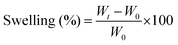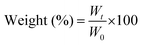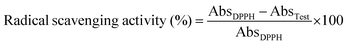A 4D printed nanoengineered super bioactive hydrogel scaffold with programmable deformation for potential bifurcated vascular channel construction†
Amit
Nain
 *ag,
Akshat
Joshi
b,
Souvik
Debnath
*ag,
Akshat
Joshi
b,
Souvik
Debnath
 a,
Saswat
Choudhury
c,
Jobin
Thomas
d,
Jitendra
Satija
a,
Saswat
Choudhury
c,
Jobin
Thomas
d,
Jitendra
Satija
 d,
Chih-Ching
Huang
d,
Chih-Ching
Huang
 ef and
Kaushik
Chatterjee
ef and
Kaushik
Chatterjee
 *ac
*ac
aDepartment of Material Engineering, Indian Institute of Science, Bangalore, Karnataka 560012, India. E-mail: amitnain@iisc.ac.in; kchatterjee@iisc.ac.in
bTerasaki Institute for Biomedical Innovation, Los Angeles, CA 90024, USA
cDepartment of Bioengineering, Indian Institute of Science, Bangalore, Karnataka 560012, India
dCentre for Nanobiotechnology, Vellore Institute of Technology, Vellore, Tamil Nadu 632014, India
eDepartment of Bioscience and Biotechnology and Centre of Excellence for the Oceans, National Taiwan Ocean University, Keelung, 202301, Taiwan
fSchool of Pharmacy, College of Pharmacy, Kaohsiung Medical University, Kaohsiung, 80708, Taiwan
gDepartment of Applied Mechanics & Biomedical Engineering, Indian Institute of Technology-Madras, 600036, Tamil Nadu, India
First published on 27th June 2024
Abstract
Four-dimensional (4D) printing of hydrogels enabled the fabrication of complex scaffold geometries out of static parts. Although current 4D fabrication strategies are promising for creating vascular parts such as tubes, developing branched networks or tubular junctions is still challenging. Here, for the first time, a 4D printing approach is employed to fabricate T-shaped perfusable bifurcation using an extrusion-based multi-material 3D printing process. An alginate/methylcellulose-based dual-component hydrogel system (with defined swelling behavior) is nanoengineered with carbonized alginate (∼100 nm) to introduce anti-oxidative, anti-inflammatory, and anti-thrombotic properties and shape-shifting properties. A computational model to predict shape deformations in the printed hydrogels with defined infill angles was designed and further validated experimentally. Shape deformations of the 3D-printed flat sheets were achieved by ionic cross-linking. An undisrupted perfusion of a dye solution through a T-junction with minimal leakage mimicking blood flow through vessels is also demonstrated. Moreover, human umbilical vein endothelial and fibroblast cells seeded with printed constructs show intact morphology and excellent cell viability. Overall, the developed strategy paves the way for manufacturing self-actuated vascular bifurcations with remarkable anti-thrombotic properties to potentially treat coronary artery diseases.
1. Introduction
Three-dimensional (3D) printing presented new opportunities for fabricating highly precise and customizable biological constructs, which have extensive applications in tissue engineering.1 However, the implantation of bulky and static 3D-printed scaffolds often entails inconvenient surgical sutures, posing a risk of additional complications and severe inflammation at/around the injured tissue site.2,3 Therefore, 4D printing gained immense attention in the rapidly emerging field of additive manufacturing as it enables the fabrication of intricate, dynamic, and programmable structures that 3D printing failed to produce.4–6 4D printing either employs smart materials, programmed design, or a combination of both.7 Smart materials respond to a wide range of stimuli like moisture, pH, light, electromagnetic field, and temperature, to name a few.8 Rapid advancements in tissue engineering combined with a pool of biomaterials have motivated researchers in 4D printing to transform regenerative medicine. Although 4D printing is a promising approach for fabricating programmable shape-morphing geometries out of soft static parts, the fabrication of complex scaffold geometries that closely mimic tissue architecture is still challenging. Among several dynamic 4D materials, hydrogels have recently gained much attention due to their ability to match tissue stiffness, water-imbibing capacity, biocompatibility, and stimuli responsiveness.9–12 Despite recent findings, the choice of hydrogels for 4D printing is limited due to their poor printability and shape fidelity.13 Hydrogels can swell/de-swell in response to moisture, a mild and cytocompatible cue, which makes them an ideal choice for aqueous medium-triggered 4D-printed biomaterials.14 Shape deformations of hydrogels are primarily induced by gradients in cross-linking densities and anisotropic swelling.15,16 Nevertheless, the fragile nature of the hydrogel makes it challenging to produce robust intricate biomimetic architectures.Tubular structures are highly in demand for the development of synthetic blood vessel substitutes.17 Existing grafts require a secondary surgical site in the patient, pose a risk of immunogenic rejection, disease transmission, and donor dependency, and are prone to thrombosis and other hyperplastic complications.18 Vascularization is one of the critical challenges that has so far hindered the clinical translation of engineered tissues.19 Researchers are devoted to developing a vascular network within the synthetic tissue construct before implantation. Typically, engineered vasculature is integrated into the host vasculature through anastomosis to supply nutrients to cells instantly after surgery. Moreover, adequate vascularization is of imminent importance in regenerative engineering, and various factors, including biomaterial/bio-scaffold design and bio-fabrication approaches, need to be understood.19,20 Due to the significant drawbacks of conventional vascular grafts and the limited availability of autologous vascular grafts, 4D fabrication has become a promising alternative to current approaches. 4D fabrication is an emerging field in which flat 3D printed constructs will undergo self-formation of tube-like structures with scalable diameters for vascular grafts.21 These constructs can mimic the complex native structural organization of tissues with superior resolutions.22 It is also possible to fabricate cell-laden constructs that allow surface patterning of different cell types in a programmed way before initiating the folding process. As a result, a scaffold can achieve homogeneous seeding of cells on the lumen of the tubular conduit.23 A popular conventional choice to fabricate tubular structures is the coaxial extrusion system.24–26 He's group designed a two-layer coaxial nozzle in which a 25G needle was inserted into an 18G needle to fabricate GelMA microfibers.24 The authors further showed the fabrication of multicompartmental perfusable microfibers through the two-in-one coaxial nozzle. A similar strategy was further modified to develop a three-layer coaxial nozzle, which can produce a double helical structure. Different mixtures, i.e., Na-Alg/GelMA/GelMA, were respectively pumped into the outer/middle/inner needles to construct straight and helical two-layered microfibers.25 The other two commonly used additive manufacturing techniques for tubular constructs include direct-ink writing and projection stereolithography.27,28 For example, a mixture of methacrylated alginate and hyaluronic acid was 3D-printed by extrusion printing and photo-crosslinked to achieve tube-like structures with tunable diameters (average internal tube diameter is ∼20 μm) and internal micro-architectures.21 On the other hand, Chatterjee's group utilized dynamic light processing (DLP)-based 3D printing for the fabrication of a cell-laden tube-shaped tissue scaffold from a mixture of gelatin methacryloyl and poly(ethylene glycol) dimethacrylate.29 DLP offers high throughput printing and is suitable for hollow structures; however, it does not allow multilayer printing with layered cells in the walls of the tubes.
Critical challenges associated with the tubular design include limited resolution, lack of structural support, and inability to perfuse through a bifurcated vascular network.30,31 For example, Zhang et al. designed Y-shaped microvascular scaffolds using gelatin methacrylamide.32 Although Y-bifurcation showed promising results as a vascular graft, the tubes were loose and leaky, and there were cracks on the tubules. Leaky structures made the hydrogel films unbridged, thus requiring a higher degree of rolling to achieve self-formed microvessels.32 Moreover, the tubular constructs lack structural integrity and programmed bifurcation. Although 4D printing has shown promise in fabricating relatively more straightforward tubes, a branched network is still challenging. The complicated element of tubular networks is perfusable bifurcations, which can be realized by developing T-shaped junctions. However, bridging two perpendicularly connected tubes from separate flat sheets connected by a junction is critical in a bifurcated design. Recently, Ionov's group was the first to demonstrate T-shaped bifurcations mimicking vascular networks.17 However, the designs were not validated using a computational model. In addition, the developed scaffold did not exhibit any biological activities necessary to alleviate post-surgical complications like thrombosis and infection, to name a few. Nevertheless, intraoperative shape deformation and deployment of hydrogel scaffolds with intricate designs like perfusable T-shaped bifurcations and superior biological activities have never been reported.
Herein, we demonstrate a nanoengineered dual-component hydrogel system to achieve complex architects for potential vascular applications. Firstly, carbonized Alg (CAlg) with anti-oxidative/inflammatory/thrombotic properties was prepared through mild pyrolysis of pristine alginate (Alg) (Fig. 1). Bioactive nano-sized CAlg was incorporated into two separately optimized alginate/methylcellulose (CAlg/Alg:MC) hydrogel inks, prepared by adjusting the composition of Alg or MC (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 or 4
9 or 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6). Two gels having different swelling ratios were overlaid over each other via extrusion-based 3D printing to fabricate dynamic structures. A computational model was also created to predict shape deformations for constructs prepared using nanoengineered hydrogel formulations. The cytocompatibility was tested, and the native gel surface was modified by depositing gelatin-based nanofibers to augment the cellular attachment. Toward clinical translation of the 4D-printed gel as a potential vascular conduit, perfusable single and multichannel/branched programmable structures are demonstrated (Fig. 1). The cell viability and morphology of endothelial and fibroblast cells, mimicking vascular channels, are also investigated. Overall, the study aimed to highlight the promise of the 4D printing approach to fabricate intricate, anatomically accurate, and relevant vascular grafts.
6). Two gels having different swelling ratios were overlaid over each other via extrusion-based 3D printing to fabricate dynamic structures. A computational model was also created to predict shape deformations for constructs prepared using nanoengineered hydrogel formulations. The cytocompatibility was tested, and the native gel surface was modified by depositing gelatin-based nanofibers to augment the cellular attachment. Toward clinical translation of the 4D-printed gel as a potential vascular conduit, perfusable single and multichannel/branched programmable structures are demonstrated (Fig. 1). The cell viability and morphology of endothelial and fibroblast cells, mimicking vascular channels, are also investigated. Overall, the study aimed to highlight the promise of the 4D printing approach to fabricate intricate, anatomically accurate, and relevant vascular grafts.
2. Experimental section
2.1. Formation of carbonized alginate (CAlg)
Polyphenolic carbonized alginate (CAlg) was prepared by heating sodium alginate (Alg), as reported previously.33 In a typical synthesis, dry powder of Alg (100 mg) was placed in a glass bottle and heated in a laboratory-grade DH300 convection oven (Dengyng, Taiwan) at 270 °C for 3 h. After heating, the as-obtained brown-black solid residues were cooled down to temperature and dissolved in 10 mL of deionized water. The larger carbonaceous aggregates were removed by centrifugation (RCF 500 g, 30 min). The purified CAlg was kept at 90 °C for 12 h to yield dry powder.2.2. Characterization of CAlg
The particle size of the CAlg before and after heating was captured using a Tecnai G2 F20 S-TWIN transmission electron microscope (TEM; Hillsboro, OG, USA) system operating at 200 kV. Purified CAlg was drop-cast onto a 300-mesh carbon-coated Cu grid and dried at ambient temperature. The UV-visible (UV-Vis) absorption and photoluminescence (PL) spectra of Alg and CAlg dispersed in sodium phosphate buffer (5 mM; pH 7.4) were analyzed using a Synergy 4 Multi-Mode monochromatic microplate spectrophotometer (BioTek Instruments, Winooski, VT, USA). Changes in the CAlg functional groups post pyrolysis were observed using a Fourier transform infrared spectrometer (FTIR, FT/IR-6100, JASCO, Easton, MD, USA) in the range of 500 to 4000 cm−1. Interference from water vapor was minimized by using high-purity nitrogen during the FTIR measurements. Furthermore, we recorded 1H-NMR spectra for pristine Alg and CAlg using the Avance 600 MHz spectrometer (Bruker, Billerica, MA, USA) and corrected the solvent signals.2.3. Preparation of nanoengineered bioactive hydrogels
Two types of hydrogels (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 and 4
9 and 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6) were prepared by adjusting the composition of Alg and methylcellulose (MC). For CAlg/Alg:MC (3
6) were prepared by adjusting the composition of Alg and methylcellulose (MC). For CAlg/Alg:MC (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9), various amounts of CAlg (0–5 wt%) and 3% (wt/vol) of Alg were dissolved in 4 ml of DI water under constant stirring at 60 °C until a clear solution is obtained. To which, 9% (wt/vol) MC was added slowly with continuous stirring at 60 °C to yield a viscous yet homogenous solution. To prepare the CAlg/Alg:MC (4
9), various amounts of CAlg (0–5 wt%) and 3% (wt/vol) of Alg were dissolved in 4 ml of DI water under constant stirring at 60 °C until a clear solution is obtained. To which, 9% (wt/vol) MC was added slowly with continuous stirring at 60 °C to yield a viscous yet homogenous solution. To prepare the CAlg/Alg:MC (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6) hydrogel, various amounts of CAlg (0–5 wt%), Alg (4% wt/vol) and MC (6% wt/vol) were dissolved in 5–20 mM of CaCl2 solution under the same condition. The resulting solutions were separately poured into cartridges and were kept at 4 °C for complete hydration of MC. After hydration, the cartridges filled with gels were stored at room temperature (RT, ≈25 °C) until further use.
6) hydrogel, various amounts of CAlg (0–5 wt%), Alg (4% wt/vol) and MC (6% wt/vol) were dissolved in 5–20 mM of CaCl2 solution under the same condition. The resulting solutions were separately poured into cartridges and were kept at 4 °C for complete hydration of MC. After hydration, the cartridges filled with gels were stored at room temperature (RT, ≈25 °C) until further use.
2.4. Characterization of hydrogels
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 and 4
9 and 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6) were analyzed using a scanning electron microscope (SEM, JEOL iT 300). The SEM sample was prepared as follows: CAlg/Alg:MC (4
6) were analyzed using a scanning electron microscope (SEM, JEOL iT 300). The SEM sample was prepared as follows: CAlg/Alg:MC (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6) was extruded at 180 kPa onto CAlg/Alg:MC (3
6) was extruded at 180 kPa onto CAlg/Alg:MC (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9) extruded at 400 kPa through a nozzle with a diameter of 0.25 mm at a scan speed of 10 mm s−1 under ambient conditions. The 3D-printed structure was lyophilized and sputter-coated with gold before observation.
9) extruded at 400 kPa through a nozzle with a diameter of 0.25 mm at a scan speed of 10 mm s−1 under ambient conditions. The 3D-printed structure was lyophilized and sputter-coated with gold before observation.
We further investigated the degradation behavior of the as-developed 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 and 4
9 and 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 hydrogels individually in phosphate-buffered saline (PBS; 1×, pH 7.4) for a period of 14 days. Firstly, the initial weight (W0; n = 3) of the 3D-printed discs was recorded. Then, the discs were soaked in PBS for two weeks, and the corresponding weight (Wt; n = 3) of the discs was monitored at regular intervals. The change in weight was determined using the equation below:
6 hydrogels individually in phosphate-buffered saline (PBS; 1×, pH 7.4) for a period of 14 days. Firstly, the initial weight (W0; n = 3) of the 3D-printed discs was recorded. Then, the discs were soaked in PBS for two weeks, and the corresponding weight (Wt; n = 3) of the discs was monitored at regular intervals. The change in weight was determined using the equation below:
The viscoelastic behavior of the gels was characterized using a rotational rheometer (Anton Paar Modular Compact Rheometer- 302) with a 10 mm parallel plate and a 1 mm gap. The viscosity of the gels was recorded at different shear rates between 1 and 100 s−1. For the angular frequency sweep, a constant strain (i.e., 1%) was selected from the linear viscoelastic region (LVR), and angular frequency was varied from 1 to 100 rad s−1. Next, the ability of the hydrogel ink to quickly recover its viscosity was evaluated using the three interval-time-thixotropy (3ITT) recovery test. Thixotropic measurement was performed in three simple steps: (a) an external low shear rate of 0.1 s−1 was applied to the gels for 60 s, (b) then the shear rate was increased to 100 s−1 for 10 s, and (c) finally, the shear rate was lowered to 0.1 s−1 for 60 s to assess the viscosity recovery.
2.5. 3D and 4D printing
The 3D models designed in SolidWorks (Dassault Systems) were converted to StereoLithography (.Stl) format and then imported into Slic3r software (Repetier Host, Cellink) to generate G-codes. The print paths were based on computational design, wherein the first layer (CAlg/Alg:MC; 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9) with 99% infill density and the rectilinear pattern was stacked by the stripes of the second and third layers (CAlg/Alg:MC; 4
9) with 99% infill density and the rectilinear pattern was stacked by the stripes of the second and third layers (CAlg/Alg:MC; 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6) with the same pattern but different infill density (40%). Various designs were created by extruding the stripes of the second and third layers (CAlg/Alg:MC; 4
6) with the same pattern but different infill density (40%). Various designs were created by extruding the stripes of the second and third layers (CAlg/Alg:MC; 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6) at different angles with respect to the first layer. The final G-codes were then input into a BioX 3D printer (CELLINK) to extrude two gels with a 0.25 mm diameter independently from two nozzles. The first layer (3
6) at different angles with respect to the first layer. The final G-codes were then input into a BioX 3D printer (CELLINK) to extrude two gels with a 0.25 mm diameter independently from two nozzles. The first layer (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 gel) was extruded at 400 kPa, while the second and third layers of 4
9 gel) was extruded at 400 kPa, while the second and third layers of 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 gel were extruded at 180–210 kPa. The 3D-printed constructs were air-dried overnight at room temperature. The 4D behavior of the printed structures was seen after immersion in 200 mM CaCl2 solution, and the resulting changes in the shapes were captured digitally.
6 gel were extruded at 180–210 kPa. The 3D-printed constructs were air-dried overnight at room temperature. The 4D behavior of the printed structures was seen after immersion in 200 mM CaCl2 solution, and the resulting changes in the shapes were captured digitally.
2.6. Material deformation study via finite element analysis (FEA) simulations
All the 3D model structures were designed similarly, as reported previously by our group.34 Fusion 360 (Autodesk) software was used to create different model structures with varying topographies and infill patterns, and these designed models were exported in the STEP (standard for the exchange of product data) format for FEA simulations. All the simulations were carried out by using the thermal expansion model in commercial software Abaqus/CAE 2017 (Simulia 2017) to predict the deformation of the designed hydrogel structures computationally.2.7. Anti-oxidant assays
Radical scavenging abilities of CAlgX/Alg: MC (where X = 0–5 wt%) were evaluated using a simple colorimetric-based 2,2-diphenyl-1-picryl-hydrazyl-hydrate (DPPH) assay. Cylindrical discs of 10 mm diameter and 1.0 mm thickness were separately printed with each hydrogel ink with a rectilinear pattern and 90% infill density. The printed discs were immersed in a DPPH (200 μM) solution prepared in 50% (vol/vol) methanol and allowed to react for 45 min at room temperature under dark conditions. Following this, the discs were removed, and 200 μL from each mixture was transferred to a 96-well flat bottom plate to record the absorbance value at 517 nm, using a monochromatic microplate spectrophotometer (Synergy 4, Biotek Instruments, Winooski, VT, USA). The free radical scavenging can be calculated as follows:where AbsDPPH is the absorbance of the control group, and ATest is the absorbance of the unknown.
A confirmatory test for the presence of phenol or polyphenol in the 3D-printed constructs was conducted with Folin–Ciocalteu (FC) reagent. In a typical assay, all the hydrogels were separately incubated with the F–C reagent (0.0125 N) dispersed in phosphate buffer (pH 10 mM; 7.4) for 5 min at room temperature, followed by the addition of Na2CO3 (0.1 N) and incubation in the dark for 1 h under orbital shaking (150 rpm). Similar to the DPPH assay, the reacted discs were removed, and 200 μL from each mixture was subjected to the absorption measurement at 765 nm (Abs765) using a monochromatic microplate spectrophotometer (Synergy 4, Biotek Instruments, Winooski, VT, USA).
2.8. Anti-inflammatory assays
The anti-inflammatory response of the CAlg/Alg:MC hydrogels was investigated using lipopolysaccharide (LPS) induced inflammation in RAW264.7. In a typical assay, RAW264.7 macrophages (1 × 105 cells per well) in DMEM (5% FBS) were seeded in a 24-well plate in 5% CO2 at 37 °C for 24 h, followed by incubation with LPS (1.0 μg mL−1) for 2 h at 37 °C to induce the inflammation and then replaced with the fresh DMEM (10% FBS) to further grow for 24 h. Cylindrical discs of 10 mm diameter and 1.0 mm thickness were separately printed with each hydrogel ink (CAlgx/Alg:MC; x = 0, 1.0, 2.5, and 5.0 wt% of Alg) with a rectilinear pattern and 90% infill density. LPS-treated and untreated (negative control) RAW264.7 macrophages were further reacted with the 3D-printed hydrogel discs in 5% CO2 at 37 °C for 24 h and 36 h. Eventually, the growth media and the printed constructs were removed from each well, and 2′-7′-dichlorofluorescein diacetate (DCFH-DA; 100 μM) to detect reactive oxygen species (ROS) and Griess reagent to quantify nitrite oxide (NO) were separately added into each well and allowed to react at 37 °C for 20 min in the dark under ambient conditions. DCF fluorescence at 530 nm was recorded with a fixed excitation at 490 nm using a monochromatic microplate spectrophotometer. On the other hand, the stable product formed by NO and Griess reagent was observed by measuring the absorbance at 540 nm. A similar procedure was performed on RAW264.7 macrophages treated with gels for 24 and 36 h, separately. The ELISA was performed to measure interleukin (IL) levels in the RAW264.7 macrophages with and without LPS treatment and in the absence and presence of CAlg/Alg:MC hydrogels to assess the inflammatory response. Each of the suspensions was then centrifuged (15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, 10 min, 4 °C), and supernatants were collected to quantify the concentrations of pro-inflammatory (IL-6, IL-8, IL-18, IFN-γ, IL-1β, and TNF-α) and (TGF-β1) and anti-inflammatory (IL-10) cytokines using ELISA kits as per the manufacturer's standard protocols. All samples were assayed in triplicates.
000g, 10 min, 4 °C), and supernatants were collected to quantify the concentrations of pro-inflammatory (IL-6, IL-8, IL-18, IFN-γ, IL-1β, and TNF-α) and (TGF-β1) and anti-inflammatory (IL-10) cytokines using ELISA kits as per the manufacturer's standard protocols. All samples were assayed in triplicates.
2.9. Anti-thrombotic assays
The anti-thrombogenic response of the CAlg was investigated by the thrombin clotting time (TCT), prothrombin time (PT), and activated-prothrombin time (aPTT) assays, as reported by Huang et al.35 The human plasma collection for these experiments was consented to and donated by a healthy male individual (age 30) at Mackay Memorial Hospital (Taipei, Taiwan). All the experiments were conducted in compliance with WHO guidelines and approved by the ethics committee of National Taiwan Ocean University.2.10. In vitro experiments
To evaluate in vitro cytotoxicity, HUVEC (1.0 × 104 cells well−1) and NIH-3T3 (1.0 × 104 cells well−1) cell lines were cultured in endothelial cell growth medium and Dulbecco's modified Eagle's medium containing 10% FBS in 5% CO2 at 37 °C for 24 h. HUVEC and NIH-3T3 cell lines were separately incubated in the presence and absence of CAlg/Alg:MC hydrogels (CAlg = 0–5 wt%) in 5% CO2 at 37 °C for 24 h and 48 h. Cell proliferation was analyzed using the AlamarBlue assay, which introduces a fluorometric growth indicator to evaluate cellular metabolic activity. Here, the Resazurin molecule is used as an indicator of cell viability and proliferation by reducing azurin through mitochondrial metabolic activity in living cells. Then, 0.5 mL of 10% AlamarBlue solution was added. The reaction was then incubated in darkness for 4 h before analysis. After incubation, four 100 μL replicates were taken from each well and transferred to a black 96-well plate for fluorescence reading using a Varioskan flash reader (Biotek Elx808, Winooski, VT, USA) (excitation 530 nm, emission 590 nm). The cell-laden scaffolds were studied for 24 and 48 h. Cell viability was calculated from the fluorescence relative to the control. The cell number was obtained using a calibration curve determined by correlating a known cell number with the fluorescent intensity of the solution. Five independent samples were tested in each group.Cell viability was assessed with a live/dead assay (Live/Dead, Invitrogen™, Carlsbad, CA, USA) according to the manufacturer's protocol. In this case, calcein AM (0.5 μL) and ethidium homodimer-1 (2.0 μL) were dissolved in 997.5 μL PBS, added to the samples, and incubated for 40 min under dark conditions at 37 °C in a humidified atmosphere with 5% CO2. The samples were evaluated with a 4× and 10× objective epifluorescence microscope with a ZEISS SCOPE.A microscope (Software ZEN LIET 2012, Carl Zeiss AG, Oberkochen, Germany). Then, images were analyzed using ImageJ software.
For further analysis, CAlg/Alg:MC hydrogels were 3D printed into discs (1 mm height × 10 mm diameter) and air-dried overnight at room temperature. The dried discs were coated with thin fibers of 6% w/v Gelatin Type-A (Sigma Aldrich) and 3% w/v of polycaprolactone (PCL, Mn: 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000; Sigma Aldrich) using a simple airbrush technique. The airbrushed hydrogels were air-dried (24 h) and crosslinked in 25% glutaraldehyde vapors for 2 h. Prior to experiments, all the discs were sterilized by UV exposure for 1 h and washed with cell culture medium (DMEM high glucose supplemented with 10% FBS). HUVEC (1.2 × 104 per disc) and NIH-3T3 (1.0 × 104 per disc) were seeded separately onto the disc-shaped scaffolds. To visualize the attached cells on the scaffolds after 24 h and 48 h of cell growth, all samples were removed from their medium, rinsed with PBS (10 mM, pH 7.4), and fixed with 4% paraformaldehyde (PFA) at room temperature. Cells were permeabilized with 1% Triton X-100 in PBS for 10 minutes at room temperature. After three washes with 0.1% Triton X-100 in DPBS, nuclei were stained for 1 h with DAPI (1
000; Sigma Aldrich) using a simple airbrush technique. The airbrushed hydrogels were air-dried (24 h) and crosslinked in 25% glutaraldehyde vapors for 2 h. Prior to experiments, all the discs were sterilized by UV exposure for 1 h and washed with cell culture medium (DMEM high glucose supplemented with 10% FBS). HUVEC (1.2 × 104 per disc) and NIH-3T3 (1.0 × 104 per disc) were seeded separately onto the disc-shaped scaffolds. To visualize the attached cells on the scaffolds after 24 h and 48 h of cell growth, all samples were removed from their medium, rinsed with PBS (10 mM, pH 7.4), and fixed with 4% paraformaldehyde (PFA) at room temperature. Cells were permeabilized with 1% Triton X-100 in PBS for 10 minutes at room temperature. After three washes with 0.1% Triton X-100 in DPBS, nuclei were stained for 1 h with DAPI (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution; 5 mg mL−1 stock solution; Sigma-Aldrich) and Alexa Fluor 488 Phalloidin (1
1000 dilution; 5 mg mL−1 stock solution; Sigma-Aldrich) and Alexa Fluor 488 Phalloidin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80 dilutions; catalog No. A12379; Invitrogen). PBS was used to remove excess coloring (10 mM, pH 7.4). The samples were evaluated with a 4× and 10× objective epifluorescence microscope with a ZEISS SCOPE system utilized to assess the dispersion of cells throughout the various 3D-printed samples (control and test groups sample size, n = 3).
80 dilutions; catalog No. A12379; Invitrogen). PBS was used to remove excess coloring (10 mM, pH 7.4). The samples were evaluated with a 4× and 10× objective epifluorescence microscope with a ZEISS SCOPE system utilized to assess the dispersion of cells throughout the various 3D-printed samples (control and test groups sample size, n = 3).
3. Results and discussion
3.1. Preparation and characterization of the bioactive nanocomponent
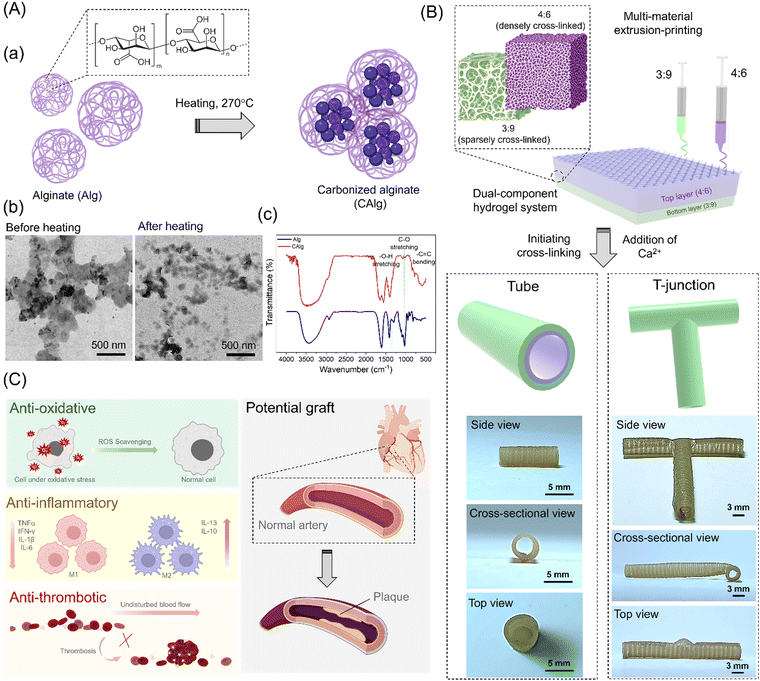
Alginate (Alg) was chosen as the base polymer to develop multifunctional hydrogel ink with high-resolution printability due to its low cost, biocompatibility, and diverse utilization in biomaterials.36 However, pristine Alg is typically a bioinert macromolecule.37 Therefore, we performed mild pyrolysis (270 °C) of pristine Alg to yield carbonized Alg (CAlg) with a variety of biological activities (Fig. 1A-a). Alginate tends to form cross-linked supramolecular structures through dehydration-based condensation processes.33 CAlg showed low viscosity even at higher concentrations (<5.2 mPa s, 50 mg mL−1). A relatively lower magnitude of zeta (ζ) potential was recorded for CAlg (−40 ± 5 mV) than Alg (−80 ± 7 mV) due to dehydration, condensation of hydroxyl groups, and carboxylic groups, and mild carbonization. The negative charge is ascribed to carboxylic groups on the C-6 position of the monomeric units.38 Transmission electron microscopy (TEM) analysis revealed that in contrast to pristine Alg, CAlg showed distinct particles with a mean diameter of 48.7 ± 6.5 nm (100 counts) (Fig. 1A-b). Furthermore, we employed Fourier-transform infrared (FTIR) spectroscopy to investigate the extent of carbonization during pyrolysis. Peaks at 1290 and 1320 cm−1 in pristine Alg correspond to the D-mannuronic (M) and L-guluronic acid (G) moieties, respectively (Fig. 1A-c).33 Upon heating at 270 °C, the intensity of the C–O stretching peak at 1020/1078 cm−1 decreased significantly, and a –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bending peak appeared at 844 cm−1. Interestingly, a tiny peak at 1325 cm−1 was attributed to the –O–H bending of phenol groups formed by mild aromatization during the heating drive dehydration process. These results suggest only partial carbonization of alginate because several functional groups are preserved, but additional bioactive functional groups are also created.33 High-resolution proton nuclear magnetic resonance (1H-NMR) spectroscopy is a useful tool to analyze the changes in the composition of phenolic groups before and after pyrolysis of Alg. CAlg displayed sp3 C–H protons (1.0–3.0 ppm) in deuterated water (D2O). Moreover, a sharp band centered at 8.4 ppm and broadband ranging from 6.5–9.0 ppm with multiple peaks are attributed to the participation of phenol groups within CAlg in proton exchange between inter-/intra-molecular –OH and –COOH groups at 298 K (Fig. S1, ESI†).33,39 The FT-IR and 1H-NMR spectroscopy analysis confirmed the formation of a phenol group via dry heating methods. Although the formation of a polyphenolic structure from Alg has been positively characterized using various tools, it is challenging to identify the exact chemical species of the phenolic groups and to detect individual sharp peaks in the 1H-NMR spectrum due to the complex nature of the CAlg structures. Furthermore, the UV-visible absorption spectra show a small hump at 280 nm for CAlg, corresponding to the π → π* transitions of the conjugated C
C bending peak appeared at 844 cm−1. Interestingly, a tiny peak at 1325 cm−1 was attributed to the –O–H bending of phenol groups formed by mild aromatization during the heating drive dehydration process. These results suggest only partial carbonization of alginate because several functional groups are preserved, but additional bioactive functional groups are also created.33 High-resolution proton nuclear magnetic resonance (1H-NMR) spectroscopy is a useful tool to analyze the changes in the composition of phenolic groups before and after pyrolysis of Alg. CAlg displayed sp3 C–H protons (1.0–3.0 ppm) in deuterated water (D2O). Moreover, a sharp band centered at 8.4 ppm and broadband ranging from 6.5–9.0 ppm with multiple peaks are attributed to the participation of phenol groups within CAlg in proton exchange between inter-/intra-molecular –OH and –COOH groups at 298 K (Fig. S1, ESI†).33,39 The FT-IR and 1H-NMR spectroscopy analysis confirmed the formation of a phenol group via dry heating methods. Although the formation of a polyphenolic structure from Alg has been positively characterized using various tools, it is challenging to identify the exact chemical species of the phenolic groups and to detect individual sharp peaks in the 1H-NMR spectrum due to the complex nature of the CAlg structures. Furthermore, the UV-visible absorption spectra show a small hump at 280 nm for CAlg, corresponding to the π → π* transitions of the conjugated C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bonds (sp2 clusters) and indicates the formation of graphitic carbon during the carbonization of Alg (Fig. S2A, ESI†). Next, the fluorescence spectra of CAlg revealed a broad fluorescence band centered at 450 nm at an excitation wavelength of 365 nm, with a low quantum yield (<1%) due to a low degree of carbonization (Fig. S2B, ESI†).
C bonds (sp2 clusters) and indicates the formation of graphitic carbon during the carbonization of Alg (Fig. S2A, ESI†). Next, the fluorescence spectra of CAlg revealed a broad fluorescence band centered at 450 nm at an excitation wavelength of 365 nm, with a low quantum yield (<1%) due to a low degree of carbonization (Fig. S2B, ESI†).
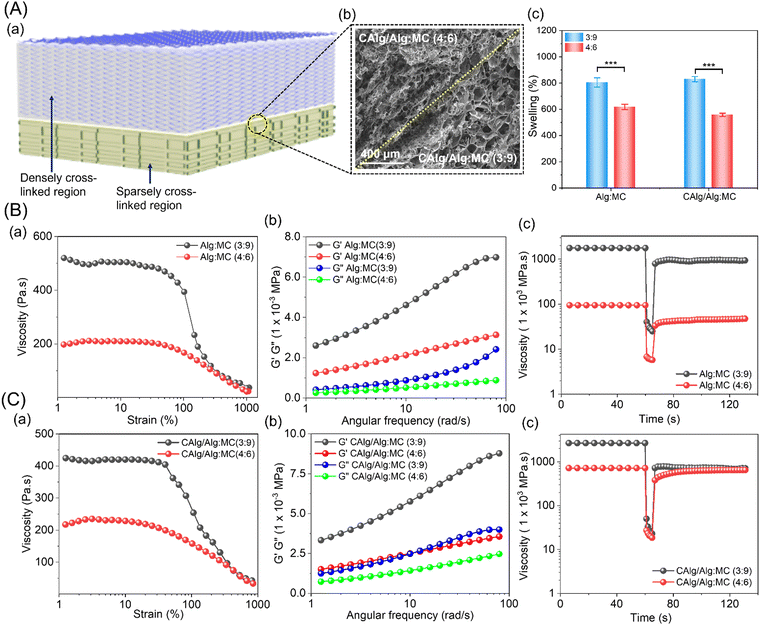
3.2. Nanoengineered dual-component hydrogel system
Two separate Alg:MC inks were developed by adjusting the composition of Alg and methylcellulose (MC) (Fig. 2). Each of the inks (Alg:MC; 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 or 4
9 or 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6) was separately blended with the newly synthesized bioactive CAlg to prepare CAlg/Alg:MC hydrogel. A strong binding affinity of Ca2+ ions with Alg resulted in a densely crosslinked region in 4
6) was separately blended with the newly synthesized bioactive CAlg to prepare CAlg/Alg:MC hydrogel. A strong binding affinity of Ca2+ ions with Alg resulted in a densely crosslinked region in 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 composition (prepared with 5 mM CaCl2), while 3
6 composition (prepared with 5 mM CaCl2), while 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 gels showed sparsely cross-linked regions (Fig. 2A-a). The scanning electron microscopy (SEM) image shows distinguishable physiological changes in the porosity of the formed gels (Fig. 2A-b). The Alg:MC (3
9 gels showed sparsely cross-linked regions (Fig. 2A-a). The scanning electron microscopy (SEM) image shows distinguishable physiological changes in the porosity of the formed gels (Fig. 2A-b). The Alg:MC (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9; ∼800%) prepared in deionized water exhibited a higher swelling rate compared to Alg:MC (4
9; ∼800%) prepared in deionized water exhibited a higher swelling rate compared to Alg:MC (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6; ∼600%) synthesized in 5 mM CaCl2 solution (Fig. 2A-c). A similar trend was observed in both types of gels, irrespective of CAlg composition (Fig. S3, ESI†). We observed insignificant degradation of the gels in a phosphate-buffered saline solution until 2 weeks (Fig. S4, ESI†). Furthermore, we conducted rheological characterization to examine the printability and viscoelastic behavior of the as-formed hydrogels (Fig. 2A and B). The shear thinning behavior of both hydrogels (3
6; ∼600%) synthesized in 5 mM CaCl2 solution (Fig. 2A-c). A similar trend was observed in both types of gels, irrespective of CAlg composition (Fig. S3, ESI†). We observed insignificant degradation of the gels in a phosphate-buffered saline solution until 2 weeks (Fig. S4, ESI†). Furthermore, we conducted rheological characterization to examine the printability and viscoelastic behavior of the as-formed hydrogels (Fig. 2A and B). The shear thinning behavior of both hydrogels (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 or 4
9 or 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6) is evident from Fig. 2B-a and C-a, rendering them ideal for extrusion-based 3D printing. As expected, the values of storage moduli are higher than loss moduli in the entire linear viscoelastic (LVR) region for both 3
6) is evident from Fig. 2B-a and C-a, rendering them ideal for extrusion-based 3D printing. As expected, the values of storage moduli are higher than loss moduli in the entire linear viscoelastic (LVR) region for both 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 and 4
9 and 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 compositions (Fig. 2B-b and C-b).40,41 Eventually, thixotropic measurement was performed to assess the viscosity recovery rates of the gels (Fig. 2B-c and C-c). Viscosities of the gels decreased under high shear rates and consequently recovered close to the original values upon releasing the stress. This property provides the gels with good printing fidelity and is a consequence of chain disentanglements and re-entanglements simultaneously under high and low shear rates, respectively. Next, we measured the mechanical strength of the pristine and nanoengineered gels. The tensile measurement revealed a relatively higher Young's modulus for CAlg/Alg:MC (0.7 kPa) compared to Alg:MC (0.4 kPa) gel (Fig. S5, ESI†). Notably, CAlg did not influence the rheological properties and printability of the hydrogels.
6 compositions (Fig. 2B-b and C-b).40,41 Eventually, thixotropic measurement was performed to assess the viscosity recovery rates of the gels (Fig. 2B-c and C-c). Viscosities of the gels decreased under high shear rates and consequently recovered close to the original values upon releasing the stress. This property provides the gels with good printing fidelity and is a consequence of chain disentanglements and re-entanglements simultaneously under high and low shear rates, respectively. Next, we measured the mechanical strength of the pristine and nanoengineered gels. The tensile measurement revealed a relatively higher Young's modulus for CAlg/Alg:MC (0.7 kPa) compared to Alg:MC (0.4 kPa) gel (Fig. S5, ESI†). Notably, CAlg did not influence the rheological properties and printability of the hydrogels.
3.3. CAlg-induced bioactivities in 3D-printed constructs
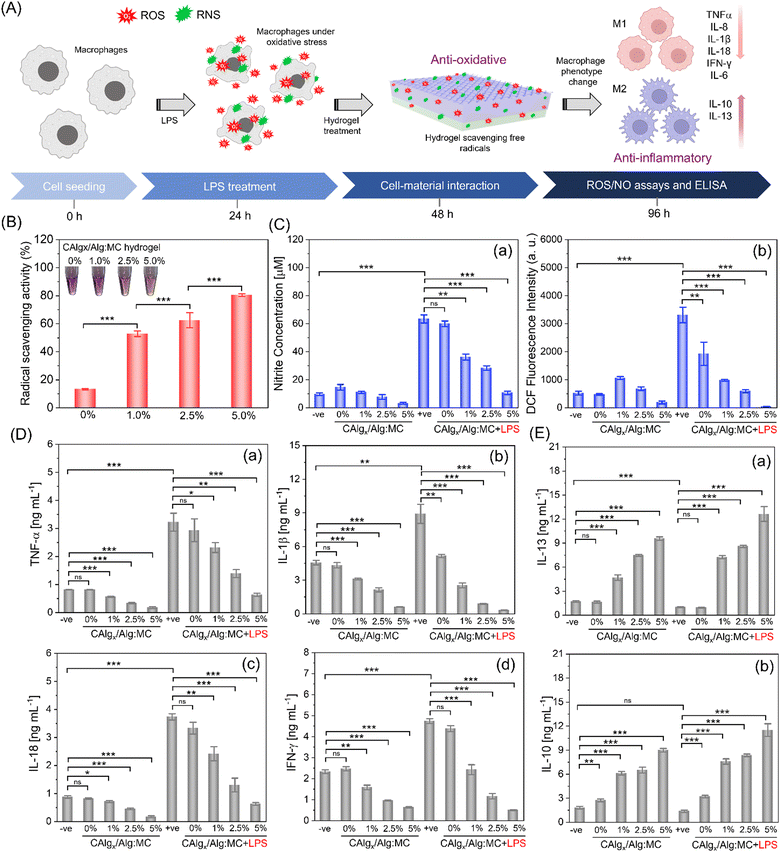
FTIR spectra of CAlg indicate a prominent peak ascribed to the phenol group (1325 cm−1, refer to Fig. 1A-c), which led us to investigate the antioxidant and anticoagulant activities of 3D-printed gels (Fig. 3A).33 To analyze the anti-oxidative behavior due to polyphenolic functional groups within CAlg, we employed the 2,2-diphenyl-1-picrylhydrazyl (DPPH) and Folin–Ciocalteu (FC) assays.42,43 Out of two, a 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 gel formulation was chosen to test the anti-oxidative properties of CAlg/Alg:MC prepared with varied amounts of CAlg (0–5%). The DPPH assay indicated that CAlg/Alg:MC gels exhibited superior free radical scavenging capabilities (50%), ca. >4-fold higher than that of Alg:MC (i.e., 0% CAlg) gel (∼12%) (Fig. 3B). An increasing trend in the radical scavenging activities (53–82%) is attributed to the increasing CAlg composition in the gel. In addition, a noticeable discoloration of 3D-printed CAlg/Alg:MC discs was observed (Fig. S6, ESI†). Furthermore, total phenolic content in the CAlg was investigated using FC reagent.43 The estimated reducing ability of the CAlg (2.5%) is equivalent to that of 40 μg mL−1 ascorbic acid, indicating the formation of phenolic polysaccharides (Fig. S7, ESI†). In contrast, pristine Alg:MC showed insignificant reducing ability in the FC assay. The anti-oxidative nature of the gels can be ascribed to polyphenolic groups. Polyphenols scavenge free radicals by hydrogen (H)-atom transfer (HAT). In HAT, the electron and proton of the H-atom are transferred to the same atomic orbital in the free radical.44 Proton-coupled electron transfer (PCET) is another mechanism in which, unlike HAT, multiple molecular orbitals are involved. In PCET, proton transfer takes place along the H-bond to the lone pair of the oxygen atom of the free radical. This transfer is coupled with electron transfer from a lone pair of antioxidants to the singly occupied molecular orbital of the free radical.45 These mechanisms are well-adapted and proven to describe HAT, from polyphenols to free radicals. However, in both mechanisms, the proton and/or electron are transferred in one step.46 There is another possible mechanism called electron transfer–proton transfer (ET–PT), which is a two-step reaction. The reaction is initiated by an electron transfer and followed by a proton release. Proton transfer is so fast that ET–PT is often considered a HAT process.47 However, the contribution of the graphitic core in facilitating fast electron transfer to promote redox reactions cannot be ruled out.33
9 gel formulation was chosen to test the anti-oxidative properties of CAlg/Alg:MC prepared with varied amounts of CAlg (0–5%). The DPPH assay indicated that CAlg/Alg:MC gels exhibited superior free radical scavenging capabilities (50%), ca. >4-fold higher than that of Alg:MC (i.e., 0% CAlg) gel (∼12%) (Fig. 3B). An increasing trend in the radical scavenging activities (53–82%) is attributed to the increasing CAlg composition in the gel. In addition, a noticeable discoloration of 3D-printed CAlg/Alg:MC discs was observed (Fig. S6, ESI†). Furthermore, total phenolic content in the CAlg was investigated using FC reagent.43 The estimated reducing ability of the CAlg (2.5%) is equivalent to that of 40 μg mL−1 ascorbic acid, indicating the formation of phenolic polysaccharides (Fig. S7, ESI†). In contrast, pristine Alg:MC showed insignificant reducing ability in the FC assay. The anti-oxidative nature of the gels can be ascribed to polyphenolic groups. Polyphenols scavenge free radicals by hydrogen (H)-atom transfer (HAT). In HAT, the electron and proton of the H-atom are transferred to the same atomic orbital in the free radical.44 Proton-coupled electron transfer (PCET) is another mechanism in which, unlike HAT, multiple molecular orbitals are involved. In PCET, proton transfer takes place along the H-bond to the lone pair of the oxygen atom of the free radical. This transfer is coupled with electron transfer from a lone pair of antioxidants to the singly occupied molecular orbital of the free radical.45 These mechanisms are well-adapted and proven to describe HAT, from polyphenols to free radicals. However, in both mechanisms, the proton and/or electron are transferred in one step.46 There is another possible mechanism called electron transfer–proton transfer (ET–PT), which is a two-step reaction. The reaction is initiated by an electron transfer and followed by a proton release. Proton transfer is so fast that ET–PT is often considered a HAT process.47 However, the contribution of the graphitic core in facilitating fast electron transfer to promote redox reactions cannot be ruled out.33
The anti-inflammatory response typically originates from intrinsic radical scavenging capabilities.48 We employed lipopolysaccharide (LPS) to induce oxidative stress in RAW264.7 macrophages and assessed anti-inflammatory properties by estimating the levels of reactive oxygen species (ROS) and nitrite oxide (NO) in the absence and presence of nanoengineered bioactive 3D-printed CAlg/Alg:MC structures at two different time intervals (36 h in Fig. 3C and 24 h in Fig. S8, ESI†). Griess reagent was utilized to measure NO levels.49 RAW264.7 macrophages pre-incubated with LPS for 2 h revealed high NO levels ca. >60 μM, relative to the control (without LPS; ∼10μM) (Fig. 3C-a). However, post-CAlg/Alg:MC treatment, the NO concentration consistently decreased (∼60 μM to ∼10 μM) as the amount of CAlg (0% to 5%) increased within CAlg/Alg:MC ink. On the other hand, pristine Alg:MC gels showed a negligible reduction in the NO levels. Then, we measured ROS levels using the DCFH-DA staining method.50 As expected, ROS levels were dramatically reduced upon CAlg/Alg:MC treatment (Fig. 3C-b). The Alg:MC gels were slightly more effective against ROS than NO. Insignificant oxidative stress in the groups without LPS treatment in the presence of printed structures is strong evidence that the gels did not exhibit a pro-inflammatory response. Due to their superior anti-inflammatory response at a cellular level, ELISA was performed to investigate the changes at the protein level by estimating a series of pro- and anti-inflammatory cytokines on two different time points (24 h and 48 h). M1 macrophages promote the secretion of proinflammatory cytokines, including tumour necrosis factor alpha (TNF-α), interleukin (IL)-6, IL-8, IL-10, IL-13, IL-1β, IL-18, and IFN-γ.51 Notably, expressions of proinflammatory markers reduced gradually as the amount of CAlg increased in the CAlg/Alg:MC gels, while pristine Alg:MC is not as effective (Fig. 3D and Fig. S9, S10A, ESI†). In addition, at the same time CAlg/Alg:MC enhanced the production of anti-inflammatory cytokines such as IL-10 and IL-13 (Fig. 3E and Fig. S10B, ESI†). Polarization from M1 (pro-inflammatory) to M2 (anti-inflammatory) phenotype can be attributed to the excellent anti-inflammatory response of CAlg.52 It has been reported that macrophage polarization can be typically driven by the activation of Toll-Like Receptors (TRLs) (a vital initiator of the innate immune response) or Janus kinase 2 (JAK2) signalling pathways.53
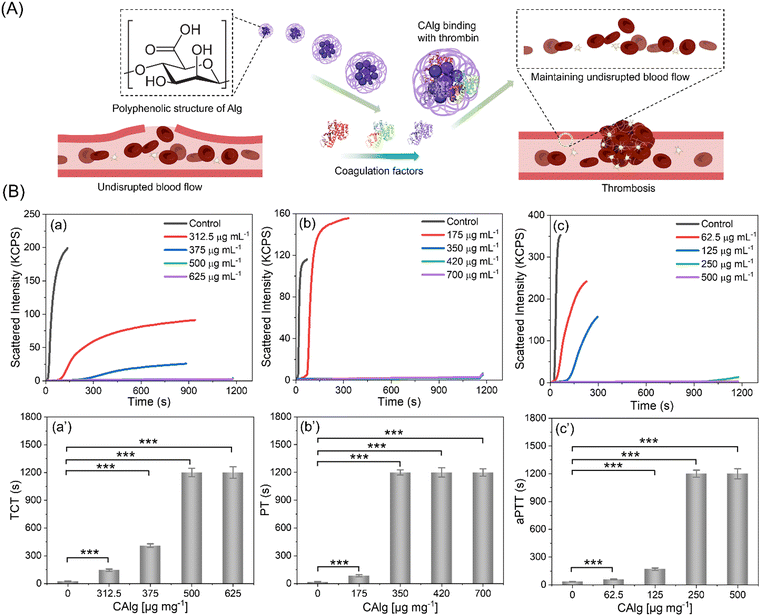
Furthermore, the anti-thrombotic performance of nanoengineered CAlg/Alg: MC-based inks was evaluated (Fig. 4A). Firstly, we estimated thrombin clotting time (TCT) (Fig. 4B). It is known that untreated Alg shows insignificant anticoagulant activities. The TCT delay time [(t − t0)/t0, where t0 and t are the time in the absence and presence of inhibitors, respectively] of the CAlg is at least 6-fold higher (144 s; 312.5 μg mL−1) than the control group (without CAlg; ∼23 s). A concentration-dependent delay in the clotting time was observed (144 s to 1200 s). Notably, CAlg showed constant TCT delay until 72 h in human plasma, indicating the long-term efficacy of this kind of anticoagulant. The prothrombin time (PT) and activated partial thromboplastin time (aPTT) of CAlg were recorded in human plasma samples (Fig. 4C and D). The PT assays evaluate the inhibition of prothrombin, fibrinogen, and factors V, VII, and X. On the other hand, the aPTT assay can determine the activity of various coagulation factors VIII, IX, XI, XII, II, V, and X, along with fibrinogen.54 The PT prolong values [(t − t0)/t0, where t0 and t are the time in the absence and presence of inhibitors, respectively] of CAlg (∼90 s) are ∼5-fold higher than the control group (without CAlg; 17 s) (Fig. 4C). The CAlg (∼1200 s) showed constant time after 250 μg mL−1. A similar trend was obtained from aPTT assessments, in which the saturation in the scattering intensity was noted ≥350 μg mL−1 (Fig. 4D). All the assays revealed the remarkable antithrombotic properties of CAlg, mainly due to their interaction with certain coagulation factors in addition to thrombin. We have previously shown that CAlg possesses a strong binding affinity towards thrombin (dissociation constant (Kd) = 2.1 × 10−10 M).33,55,56 Interestingly, the Kd value for a well-known anticoagulant such as heparin (with thrombin) is only 1.0 × 10−7 M, indicating 200-fold lower affinity than CAlg.55 In addition, stronger inhibitory activities can also be ascribed to phenol or phenolic acid groups self-preserved within CAlg under mild pyrolysis. These functional groups facilitate a multivalent phenolic acid-like interaction with thrombin. These results corroborate the aforementioned DPPH and FC assays, supporting polyphenolic and phenolic acid structure facilitated anti-oxidant activities (Fig. 3B and Fig. S7, ESI†). Overall, the results suggest that CAlg suppresses thrombin and other coagulation factors in different pathways.
3.4. 4D printing of dynamic and programmable structures
A computational model was designed to predict the changes in the shapes of the 3D-printed structures. We used a thermal expansion model to simulate the deformations of the hydrogels after establishing a quantitative correlation between the swelling model. Primarily, the type and direction of deformations were assessed by modeling a rectangular geometry structure with different topographies of the top layer (Fig. S11, ESI†). The gel1 (CAlg/Alg:MC; 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9) or the first layer is a solid base material (thickness = 0.25 mm) with a higher expansion coefficient. It is decorated with stripes of gel2 (CAlg/Alg:MC; 4
9) or the first layer is a solid base material (thickness = 0.25 mm) with a higher expansion coefficient. It is decorated with stripes of gel2 (CAlg/Alg:MC; 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6) (width = 0.25 mm, thickness = 0.10 mm), which has a lower expansion coefficient, and these stripes are accurately placed parallel to each other with precise interspacing (0.20 mm) and at a specific angle (i.e. 0°, 45° and 90°) with respect to the base material. The calculation of the expansion coefficient for both materials is based on the swelling ratios of the hydrogel layers, as shown previously.34 The FEA simulations inferred that the bilayer structure can be deformed in different orientations with respect to the different patterns of the stripes on the base material. The deformation of the material is governed by the orientation of the stripes (gel2) on the base material, i.e., bending along the longer axis (0° stripes arrangement), twisting (45° stripes arrangement), and bending along the shorter axis (90° stripes arrangement). For experimental validations, we created G-codes to match the computational design. An extrusion-based 3D printer was utilized to print various 3D constructs. Both the formulations (3
6) (width = 0.25 mm, thickness = 0.10 mm), which has a lower expansion coefficient, and these stripes are accurately placed parallel to each other with precise interspacing (0.20 mm) and at a specific angle (i.e. 0°, 45° and 90°) with respect to the base material. The calculation of the expansion coefficient for both materials is based on the swelling ratios of the hydrogel layers, as shown previously.34 The FEA simulations inferred that the bilayer structure can be deformed in different orientations with respect to the different patterns of the stripes on the base material. The deformation of the material is governed by the orientation of the stripes (gel2) on the base material, i.e., bending along the longer axis (0° stripes arrangement), twisting (45° stripes arrangement), and bending along the shorter axis (90° stripes arrangement). For experimental validations, we created G-codes to match the computational design. An extrusion-based 3D printer was utilized to print various 3D constructs. Both the formulations (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 and 4
9 and 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6) were extruded as per the G-codes under different pressures and using two different 0.25 Gauze nozzles. The printed structures were air-dried overnight and subsequently immersed in 200 mM CaCl2 solution to observe shape deformation (Fig. S12, ESI†). The real-time shape changes were digitally recorded (Videos V1–V3, ESI†). The resulting deformation matched closely with the predictive computational model. Moreover, G-codes were designed to print several complex geometries, including a hollow rectangle, and to validate the accuracy of the model (Fig. S13, ESI†). A hollow rectangle (Videos V4 and V5, ESI†) and plus (Videos V6 and V7, ESI†) shaped 3D constructs were designed and printed in a way that strands of the top layer were placed at different angles (0° or 45°) with respect to the base layer. Resultant deformations were predetermined to bend or twist depending upon the arrangement of the stripes.
6) were extruded as per the G-codes under different pressures and using two different 0.25 Gauze nozzles. The printed structures were air-dried overnight and subsequently immersed in 200 mM CaCl2 solution to observe shape deformation (Fig. S12, ESI†). The real-time shape changes were digitally recorded (Videos V1–V3, ESI†). The resulting deformation matched closely with the predictive computational model. Moreover, G-codes were designed to print several complex geometries, including a hollow rectangle, and to validate the accuracy of the model (Fig. S13, ESI†). A hollow rectangle (Videos V4 and V5, ESI†) and plus (Videos V6 and V7, ESI†) shaped 3D constructs were designed and printed in a way that strands of the top layer were placed at different angles (0° or 45°) with respect to the base layer. Resultant deformations were predetermined to bend or twist depending upon the arrangement of the stripes.
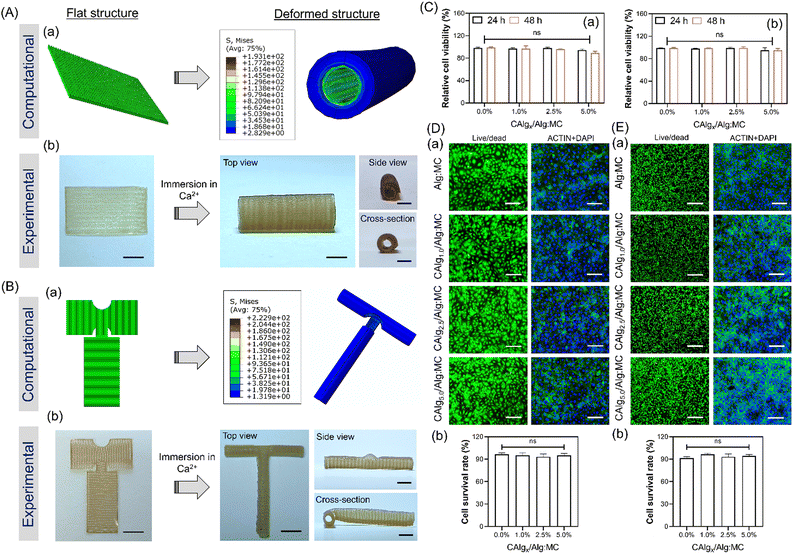
While there have been many reports on the fabrication of hollow tubular constructs using 4D printing, developing complicated designs like bifurcating T-junctions, which are perfusable, is highly in demand. Such designs like T and Y junctions mimic the physiology of vascular networks, yet achieving such shapes from design-encoded 2D sheets is challenging to realize. Also, since these bifurcations are more prone to blockage, it highlights the need to develop an approach to target such designs using 4D fabrication.57,58 Formulation of bioactive alginate (i.e., CAlg) blended with highly thixotropic Alg:MC composition was demonstrated to fabricate tubes with intricate designs (Fig. S12, ESI†). We used the predictive modeling-based approach to computationally evaluate how different designs can lead to tubes of different shapes, including T junctions (Fig. 5A and B). The T junction structure was designed with a 90° stripes arrangement of gel2 (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6; both in the vertical and horizontal part of the structure) with respect to the base layer (gel; 3
6; both in the vertical and horizontal part of the structure) with respect to the base layer (gel; 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9). The FEA simulations revealed that the horizontal and vertical sections of the T-shaped structure bent to form a cylindrical structure, resulting in an overall curved T-shaped structure with hollow cylindrical tubes (Fig. S14, ESI†). These predicted deformations are in accordance with the experimental results (Video V8, ESI†). As demonstrated, computational predictions were utilized to fabricate and design encoded 2D sheets that can deform into tubes such as single channels and bifurcations upon simple swelling in a calcium chloride solution (Fig. 5A-b and B-b). We also confirmed that fabricated designs (single channel and T-junction) were perfusable, presenting a promising candidate for vascular tissue engineering (Video V9, ESI†). To confirm that, the biocompatibility of the hydrogel composition was evaluated for two important cells, HUVECs and fibroblasts, the two major cell populations in the vascular channel (Fig. 5C).59 For control experiments, pristine Alg:MC based hydrogel was utilized. Both the cells were highly viable as estimated through normalized Alamar Blue reduction values at 24 and 48 hours, respectively (Fig. 5Ca and b). No significant impact on viability was observed by varying the concentrations of bioactive CAlg as seen from live/dead stained images, which show a substantial fraction of live cells (Fig. 5D and E-a; left panel). Furthermore, we performed F-actin staining to observe the cellular arrangement of hydrogel compositions after five days of culture (Fig. 5D and E-a; right panel). As expected, both cell types were nicely spread and formed a monolayer at day 5 with no cytotoxic effects (Fig. 5D and E-b). Overall, current work demonstrated how bioactivity can be incorporated into pristine hydrogel inks without compromising shape deformations. Furthermore, the freedom to predict shape deformation has tremendous potential to target more complicated designs in the future, such as a vascular tree. Biocompatibility for two cells further confirms the immense potential of this approach to target cardiovascular diseases such as atherosclerosis, which primarily impacts bifurcations. Although we have demonstrated the utility of 4D fabrication for targeting complicated designs, further evaluations are warranted for their application as vascular grafts.
9). The FEA simulations revealed that the horizontal and vertical sections of the T-shaped structure bent to form a cylindrical structure, resulting in an overall curved T-shaped structure with hollow cylindrical tubes (Fig. S14, ESI†). These predicted deformations are in accordance with the experimental results (Video V8, ESI†). As demonstrated, computational predictions were utilized to fabricate and design encoded 2D sheets that can deform into tubes such as single channels and bifurcations upon simple swelling in a calcium chloride solution (Fig. 5A-b and B-b). We also confirmed that fabricated designs (single channel and T-junction) were perfusable, presenting a promising candidate for vascular tissue engineering (Video V9, ESI†). To confirm that, the biocompatibility of the hydrogel composition was evaluated for two important cells, HUVECs and fibroblasts, the two major cell populations in the vascular channel (Fig. 5C).59 For control experiments, pristine Alg:MC based hydrogel was utilized. Both the cells were highly viable as estimated through normalized Alamar Blue reduction values at 24 and 48 hours, respectively (Fig. 5Ca and b). No significant impact on viability was observed by varying the concentrations of bioactive CAlg as seen from live/dead stained images, which show a substantial fraction of live cells (Fig. 5D and E-a; left panel). Furthermore, we performed F-actin staining to observe the cellular arrangement of hydrogel compositions after five days of culture (Fig. 5D and E-a; right panel). As expected, both cell types were nicely spread and formed a monolayer at day 5 with no cytotoxic effects (Fig. 5D and E-b). Overall, current work demonstrated how bioactivity can be incorporated into pristine hydrogel inks without compromising shape deformations. Furthermore, the freedom to predict shape deformation has tremendous potential to target more complicated designs in the future, such as a vascular tree. Biocompatibility for two cells further confirms the immense potential of this approach to target cardiovascular diseases such as atherosclerosis, which primarily impacts bifurcations. Although we have demonstrated the utility of 4D fabrication for targeting complicated designs, further evaluations are warranted for their application as vascular grafts.
4. Conclusion
In this work, we have demonstrated 4D-printing assisted construction of complex scaffold geometries prepared from static hydrogel parts in an aqueous calcium chloride solution. Bioinert Alg was transformed into nanosized CAlg with superior anti-oxidative/inflammatory/thrombotic properties through a solid-state synthesis process. Furthermore, an Alg/MC dual-component hydrogel system is nanoengineered with CAlg to optimize two separate hydrogel inks (CAlg/Alg:MC; 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 or 4
9 or 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6) by simply adjusting the composition of Alg and MC. Two gels with predefined swelling behaviour were overlaid over each using an extrusion-based multi-material 3D printing process to achieve dynamic structures. This is the first report of utilizing a 4D printing approach to manufacture T-shaped perfusable bifurcation. A predictive computational model was designed to realize shape deformations in the printed hydrogels with defined infill angles and subsequently validated experimentally. A perfusion test was conducted using a dye solution through a T-junction, mimicking blood flow through vessels, which showed minimal leakage. Moreover, both HUVECs and NIH3T3 cells incubated with 3D-printed constructs show intact morphology and excellent cell viability. Our strategy paves the way for the fabrication of fully automatic self-folding vascular bifurcations with remarkable anti-thrombotic properties to treat coronary artery diseases potentially. Overall, the study aimed to highlight the potential of the 4D printing approach to fabricate intricate and anatomically accurate tissue scaffolds.
6) by simply adjusting the composition of Alg and MC. Two gels with predefined swelling behaviour were overlaid over each using an extrusion-based multi-material 3D printing process to achieve dynamic structures. This is the first report of utilizing a 4D printing approach to manufacture T-shaped perfusable bifurcation. A predictive computational model was designed to realize shape deformations in the printed hydrogels with defined infill angles and subsequently validated experimentally. A perfusion test was conducted using a dye solution through a T-junction, mimicking blood flow through vessels, which showed minimal leakage. Moreover, both HUVECs and NIH3T3 cells incubated with 3D-printed constructs show intact morphology and excellent cell viability. Our strategy paves the way for the fabrication of fully automatic self-folding vascular bifurcations with remarkable anti-thrombotic properties to treat coronary artery diseases potentially. Overall, the study aimed to highlight the potential of the 4D printing approach to fabricate intricate and anatomically accurate tissue scaffolds.
Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
The authors are grateful to Mr V. S. Baghel for his help in the computational studies. This work was supported by the Department of Science and Technology (DST) sponsored Innovation in Science Pursuit for Inspired Research (INSPIRE) Faculty Programme (DST/INSPIRE/04/2021/001535). The authors acknowledge support from the Science and Engineering Research Board (SERB), Government of India (IPA/2020/000025).References
- A. Joshi, S. Choudhury, S. B. Gugulothu, S. S. Visweswariah and K. Chatterjee, Biomacromolecules, 2022, 23, 2730–2751 CrossRef CAS PubMed
.
- Z. U. Arif, M. Y. Khalid, R. Noroozi, A. Sadeghianmaryan, M. Jalalvand and M. Hossain, Int. J. Biol. Macromol., 2022, 218, 930–968 CrossRef CAS
.
- S. Mohandesnezhad, M. H. Monfared, S. Samani, A. Farzin, S. A. Poursamar, J. Ai, S. Ebrahimi-barough and M. Azami, J. Non-Cryst. Solids, 2023, 609, 122261 CrossRef CAS
.
- A. Ding, O. Jeon, R. Tang, Y. Bin Lee, S. J. Lee and E. Alsberg, Adv. Sci., 2021, 8, 1–9 Search PubMed
.
- C. M. González-Henríquez, M. A. Sarabia-Vallejos and J. Rodriguez-Hernandez, Prog. Polym. Sci., 2019, 94, 57–116 CrossRef
.
- A. Mandal and K. Chatterjee, J. Mater. Chem. B, 2024, 12, 2985–3005 RSC
.
- S. Parimita, A. Kumar, H. Krishnaswamy and P. Ghosh, J. Manuf. Process., 2023, 85, 875–884 CrossRef
.
- E. M. White, J. Yatvin, J. B. Grubbs, J. A. Bilbrey and J. Locklin, J. Polym. Sci., Part B: Polym. Phys., 2013, 51, 1084–1099 CrossRef CAS
.
- D. Qureshi, S. K. Nayak, S. Maji, A. Anis, D. Kim and K. Pal, Eur. Polym. J., 2019, 120, 109220 CrossRef CAS
.
- A. Ding, O. Jeon, D. Cleveland, K. L. Gasvoda, D. Wells, S. J. Lee and E. Alsberg, Adv. Mater., 2022, 34, 389–396 Search PubMed
.
- C. Cui, D.-O. Kim, M. Y. Pack, B. Han, L. Han, Y. Sun and L.-H. Han, Biofabrication, 2020, 12, 045018 CrossRef CAS PubMed
.
- A. Nain, S. Chakraborty, N. Jain, S. Choudhury, S. Chattopadhyay, K. Chatterjee and S. Debnath, Biomater. Sci., 2024 10.1039/D3BM02044D
.
- M. Champeau, D. A. Heinze, T. N. Viana, E. R. de Souza, A. C. Chinellato and S. Titotto, Adv. Funct. Mater., 2020, 30, 1–22 CrossRef
.
- J. Li, G. Zhang, Z. Cui, L. Bao, Z. Xia, Z. Liu and X. Zhou, Small, 2023, 19, 1–11 Search PubMed
.
- M. T. I. Mredha and I. Jeon, Prog. Mater. Sci., 2022, 124, 100870 CrossRef CAS
.
- S. Ma, B. Yu, X. Pei and F. Zhou, Polymer, 2016, 98, 516–535 CrossRef CAS
.
- W. Kitana, I. Apsite, J. Hazur, A. R. Boccaccini and L. Ionov, Adv. Mater. Technol., 2023, 8, 2200429 CrossRef CAS
.
-
S. Ozturk, F. B. Ayanoğlu, M. Parmaksiz, A. E. Elçin and Y. M. Elçin, Handbook of Biomaterials Biocompatibility, Elsevier, 2020, vol. 23, pp. 219–250 Search PubMed
.
- G. Yang, B. Mahadik, J. Y. Choi and J. P. Fisher, Prog. Biomed. Eng., 2020, 2, 012002 CrossRef
.
- L. Zeenat, A. Zolfagharian, Y. Sriya, S. Sasikumar, M. Bodaghi and F. Pati, Adv. Mater. Technol., 2023, 8, 6671–6677 Search PubMed
.
- A. Kirillova, R. Maxson, G. Stoychev, C. T. Gomillion and L. Ionov, Adv. Mater. Chem. B, 2019, 7, 1597–1624 RSC
.
- L. Zhang, Y. Xiang, H. Zhang, L. Cheng, X. Mao, N. An, L. Zhang, J. Zhou, L. Deng, Y. Zhang, X. Sun, H. A. Santos and W. Cui, Adv. Sci., 2020, 7, 255–305 Search PubMed
.
- S. Zakharchenko, E. Sperling and L. Ionov, Biomacromolecules, 2011, 12, 2211–2215 CrossRef CAS PubMed
.
- L. Shao, Q. Gao, C. Xie, J. Fu, M. Xiang and Y. He, Adv. Healthcare Mater., 2019, 8, 308–320 Search PubMed
.
- L. Shao, Q. Gao, H. Zhao, C. Xie, J. Fu, Z. Liu, M. Xiang and Y. He, Small, 2018, 14, 1–15 CrossRef
.
- D. Podstawczyk, M. Nizioł, P. Śledzik, J. Simińska-Stanny, A. Dawiec-Liśniewska and A. Shavandi, Adv. Funct. Mater., 2023, 110, 23–26 Search PubMed
.
- W. Jia, P. S. Gungor-Ozkerim, Y. S. Zhang, K. Yue, K. Zhu, W. Liu, Q. Pi, B. Byambaa, M. R. Dokmeci, S. R. Shin and A. Khademhosseini, Biomaterials, 2016, 106, 58–68 CrossRef CAS PubMed
.
- B. Grigoryan, S. J. Paulsen, D. C. Corbett, D. W. Sazer, C. L. Fortin, A. J. Zaita, P. T. Greenfield, N. J. Calafat, J. P. Gounley, A. H. Ta, F. Johansson, A. Randles, J. E. Rosenkrantz, J. D. Louis-Rosenberg, P. A. Galie, K. R. Stevens and J. S. Miller, Science, 2019, 364, 458–464 CrossRef CAS PubMed
.
- S. B. Gugulothu and K. Chatterjee, ACS Macro Lett., 2023, 12, 494–502 CrossRef CAS PubMed
.
- V. Stroganov, J. Pant, G. Stoychev, A. Janke, D. Jehnichen, A. Fery, H. Handa and L. Ionov, Adv. Funct. Mater., 2018, 28, 1–8 CrossRef
.
- A. Kirillova and L. Ionov, J. Mater. Chem. B, 2019, 7, 1597–1624 RSC
.
- L. Zhang, Y. Xiang, H. Zhang, L. Cheng, X. Mao, N. An, L. Zhang, J. Zhou, L. Deng, Y. Zhang, X. Sun, H. A. Santos and W. Cui, Adv. Sci., 2020, 7, 1903553 CrossRef CAS
.
- J. Y. Mao, B. Unnikrishnan, H. W. Chu, S. G. Harroun, Y. R. Chen, A. T. Wu, H. T. Chang, H. J. Lin and C. C. Huang, Biomater. Sci., 2021, 9, 4679–4690 RSC
.
- A. Joshi, S. Choudhury, V. S. Baghel, S. Ghosh, S. Gupta, D. Lahiri, G. K. Ananthasuresh and K. Chatterjee, Adv. Healthcare Mater., 2023, 12, 2300701 CrossRef CAS PubMed
.
- S. S. Huang, S. C. Wei, H. T. Chang, H. J. Lin and C. C. Huang, J. Controlled Release, 2016, 221, 9–17 CrossRef CAS PubMed
.
- M. M. Capeling, M. Czerwinski, S. Huang, Y. H. Tsai, A. Wu, M. S. Nagy, B. Juliar, N. Sundaram, Y. Song, W. M. Han, S. Takayama, E. Alsberg, A. J. Garcia, M. Helmrath, A. J. Putnam and J. R. Spence, Stem Cell Rep., 2019, 12, 381–394 CrossRef CAS
.
- M. I. Neves, L. Moroni and C. C. Barrias, Front. Bioeng. Biotechnol., 2020, 8, 665 CrossRef PubMed
.
- R. Kadri, G. Ben Messaoud, A. Tamayol, B. Aliakbarian, H. Y. Zhang, M. Hasan, L. Sánchez-González and E. Arab-Tehrany, RSC Adv., 2016, 6, 27879–27884 RSC
.
- P. Charisiadis, V. Kontogianni, C. Tsiafoulis, A. Tzakos, M. Siskos and I. Gerothanassis, Molecules, 2014, 19, 13643–13682 CrossRef PubMed
.
- D. Petta, D. W. Grijpma, M. Alini, D. Eglin and M. D’Este, ACS Biomater. Sci. Eng., 2018, 4, 3088–3098 CrossRef CAS PubMed
.
- Y. J. Li, S. C. Wei, H. W. Chu, H. J. Jian, A. Anand, A. Nain, Y. F. Huang, H. T. Chang, C. C. Huang and J. Y. Lai, Chem. Eng. J., 2022, 437, 135315 CrossRef CAS
.
- M. Shin, J. H. Galarraga, M. Y. Kwon, H. Lee and J. A. Burdick, Acta Biomater., 2019, 95, 165–175 CrossRef CAS
.
- A. Nain, Y. T. Tseng, A. Gupta, Y. F. Lin, S. Arumugam, Y. F. Huang, C. C. Huang and H. T. Chang, Nanoscale Horiz., 2023, 8, 1652–1664 RSC
.
- S. V. Jovanovic, S. Steenken, M. Tosic, B. Marjanovic and M. G. Simic, J. Am. Chem. Soc., 1994, 116, 4846–4851 CrossRef CAS
.
- G. A. DiLabio and E. R. Johnson, J. Am. Chem. Soc., 2007, 129, 6199–6203 CrossRef CAS PubMed
.
- A. A. Gentile, B. Flynn, S. Knauer, N. Wiebe, S. Paesani, C. E. Granade, J. G. Rarity, R. Santagati and A. Laing, Nat. Phys., 2021, 17, 837–843 Search PubMed
.
- F. Di Meo, V. Lemaur, J. Cornil, R. Lazzaroni, J.-L. Duroux, Y. Olivier and P. Trouillas, J. Phys. Chem. A, 2013, 117, 2082–2092 CrossRef CAS PubMed
.
- C. J. Lin, T. L. Hwang, R. Y. L. Wang, A. Nain, R. H. Shih, L. Chang, H. J. Lin, S. G. Harroun, H. T. Chang and C. C. Huang, Small, 2024, 2307210, 1–14 Search PubMed
.
- D. Giustarini, R. Rossi, A. Milzani and I. Dalle-Donne, Methods Enzymol., 2008, 440, 361–380 CAS
.
- Y. J. Hsu, A. Nain, Y. F. Lin, Y. T. Tseng, Y. J. Li, A. Sangili, P. Srivastava, H. L. Yu, Y. F. Huang, C. C. Huang and H. T. Chang, J. Nanobiotechnol., 2022, 20, 1–15 CrossRef PubMed
.
- L. Y. Lu, F. Loi, K. Nathan, T. H. Lin, J. Pajarinen, E. Gibon, A. Nabeshima, L. Cordova, E. Jämsen, Z. Yao and S. B. Goodman, J. Orthop. Res., 2017, 35, 2378–2385 CrossRef CAS PubMed
.
- M. Zeyda, D. Farmer, J. Todoric, O. Aszmann, M. Speiser, G. Györi, G. J. Zlabinger and T. M. Stulnig, Int. J. Obes., 2007, 31, 1420–1428 CrossRef CAS PubMed
.
- C. Kerneur, C. E. Cano and D. Olive, Front. Immunol., 2022, 13, 1026954 CrossRef CAS PubMed
.
- J. H. Levy, F. Szlam, A. S. Wolberg and A. Winkler, Clin. Lab. Med., 2014, 34, 453–477 CrossRef PubMed
.
- J. Deadman, G. Claeson and M. F. Scully, J. Enzyme Inhib. Med. Chem., 1995, 9, 29–41 CAS
.
- S. R. Stone and J. Hofsteenge, Eur. J. Biochem., 1987, 169, 373–376 CrossRef CAS PubMed
.
- P. Gupta and B. B. Mandal, Adv. Funct. Mater., 2021, 31, 1–28 Search PubMed
.
- J. C. Townsend, D. H. Steinberg, C. D. Nielsen, T. M. Todoran, C. P. Patel, R. A. Leonardi, B. J. Wolf, E. S. Brilakis, K. A. Shunk, J. A. Goldstein, M. J. Kern and E. R. Powers, Am. J. Cardiol., 2013, 112, 369–372 CrossRef PubMed
.
- M. Malvè, A. M. Gharib, S. K. Yazdani, G. Finet, M. A. Martínez, R. Pettigrew and J. Ohayon, Ann. Biomed. Eng., 2015, 43, 82–93 CrossRef PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4tb00498a |
| This journal is © The Royal Society of Chemistry 2024 |