, Y. Zhang, Y. Luo, H. Li, J. Zhai and X. Sun, Analyst, 2011, 136, 2221 RSC; (k) H. Li and X. Sun, Chem. Commun., 2011, 47, 2625 RSC; (l) H. Li, L. Wang, Y. Zhang, J. Tian and X. Sun, Macromol. Rapid Commun., 2011, 32, 899 CrossRef CAS; (m) Y. Luo, F. Liao, W. Lu, G. Chang and X. Sun, Nanotechnol., 2011, 22, 195502 CrossRef; (n) Y. Zhang, L. Wang, J. Tian, H. Li, Y. Luo and X. Sun, Langmuir, 2011, 27, 2170 CrossRef CAS; (o) H. Li, L. Wang, J. Zhai, Y. Luo, Y. Zhang, J. Tian and X. Sun, Anal. Methods, 2011, 3, 1051 RSC; (p) H. Li, J. Zhai and X. Sun, PLoS ONE, 2011, 6, e18958 CrossRef CAS.
- C. J. Medforth, Z. Wang, K. E. Martin, Y. Song, J. L. Jacobsenc and J. A. Shelnutt, Chem. Commun., 2009, 7261 RSC.
- Porphyrin handbook, applications: past, present and future, vol. 6, Academic Press, San Diego, CA, 2000 Search PubMed.
- J. Hu, Y. Guo, H. Liang, L. Wan and L. Jiang, J. Am. Chem. Soc., 2005, 127, 17090 CrossRef CAS.
- N. Varghese, U. Mogera, A. Govindaraj, A. Das, P. K. Maiti, A. K. Sood and C. N. R. Rao, ChemPhysChem, 2009, 10, 206 CrossRef CAS.
Footnotes |
| † Electronic Supplementary Information (ESI) available: Experimental section and supplementary figures. See DOI: 10.1039/c1ra00026h/ |
| ‡ J. Zhai and H. Li made an equal contribution to this work. |
| This journal is © The Royal Society of Chemistry 2011 |
