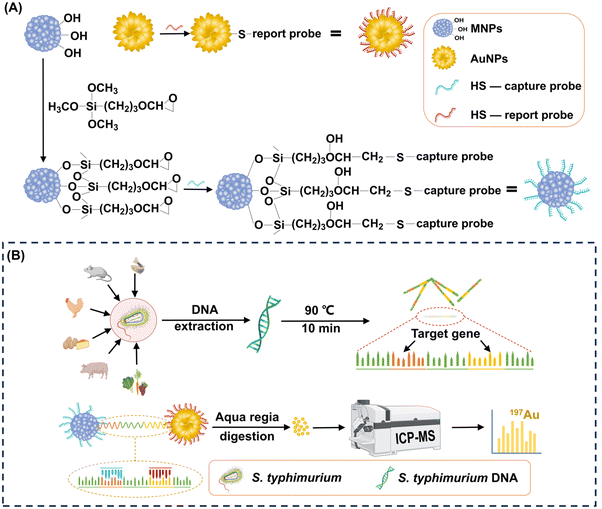
Highly sensitive detection of Salmonella typhimurium via gold and magnetic nanoparticle-mediated sandwich hybridization coupled with ICP-MS†
Yujie
Zhou
 a,
Zhihui
Tang
b,
Lei
Li
a,
Zhihui
Tang
b,
Lei
Li
 a,
Yuzuo
Chen
b,
Yuanyuan
Xu
a,
Yuzuo
Chen
b,
Yuanyuan
Xu
 a,
Renjie
Liu
b,
Yanrong
Zhang
b,
Xiaoyan
Liu
c,
Wenjuan
Yang
d,
Baoning
Wang
b,
Jieyu
Zhang
a,
Renjie
Liu
b,
Yanrong
Zhang
b,
Xiaoyan
Liu
c,
Wenjuan
Yang
d,
Baoning
Wang
b,
Jieyu
Zhang
 *a,
Qing
Jiang
*a and
Yunbing
Wang
*a,
Qing
Jiang
*a and
Yunbing
Wang
 a
a
aNational Engineering Research Center for Biomaterials, Sichuan University, Chengdu, Sichuan 610065, China. E-mail: jieyu@scu.edu.cn; jiangq@scu.edu.cn; Fax: +86 028 85410246; Tel: +86 028 85410537
bWest China School of Basic Medical Sciences and Forensic Medicine, Sichuan University, Chengdu, Sichuan 610041, China
cDepartment of Orthopedic Surgery, West China Hospital, Sichuan University/West China School of Nursing, Chengdu, Sichuan 610041, China
dDepartment of Gastroenterology, West China Hospital, Sichuan University, Chengdu, Sichuan 610041, China
First published on 25th June 2024
Abstract
Foodborne pathogens including Salmonella typhimurium (S. typhimurium) are responsible for over 600 million global incidences of illness annually, posing a significant threat to public health. Inductively coupled plasma mass spectrometry (ICP-MS), coupled with element labeling strategies, has emerged as a promising platform for multivariate and accurate pathogen detection. However, achieving high specificity and sensitivity remains a critical challenge. Herein, we synthesize clustered magnetic nanoparticles (MNPs) and popcorn-shaped gold nanoparticles (AuNPs) to conjugate capture and report DNA probes for S. typhimurium, respectively. These engineered nanoparticles facilitate the identification of S. typhimurium DNA through a sandwich hybridization technique. ICP-MS quantification of Au within the sandwich-structure complexes allows for precise S. typhimurium detection. The unique morphology of the AuNPs and MNPs increases the available sites for probe attachment, enhancing the efficiency of S. typhimurium DNA capture, broadening the detection range to 101–1010 copies mL−1, and achieving a low detection limit of 1 copy mL−1, and the overall assay time is 70 min. The high specificity of this method is verified by anti-interference assays against ten other pathogens. The recovery was 96.8–102.8% for detecting S. typhimurium DNA in biological samples. As these specially designed nanoparticles may facilitate the attachment of various proteins and nucleic acid probes, they may become an effective platform for detecting multiple pathogens.
1. Introduction
The World Health Organization has identified foodborne illness as an escalating global issue, with over 600 million cases annually, mainly caused by pathogen-contaminated food.1Salmonella typhimurium (S. typhimurium) is a major contributor to foodborne illness and a significant health hazard.2 The Centers for Disease Control and Prevention highlighted that S. typhimurium infections cause about 450 deaths and 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 hospitalizations each year in the USA, with associated direct medical costs amounting to $365 million.3 Therefore, the development of rapid, sensitive, and reliable strategies for S. typhimurium detection is critical to enhancing food safety, reducing the risk of infections, and guiding the timely clinical response alongside effective pharmacological interventions.
000 hospitalizations each year in the USA, with associated direct medical costs amounting to $365 million.3 Therefore, the development of rapid, sensitive, and reliable strategies for S. typhimurium detection is critical to enhancing food safety, reducing the risk of infections, and guiding the timely clinical response alongside effective pharmacological interventions.
Current methods for S. typhimurium detection mainly include the traditional plate culture method,4 enzyme-linked immunosorbent assays,5,6 and polymerase chain reaction.7–10 Despite their widespread adoption, these methods suffer from disadvantages, such as being time-consuming,11 and in certain contexts, compromised accuracy.12 Consequently, there is an imperative need to construct more convenient, easy-to-operate, and accurate detection methods for S. typhimurium detection. Recent advancements have led to the development of new biosensors, including optical and electrochemical biosensors,13–17 which involve quantum dots18–21 and lanthanide metals22,23 and offer advantages of fast response, ease of use, and good sensitivity.2 However, these new methods are not without limitations. For example, the color change or colorimetric signals of optical biosensors are easily interfered with by impurities in the sample,22 and most electrochemical biosensors rely on labeled probes or complex materials to assist the signal output, which increases the cost of detection.14
Inductively coupled plasma mass spectrometry (ICP-MS) in conjunction with the element labeling technique represents an advancement in pathogen detection. This combination effectively overcomes the limitations of optical and electrochemical biosensors, such as spectral overlap, background scattering, and interference from the biological matrix. ICP-MS is capable of precise chemical characterization and quantification of trace and ultra-trace elements in biological materials,24 establishing itself as a superior tool for multivariate and accurate biological analysis with remarkable sensitivity and wide detection range. Recent studies have demonstrated the combination of specially designed metal nanoparticles with ICP-MS for pathogen detection.25–28 Protein probe-modified nanoparticles can target the specific surface proteins on pathogens via immunological antibody–antigen recognition,29,30 facilitating accurate pathogen detection. However, this method relies on the availability of highly specific antibodies or probes for accurate pathogen identification, with diagnostic precision depending on the affinity and specificity of these antibodies or probes. Cross-reactivity with similar pathogens may result in false positive or negative signals.31 In contrast, nucleic acid probe-modified nanoparticles may offer improved specificity.32,33 Nonetheless, designing highly specific nucleic acid probes and developing the nanocarriers with optimal specific surface area presents significant challenges.
In this study, we introduced a highly sensitive S. typhimurium detection method that utilizes a novel combination of popcorn-sharped AuNPs and magnetic nanoparticles (MNPs) with a cluster structure to target S. typhimurium DNA via sandwich hybridization (Fig. 1). We designed capture and report probes to bind conserved sequences of S. typhimurium DNA, allowing specific recognition. The large surface area of the popcorn-shaped AuNPs, functionalized with reporter probes, provides efficient capture of target S. typhimurium DNA. Each popcorn-shaped AuNP contains hundreds of millions of Au atoms. These atoms contribute to the generation of robust signals in ICP-MS, thereby significantly enhancing the sensitivity. Simultaneously, cluster-structured MNPs, functionalized with capture probes, enable magnetic separation of the formed MNPs-S. typhimurium DNA-AuNPs sandwich-structure complexes. Quantitative detection of S. typhimurium is achieved by analyzing the Au content within these complexes using ICP-MS. This approach offers rapid detection, high sensitivity, wide detection range, and enhanced specificity. It advances the S. typhimurium detection strategy and provides a versatile framework for the development of prompt and accurate pathogen detection technologies, crucial for managing emergent pathogen infection events.
2. Experimental
2.1. Chemicals and materials
Anhydrous ferric chloride (FeCl3), ethylene glycol (EG), diethylene glycol (DEG), sodium acetate (NaOAc), polyvinylpyrrolidone (PVP, K-30), 3-glycidoxypropyltrimethoxylsilane (GPTMS), chloroauric acid tetrahydrate (HAuCl·4H2O), ascorbic acid (AA), hydroxylammonium chloride (NH2OH·HCl), potassium carbonate anhydrous (K2CO3) and dithiothreitol (DTT) were purchased from Sigma-Aldrich Chemical Co.2.2. Design of capture probe and report probe for the detection of S. typhimurium DNA
The genome-wide information of S. typhimurium was searched in the National Center for Biotechnology Information (NCBI) database, and the genome file was downloaded (GenBank assembly accession: GCA_016864495.1), which yielded a total of 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 870
870![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 244 bp of genome-wide information. The whole genome information was imported into Geneious for the invA gene sequence. The invA gene sequences (5′-TCCTCAACTTCAGCAGATACCATTACTGCTCGTAATTCGCCGCCATTGGCGAATTTATGACAAATATAACGCGCCATTGCTCCACGAATATGCTCCACAAGGT-3′) was selected as the target gene for the detection of this experiment.
244 bp of genome-wide information. The whole genome information was imported into Geneious for the invA gene sequence. The invA gene sequences (5′-TCCTCAACTTCAGCAGATACCATTACTGCTCGTAATTCGCCGCCATTGGCGAATTTATGACAAATATAACGCGCCATTGCTCCACGAATATGCTCCACAAGGT-3′) was selected as the target gene for the detection of this experiment.
The obtained invA gene sequences were imported into Primer Premier 6.0 to design the capture probe and report probe. The thiolated capture probe (5′-HS-(CH2)6-(A)12-ACCTTGTGGAGCATATTCG-3′) and report probe (5′-GTATCTGCTGAAGTTGAGGA-(A)12-(CH2)6-SH-3′) were designed to be complementary to the target. All oligonucleotide sequences were synthesized and purified by high-performance liquid chromatography (HPLC). All oligonucleotides were dissolved in Tris-EDTA (TE) buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8) and kept at −20 °C.
2.3. Preparation of capture probe-grafted MNPs
MNPs were synthesized following a previously reported method.34 Briefly, 2 mmol FeCl3 was added into a mixture of 4 mL EG and 16 mL DEG in a 50 mL two-necked flask followed by continuous stirring for 30 min at room temperature (RT). After adding 2 g PVP (K-30), the two-necked flask was placed in a 125 °C oil bath, and the mixture was stirred for 1 h. Subsequently, 1.5 g NaOAc was added into the flask and stirred for 30 min at RT. The mixture in the two-necked flask was poured into a 50 mL stainless-steel autoclave lined with Teflon, and this autoclave was subjected to heating in an oven at 200 °C for 12 h. The autoclave content was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 621 g for 15 min to separate the supernatant from the MNPs. The collected MNPs were washed three times with ethanol and Milli-Q water, followed by drying in a vacuum oven for 12 h.
621 g for 15 min to separate the supernatant from the MNPs. The collected MNPs were washed three times with ethanol and Milli-Q water, followed by drying in a vacuum oven for 12 h.
The obtained MNPs were functionalized with epoxy groups utilizing a silanization agent. Specifically, 0.5 mL of GPTMS was added into 50 mL of a 0.4% (w/v) MNP suspension in toluene. The mixture was sonicated for 30 min to ensure homogeneity and then refluxed for 12 h under a N2 atmosphere. The epoxy-functionalized MNPs were isolated from the solution by magnetic attraction and purified by sequential washing with toluene, ethanol, and Milli-Q water. The epoxy-functionalized MNPs were finally obtained after 12 h of vacuum drying.
Before coupling the thiolated capture probes to the epoxy-functionalized MNPs, a reductive cleavage step was performed to cleave the disulfide bonds. In brief, 50 μL of a 2 nM thiolated capture probe solution was treated with 100 μL of a 0.1 M DTT solution in TE buffer for 3 h. The resulting probe solution was purified using a NAP-5 column (Cytiva, USA) to remove excess DTT and salt matrix following the manufacturer's protocol. The concentration of the purified probe solution was measured using a Nanodrop spectrophotometer (Thermo, USA).
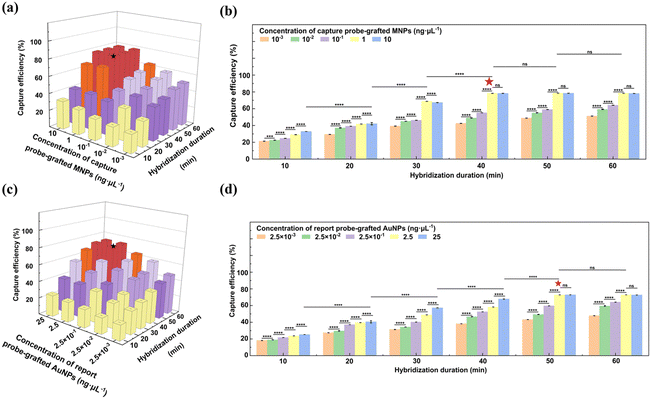
To graft the thiolated capture probes onto the epoxy-functionalized MNPs, 0.39 mg of the epoxy-functionalized MNPs were initially dispersed in 200 μL of a 50 mM Tris-HCl buffer (pH 8). Then, 480 μL of the capture probe solution at different concentrations was introduced into the suspension, and the mixture was incubated at RT for different durations. The resulting capture probe-grafted MNPs were isolated by magnetic separation, washed three times with TE buffer, and re-suspended in 200 μL of TE buffer. The grafting parameters, including capture probe concentration (0.23, 0.45, 0.66, 0.90, and 1.13 ng μL−1), and reaction duration (0.17, 0.5, 1, 2, 4, 6, 8, and 24 h) were optimized to achieve the maximum grafting density. The optimal grafting density of the capture probes on the epoxy-functionalized MNPs was achieved with a capture probe concentration of 0.66 ng μL−1 and an incubation time of 8 h. Detailed calculations regarding the grafting density of the capture probes on the epoxy-functionalized MNPs are provided in the ESI.†
2.4. Preparation of report probe-grafted AuNPs
Popcorn-shaped AuNPs were synthesized using a template-free method.35 Initially, 1 mL of a 0.2 M K2CO3 solution was mixed with 25 mL of a 2.4 × 10−4 M HAuCl4 solution. Subsequently, 5 μL of a 10 mM AA solution was introduced to reduce Au3+ to generate Au seeds. After 5 min of reaction, different volumes of a 10 mM NH2OH solution (0, 500, 1000, and 2000 μL) were rapidly injected to induce the formation of popcorn-shaped AuNPs. This transformation was indicated by an immediate change in the color of the suspension from light yellow to dark blue. The popcorn-shaped AuNPs were harvested by centrifugation at 956 g for 6 min, followed by washing twice with Milli-Q water and drying in a vacuum oven for 12 h.The thiolated report probes were pretreated by DTT to cleave disulfide bonds before grafting onto the AuNPs. For grafting, 0.74 mg AuNPs were ultrasonically dispersed in 200 μL of TE buffer. Subsequently, 480 μL of the capture probe solution at different concentrations was introduced into the AuNPs suspension. The mixtures were incubated at RT for different durations. The resulting report probe-grafted AuNPs were isolated by centrifugation, washed three times with TE buffer, and re-suspended in 200 μL of TE buffer. The grafting parameters, including the report probe concentration (0.23, 0.46, 0.69, 0.93, and 1.16 ng μL−1), and reaction duration (0.17, 0.5, 1, 2, 4, 6, 8, and 24 h) was optimized to maximize grafting density. The optimal condition was identified when 0.93 ng μL−1 of report probes were used with a reaction duration of 8 h. The calculation of the grafting density of the report probes on the AuNPs is detailed in the ESI.†
2.5. Nanoparticle characterization
The morphology and zeta potential of the synthesized nanoparticles were analyzed by transmission electron microscopy (TEM; Zeiss, Germany) and Zetasizer Nano (Malvern, UK), respectively. The crystal structures of the MNPs and epoxy-functionalized MNPs were analyzed by X-ray diffraction (XRD; Bruker, Germany) at a voltage of 40 kV, a current of 300 mA, and a scanning rate of 2° min−1 with step size 0.01°. The magnetic properties of the MNPs and epoxy-functionalized MNPs were measured using vibrating sample magnetometry (VSM; Lake Shore, USA). The chemical structure of the MNPs and epoxy-functionalized MNPs was characterized using attenuated total reflectance Fourier transform infrared spectroscopy (ATR-FTIR Spectrometer; Thermo, USA) and X-ray photoelectron spectroscopy (XPS; Kratos, UK).2.6. S. typhimurium DNA extraction
S. typhimurium bacteria were cultured in lysogeny broth for 48 h, and harvested by centrifugation at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 294 g for 1 min. S. typhimurium DNA was extracted according to the protocol of the TIANamp Bacteria DNA Kit (TIANGEN, Beijing), and stored at −20 °C for further use.
294 g for 1 min. S. typhimurium DNA was extracted according to the protocol of the TIANamp Bacteria DNA Kit (TIANGEN, Beijing), and stored at −20 °C for further use.
2.7. Capture of S. typhimurium DNA by capture probe-grafted MNPs
The stock solution of S. typhimurium DNA was heated at 90 °C for 10 min to unwind the double helix. Then, 10 μL of the resultant DNA solution at a concentration of 1011 copies mL−1 was incubated with 45 μL of the capture probe-grafted MNP suspension at different concentrations (10−3, 10−2, 10−1, 1, and 10 ng μL−1). The hybridization was conducted over various durations (10, 20, 30, 40, 50, and 60 min) at 37 °C under continuous shaking at 200 rpm. After hybridization, the S. typhimurium DNA-MNP complexes were harvested by magnetic separation. The concentration of unbound S. typhimurium DNA in the supernatant was quantified with the Nanodrop spectrophotometer, enabling the determination of the capture efficiency of the capture probe-grafted MNPs for S. typhimurium DNA. The methodology for this calculation is elaborated in the ESI.† Optimal capture efficiency, 78.3%, was achieved with the capture probe-grafted MNP suspension at 1 ng μL−1 and a hybridization period of 40 min.2.8. Capture of S. typhimurium DNA by report probe-grafted AuNPs
The hybridization protocol for the report probe-grafted AuNPs with S. typhimurium DNA paralleled that of the capture probe-grafted MNPs, with the exception the report probe-grafted AuNPs concentration due to their different probe grafting density. To optimize the capture efficiency, the report probe-grafted AuNP suspension at different concentrations (2.5 × 10−3, 2.5 × 10−2, 2.5 × 10−1, 2.5, and 25 ng μL−1) were explored. After hybridization, the S. typhimurium DNA-AuNP complexes were isolated by centrifugation at 956 g for 5 min. Optimal capture efficiency, 72.8%, was obtained with the report probe-grafted AuNP suspension at 2.5 ng μL−1 over a hybridization period of 50 min. The methodology for efficiency calculation is detailed in the ESI.†2.9. Formation of sandwich-structure complexes of MNPs-S. typhimurium DNA-AuNPs
For the formation of the sandwich-structure complexes, 45 μL of the capture probe-grafted MNP suspension at 1 ng μL−1 and 45 μL of the report probe-grafted AuNP suspension at 2.5 ng μL−1 were incubated with 10 μL of a S. typhimurium DNA solution for 50 min at 37 °C under shaking at 200 rpm. The concentration of S. typhimurium DNA in the hybridization assay ranged from 101 to 1010 copies mL−1. The resultant composites were collected by magnetic separation and washed with 5 mL of TE buffer.2.10. ICP-MS analysis
The sandwich-structure complexes were digested with aqua regia, after which the sample volume was adjusted to 10 mL. The sample solution containing the dissolved Au ions were introduced into the Agilent 7800 ICP-MS (Agilent Technologies, Japan) to detect Au at m/z 197. The specific models and operating parameters used for Au determination are detailed in Table S1 in the ESI.†2.11. Anti-interference assay
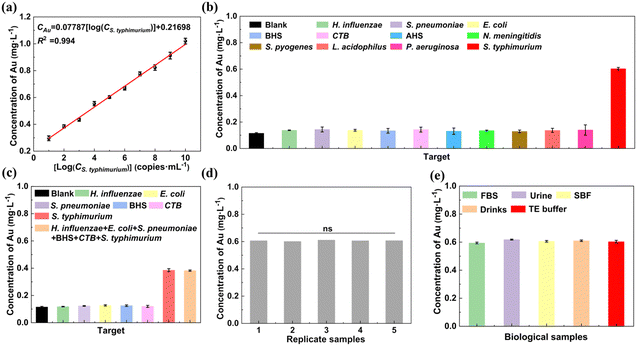
To evaluate the specificity of our method for S. typhimurium DNA detection, 45 μL of the capture probe-grafted MNPs at 1 ng μL−1 and 45 μL of the report probe-grafted AuNP suspension at 2.5 ng μL−1 were incubated with 10 μL of DNA from ten different pathogens Haemophilus influenzae (H. influenzae), Streptococcus pneumoniae (S. pneumoniae), Escherichia coli (E. coli), β-hemolytic streptococcus (BHS), CTB, α-hemolytic streptococcus (AHS), Neisseria meningitidis (N. meningitidis), Streptococcus pyogenes (S. pyogenes), Lactobacillus acidophilus (L. acidophilus), and Pseudomonas aeruginosa (P. aeruginosa), each at a concentration of 105 copies mL−1, for 50 min at 37 °C under shaking at 200 rpm. After that, the composites were harvested from the mixture via magnetic separation and washed with 5 mL of TE. We also tested the performance of our method in detecting a low concentration of target S. typhimurium from high concentrations of interference bacterial DNA (H. influenzae, S. pneumoniae, E. coli, BHS, and CTB). In detail, we measured 102 copies mL−1 of S. typhimurium DNA in the presence of 109 copies mL−1 of DNA from each of the five aforementioned interference bacteria. In addition, signals from a mixed sample containing both high concentrations of all the five interference bacterial DNAs and the low concentration of S. typhimurium DNA were analyzed. Detailed information on the five interference bacteria is provided in ESI† Table S2.2.12. Repeatability assay
To evaluate the repeatability of our method for S. typhimurium DNA detection, five samples containing the same concentration (105 copies mL−1) of S. typhimurium DNA were assessed. Specifically, 45 μL of the capture probe-grafted MNPs (1 ng μL−1) and 45 μL of the report probe-grafted AuNPs (2.5 ng μL−1) were incubated with 10 μL of S. typhimurium DNA (105 copies mL−1) for 50 min at 37 °C under shaking at 200 rpm. After that, the sandwich-structure complexes were harvested from the mixture via magnetic separation and washed with 5 mL of TE buffer. After 5 cycles of washing, the Au concentration in the complexes was measured by ICP-MS.2.13. Performance in biological samples
To further investigate the practical application, the detection assay was conducted in biological samples. The S. typhimurium DNA was dissolved in fetal bovine serum (FBS; Gibco, New Zealand), urine, simulated body fluid (SBF; LEAGENE Biotechnology, China), or drink (Mizone®, Danone, China). The detection assay was performed following the procedure described in Section 2.12.2.14. Statistical analysis
One-way analysis of variance (ANOVA) and two-way ANOVA were used to analyze the differences among groups. Significance levels were defined as *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.001. All statistical analyses were performed using Graph Pad. Prism 10.1.3. Results and discussion
3.1. Characterization of capture probe-grafted MNPs
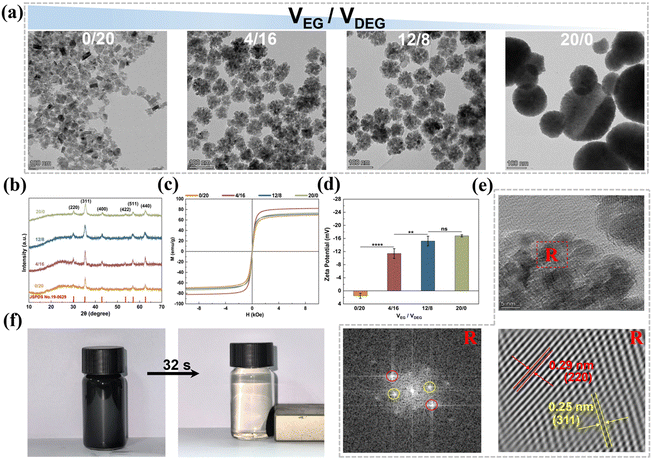
Fig. 2b shows the XRD patterns of the MNPs prepared with different VEG/VDEG ratios. These patterns were consistent with the cubic-phase Fe3O4 spectrum (PDF#88-0866). The diffraction peaks at 30.1°, 35.4°, 43.1°, 53.4°, 57°, and 62.6° corresponded to the (220), (311), (400), (422), (511), and (440) crystal planes of magnetite, respectively.37 The average crystallite size of Fe3O4 MNPs prepared at VEG/VDEG = 0/20 was calculated to be 18.9 nm using the Debye–Scherrer equation (Dhkl = kλ/β![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cos
cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ),36 with the full width at half maximum of the primary (311) diffraction peak. Furthermore, the diffraction peaks of the MNPs broadened with increasing VEG/VDEG ratio, implying the aggregation of the Fe3O4 seeds.36 The crystallite sizes estimated from XRD are in agreement with the dimensional changes observed by TEM (Fig. 2a).
θ),36 with the full width at half maximum of the primary (311) diffraction peak. Furthermore, the diffraction peaks of the MNPs broadened with increasing VEG/VDEG ratio, implying the aggregation of the Fe3O4 seeds.36 The crystallite sizes estimated from XRD are in agreement with the dimensional changes observed by TEM (Fig. 2a).
The magnetic absorption performance of the MNPs was evaluated by RT hysteresis loop measurements using a vibrating sample magnetometer (Fig. 2c). All the MNPs, synthesized with different VEG/VDEG ratios, exhibited hysteresis loops without coercivity or remanence, indicating their superparamagnetic characteristics.38 The magnetic saturation (Ms) values of the MNPs synthesized with VEG/VDEG ratios of 0/20, 4/16, 12/8, and 20/0, were recorded as 69.2, 82.0, 71.3, and 73.7 emu g−1, respectively. The MNPs obtained with a VEG/VDEG ratio of 4/16 achieved the highest Ms value, indicating a superior magnetic response suitable for enhanced magnetic separation applications.
In addition to magnetic separation capability, the colloidal stability of the MNPs in aqueous solution is important for their biomedical application. To evaluate this, the zeta potential of the MNPs was assessed. As shown in Fig. 2d, the MNPs produced with a VEG/VDEG ratio of 0/20 exhibited a positive zeta potential of 1.49 mV, which is indicative of a greater tendency for aggregation. Conversely, increasing the VEG/VDEG ratio from 4/16 to 20/0 resulted in a negative shift in zeta potential from −11.4 mV to −16.8 mV, implying improved colloidal stability.39
The MNPs obtained with a VEG/VDEG ratio of 4/16 were selected for subsequent investigation, due to their optimal magnetic response, high specific surface area, and enhanced colloidal stability. Morphological examination of the MNPs prepared from three independent batches at this VEG/VDEG ratio revealed uniformly clustered structures with a particle size of 64 ± 12 nm (Fig. S10, ESI†), indicating reproducible synthesis with consistent structural and dimensional characteristics across batches. High-resolution TEM (HRTEM) further confirmed their crystallinity, with interplanar spacings of 0.25 and 0.29 nm aligning with the (311) and (220) lattice planes of cubic Fe3O4 magnetite, respectively (Fig. 2e).37 Additionally, these MNPs displayed a rapid and efficient magnetic response, achieving complete alignment in 32 s upon application of an external magnetic field (Fig. 2f).
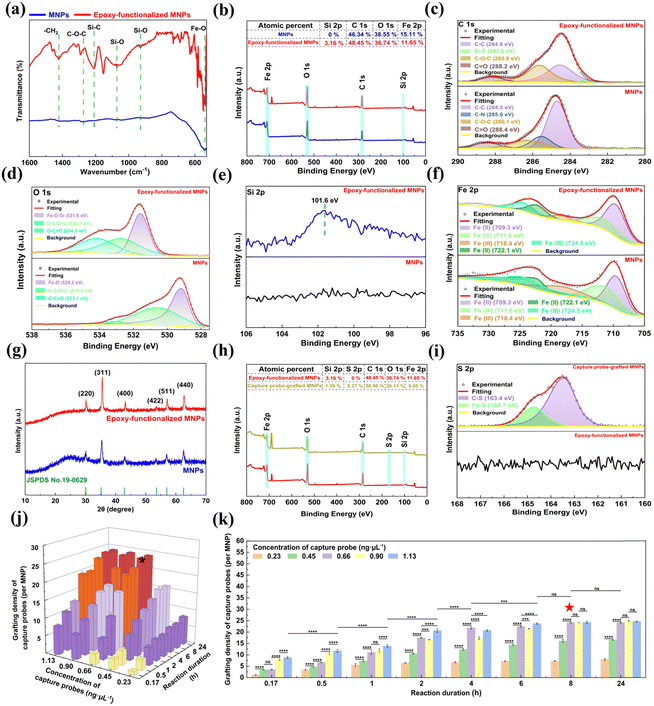
The chemical state and surface groups of the epoxy-functionalized MNPs were further verified by XPS analysis (Fig. 3b–f). The wide scan XPS spectra revealed the presence of Fe, O, and C elements in the MNPs, with an additional Si signal detected in the epoxy-functionalized MNPs (Fig. 3b). High-resolution XPS spectra deconvolution of C 1s (Fig. 3c) and O 1s (Fig. 3d) provided insight into the molecular structure changes following GPTMS silanization of the MNPs. The asymmetric and broad C 1s spectrum of the MNPs was deconvoluted into four peaks at binding energies of 284.5, 285.6, 286.1, and 288.4 eV, which can be attributed to the C–C, C–N, C–O–C, and C = O bonds, respectively (Fig. 3c).43 The presence of these peaks confirms the incorporation of PVP as a stabilizer during MNP synthesis. After epoxy-functionalization, the high-resolution C 1s XPS spectrum showed four deconvoluted peaks at 284.8, 283.2, 285.9, and 288.2 eV (Fig. 3c).43 These peaks correspond to the C–C bonds in the alkane chain of the silane layer, Si–C bonds indicative of silane coupling, the C–O–C ether linkages of the epoxy groups, and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bonds in the silane layer, respectively.42 For the O 1s high-resolution spectrum, the MNPs displayed three sub-peaks at 529.2, 530.6, and 533.1 eV, which are attributed to the Fe–O, O–C/O
O bonds in the silane layer, respectively.42 For the O 1s high-resolution spectrum, the MNPs displayed three sub-peaks at 529.2, 530.6, and 533.1 eV, which are attributed to the Fe–O, O–C/O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and O–C
C and O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bonds, respectively. GPTMS silanization led to a Fe–O–Si peak at 531.6 eV and shifted the binding energy of the O–C/O
O bonds, respectively. GPTMS silanization led to a Fe–O–Si peak at 531.6 eV and shifted the binding energy of the O–C/O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and O–C
C and O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bonds by approximately 1.1 eV (Fig. 3d),44 suggesting that silanization induced the conductivity reduction and electron cloud density changes of the MNPs.45 Furthermore, the Si 2p peak at 101.6 eV was detected in the spectrum of the epoxy-functionalized MNPs (Fig. 3e), which is attributed to the Si–O bonds in the silane layer.46 For the Fe 2p high-resolution spectra (Fig. 3f), the fitting peaks at 722.1 and 709.3 eV were consistent with the Fe–O bonds of ferrite, suggesting that the epoxy-functionalization did not alter the MNP core.43 XRD analysis further supports this preservation, the diffraction peaks at 2θ values of 30.1°, 35.4°, 43.1°, 53.4°, 57.0°, and 62.6° remained consistent for the MNPs after epoxy-functionalization (Fig. 3g). These peaks corresponded to the crystalline planes (220), (311), (400), (422), (511) and (440) of the Fe3O4 crystal, indicating core structure integrity.37
O bonds by approximately 1.1 eV (Fig. 3d),44 suggesting that silanization induced the conductivity reduction and electron cloud density changes of the MNPs.45 Furthermore, the Si 2p peak at 101.6 eV was detected in the spectrum of the epoxy-functionalized MNPs (Fig. 3e), which is attributed to the Si–O bonds in the silane layer.46 For the Fe 2p high-resolution spectra (Fig. 3f), the fitting peaks at 722.1 and 709.3 eV were consistent with the Fe–O bonds of ferrite, suggesting that the epoxy-functionalization did not alter the MNP core.43 XRD analysis further supports this preservation, the diffraction peaks at 2θ values of 30.1°, 35.4°, 43.1°, 53.4°, 57.0°, and 62.6° remained consistent for the MNPs after epoxy-functionalization (Fig. 3g). These peaks corresponded to the crystalline planes (220), (311), (400), (422), (511) and (440) of the Fe3O4 crystal, indicating core structure integrity.37
The epoxy-functionalization resulted in a more negative zeta potential (−28.1 mV) for the MNPs (Fig. S2, ESI†). This alternation enhanced the colloidal stability and dispersibility of the epoxy-functionalized MNPs, beneficial to subsequent conjugation with thiolated capture probes for the detection of S. typhimurium DNA.39 Besides, the magnetic properties of the MNPs were retained after the epoxy-functionalization, as evidenced by the similar RT hysteresis loops before and after GPTMS modification, and the high Ms value of the epoxy-functionalized MNPs (73.4 emu g−1) (Fig. S3, ESI†). This feature is critical to ensure rapid and efficient magnetic separation during the detection of S. typhimurium.
To optimize the conjugation process of the capture probes onto the epoxy-functionalized MNPs, the effect of the capture probe concentration and reaction duration on the grafting density of the capture probes on the epoxy-functionalized MNPs was systematically investigated (Fig. 3j and k). For reaction duration less than 8 h, the grafting density on the MNPs increased with the capture probe concentration up to 1.13 ng μL−1. When the reaction duration ≥8 h, the grafting density also increased but reached a plateau at a capture probe concentration of 0.66 ng μL−1. At a fixed capture probe concentration of 0.66 ng μL−1, the grafting density augmented as the reaction duration extended, plateauing at 8 h. This plateau indicates a saturation of the available epoxy sites on the MNPs. Therefore, the optimal conjugation conditions were determined to be a capture probe concentration of 0.66 ng μL−1 and a reaction time of 8 h, which corresponded to an average of approximately 24 capture probes grafted onto each MNP. Further details on the grafting density calculations of the capture probes on the epoxy-functionalized MNPs were provided in the ESI.†
3.2. Characterization of report probe-grafted AuNPs
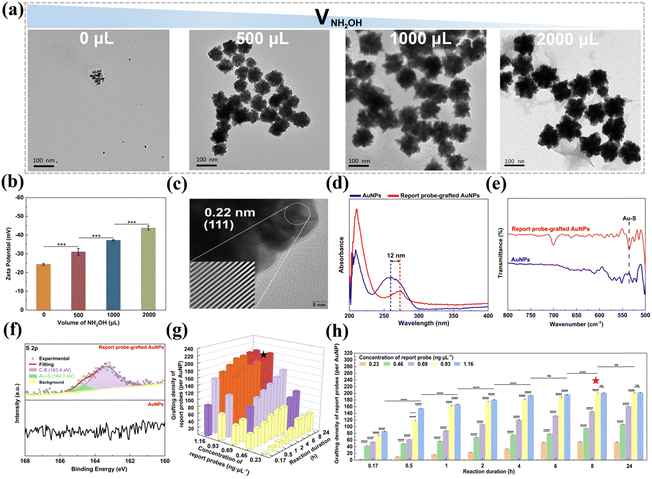
Herein, we synthesized AuNPs via a facile and efficient method, which involves a two-step reduction process. Initially, AuCl4− ions are reduced by AA to generate small Au seeds. Subsequently, these Au seeds serve as nucleation sites for further reduction and deposition of Au atoms by NH2OH, ultimately yielding popcorn-shaped AuNPs.35 Notably, this protocol is surfactant-free and operates at RT, with a rapid reaction time of 18 min. The size of the resultant AuNPs can be easily adjusted by varying the volume of the 10 mM NH2OH solution added (Fig. 4a). In the absence of NH2OH, only the initial Au seeds with a diameter of 10 ± 2 nm were observed. The introduction of NH2OH triggers a reduction of Au atoms and their subsequent deposition onto the seeds, promoting their aggregation into the unique popcorn-like structures. Increasing the volume of the NH2OH solution from 500 μL to 2000 μL led to the growth of the AuNPs from 60 ± 7 nm to 94 ± 14 nm in diameter. The specific surface area of the prepared popcorn-shaped AuNPs was quantified using Brunauer–Emmett–Teller (BET) analysis (Fig. S5, ESI†). The popcorn-shaped AuNPs had a larger specific surface area (10.64 m2 g−1), compared to their spherical counterparts (3.53 m2 g−1) with the same diameter (88 nm). This enhanced surface area provided additional binding sites for the thiolated reporter probes, allowing the grafting of a larger number of reporter probes to the AuNPs. This configuration broadened the detection range and lowered the detection limit for S. typhimurium DNA.
The zeta potential of AuNPs synthesized with different volumes of a 10 mM NH2OH solution was analyzed to assess their colloidal stability. Fig. 4b shows that the AuNPs were negatively charged, and their zeta potential became more negative as the NH2OH volume increased. This trend suggests that the colloidal stability was enhanced with higher NH2OH volumes, attributed to stronger electrostatic repulsion between the AuNPs that reduced the likelihood of aggregation, and thus improved the stability of the AuNP dispersion.39 Specifically, the AuNPs generated with 1000 μL of the 10 mM NH2OH solution exhibited a zeta potential of −37.3 ± 0.35 mV, less negative than the zeta potential of −43.8 ± 1.10 mV observed for the AuNPs synthesized with 2000 μL. Despite the reduced zeta potential, the AuNPs produced with 1000 μL of the 10 mM NH2OH solution displayed a smaller diameter (88 ± 12 nm) than those prepared with 2000 μL (94 ± 14 nm). Given the importance of maximizing specific surface area alongside maintaining colloidal stability for the efficient and sensitive detection of S. typhimurium, the AuNPs synthesized with 1000 μL of the 10 mM NH2OH solution were selected for further exploration due to their smaller size characteristics and acceptable stability. HRTEM analysis further confirmed the crystalline nature of the AuNPs, with interplanar spacings of 0.22 nm aligning with the (111) lattice planes of face-centered cubic Au (Fig. 4c).51
The conjugation process of the report probes onto the AuNPs was refined by assessing the impact of the report probe concentration and reaction duration on the grafting density of the report probes on the AuNPs (Fig. 4g and h). For reaction duration shorter than 8 h, the grafting density was positively correlated with increased reporter probe concentration, indicating an undersaturation of reporter probes on the AuNP surface. However, when the reaction duration was prolonged to 8 h or beyond, the grafting density continued to rise but reached a steady state at a report probe concentration of 0.93 ng μL−1, indicating that the maximal surface capacity of the AuNPs had been saturated. Based on these observations, an optimal protocol of 0.93 ng μL−1 report probes and 8 h of reaction duration was implemented, which was found to achieve a grafting density of 201 report probes per AuNP, enhancing the sensitivity and detection range for S. typhimurium DNA. Calculations supporting these findings were provided in the ESI.†
3.3. ICP-MS analysis for the detection of S. typhimurium
The capture probe-grafted MNPs play a crucial role in the formation and isolation of the sandwich-structure complexes. To verify this aspect, a control experiment was designed, where 45 μL of TE buffer and 45 μL of the report probe-grafted AuNPs (2.5 ng μL−1) were incubated with 10 μL of S. typhimurium DNA (105 copies mL−1). The absence of the capture probe-grafted MNPs in this configuration prevented the recovery of the report probe-grafted AuNPs that bound to S. typhimurium DNA, resulting in a negligible detection signal (only 15.7% of that of the blank control where the capture probe-grafted MNPs were incubated with report probe-grafted AuNPs in the absence of S. typhimurium DNA, Fig. S7, ESI†).
To verify whether the size and number of sandwich-structure complexes affect the assay accuracy, 45 μL of different concentrations (0.25, 0.5, 1, 2, and 3 ng μL−1) of the capture probe-grafted MNPs were incubated with 45 μL of the report probe-grafted AuNPs (2.5 ng μL−1) and 10 μL of S. typhimurium DNA (105 copies mL−1). As shown in Fig. S8b (ESI†), with increasing mass ratios of MNPs to AuNPs, there was no significant change in the detection signals as tested by ICP-MS (RSD = 0.38%). This suggests that the size and number of the sandwich-structure complexes do not affect the assay sensitivity, given that sufficient capture probe-grafted MNPs and report probe-grafted AuNPs are present for S. typhimurium DNA binding.
To investigate the formation of the sandwich-structure complexes, we employed atomic force microscopy (AFM) and dynamic light scattering (DLS) for characterization. The AFM images (Fig. S8c, ESI†) confirmed the formation of the sandwich-structure complexes involving the capture probe-grafted MNPs, the report probe-grafted AuNPs, and S. typhimurium. Further analysis using DLS (Fig. S8d, ESI†) supports this finding, with the capture probe-grafted MNPs and report probe-grafted AuNPs displaying particle sizes of 67 ± 16 nm and 84 ± 17 nm, respectively. The addition of both types of nanoparticles in the absence of S. typhimurium did not result in a significant increase in the particle size but broadened the size distribution. However, upon exposure of these nanoparticles to S. typhimurium DNA, a primary peak at 165 ± 72 nm was observed, confirming the formation of MNPs-S. typhimurium-AuNPs complexes.
To assess the repeatability of our detection system, five samples, each containing 105 copies mL−1 of S. typhimurium DNA, were analyzed. The resulting signals were highly consistent across all samples (Fig. 6d), confirming the good repeatability. To evaluate the potential application, S. typhimurium DNA in biological samples such as FBS, urine, SBF, Mizone® drink, and TE buffer was detected, respectively. The results in Fig. 6e and Table 1 indicate that our strategy yielded recovery of 96.8% to 102.8%, with RSDs ranging from 0.75% to 1.61% across all tested biological samples. Comparative analysis with existing detection protocols for S. typhimurium DNA (Table S3, ESI†), revealed that both the recovery and RSDs achieved with our assay were superior to previously reported methods.
| Biological samples | Added Salmonella typhimurium (log10 CFU mL−1) | Detected Salmonella typhimurium (log10 CFU mL−1) | Recovery (%) | RSD (%, n = 3) |
|---|---|---|---|---|
| FBS | 5 | 4.74 | 96.8 | 1.61 |
| 4.93 | ||||
| 4.88 | ||||
| Urine | 5 | 5.18 | 102.8 | 0.90 |
| 5.09 | ||||
| 5.19 | ||||
| SBF | 5 | 4.96 | 99.2 | 1.54 |
| 5.10 | ||||
| 4.92 | ||||
| Mizone® Drink | 5 | 4.97 | 101.0 | 1.43 |
| 5.02 | ||||
| 5.14 | ||||
| TE buffer | 5 | 4.94 | 100.0 | 0.75 |
| 5.00 | ||||
| 5.03 |
Moreover, our method outperforms previously reported methods for detecting S. typhimurium, as shown in Table S3 (ESI†). It achieves a broader linear detection and an outstanding low LOD. Such excellent performance can be largely attributed to the popcorn-shaped AuNPs and the clustered MNP. These structures provide a high specific surface area which facilitates rapid and stable conjugation of a large number of oligonucleotide probes, thereby enhancing the capture efficiency of S. typhimurium.
4. Conclusion
We have successfully engineered MNPs and AuNPs of unique morphology, followed by functionalizing with selective probes to bind with S. typhimurium DNA. Special sandwich-structured complexes were formed during the detection. The MNPs confer the benefit of easy magnetic separation of the composites, and the subsequent ICP-MS analysis of Au element results in a substantial amplification of detection signals. Thus, the accuracy and sensitivity were simultaneously improved, offering a straightforward and rapid operational protocol for pathogen detection.Author contributions
Yujie Zhou: investigation, data curation, formal analysis, writing – original draft; Zhihui Tang: investigation, data curation, formal analysis, writing – review & editing; Lei Li: investigation, formal analysis, validation, writing – original draft; Yuzuo Chen: investigation, formal analysis; Yuanyuan Xu: investigation, formal analysis; Renjie Liu: investigation, formal analysis; Yanrong Zhang: investigation, formal analysis; Xiaoyan Liu: resources, supervision; Wenjuan Yang: resources, supervision; Baoning Wang: conceptualization, resources, supervision; Jieyu Zhang: conceptualization, resources, supervision, writing – review & editing; Qing Jiang: conceptualization, resources, supervision, writing – review & editing; Yunbing Wang: conceptualization, resources, supervision.Data availability
The data supporting this article have been included within the article and the ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
We gratefully acknowledge the financial support from the Key Research and Development Program of Sichuan Province (2022YFS0525 and 2023YFS0012), the Major Science and Technology Project of Sichuan Province (2022ZDZX0032), the West China Nursing Discipline Development Special Fund Project of Sichuan University (HXHL21005) and the Natural Science Foundation of Sichuan Province (2022NSFSC0819).References
- A. Prasad, S. Khan, J. K. Monteiro, J. Li, F. Arshad, L. Ladouceur, L. Tian, A. Shakeri, C. D. M. Filipe, Y. Li and T. F. Didar, Adv. Mater., 2023, 35, 2302641 CrossRef CAS PubMed.
- S. Wei, Z. Su, X. Bu, X. Shi, B. Pang, L. Zhang, J. Li and C. Zhao, npj Sci. Food, 2022, 6, 48 CrossRef PubMed.
- J. Li, S. Khan, J. Gu, C. D. M. Filipe, T. F. Didar and Y. Li, Angew. Chem., Int. Ed., 2023, 62, e202300828 CrossRef CAS PubMed.
- L. Chui, C. Ferrato, V. Li and S. Christianson, Microorganisms, 2021, 9, 955 CrossRef CAS PubMed.
- M. G. Aruta, E. Lari, D. De Simone, B. Semplici, C. Semplici, H. Dale, E. Chirwa, I. Kadwala, M. Mbewe, H. Banda, M. Iturriza-Gomara, M. Gordon, T. Nyirenda, P. Piu, M. Pizza, F. Berlanda Scorza, S. Grappi, R. Canals, O. Rossi and B. O. T. V. On, Biotech, 2023, 12, 54 CAS.
- M. Arnold, R. P. Smith, F. Martelli and R. Davies, Zoonoses Public Health, 2024, 00, 1–12 Search PubMed.
- Y. Ding, Z. Li, C. Huang, Y. Zhang, J. Wang and X. Wang, J. Food Saf., 2023, 43, e13091 CrossRef CAS.
- R. Heymans, A. Vila, C. A. M. van Heerwaarden, C. C. C. Jansen, G. A. A. Castelijn, M. van der Voort and E. G. Biesta-Peters, PLoS One, 2018, 13, e0206316 CrossRef PubMed.
- J. Gao, Y. Jiao, J. Zhou and H. Zhang, Talanta, 2024, 270, 125553 CrossRef CAS PubMed.
- E. Kim, S. Yang and H. Kim, Food Biosci., 2023, 56, 103242 CrossRef CAS.
- K. G. Maciorowski, P. Herrera, F. T. Jones, S. D. Pillai and S. C. Ricke, Vet. Res. Commun., 2006, 30, 127–137 CrossRef CAS PubMed.
- H. Ahari, S. Kakoolaki and S. A. A. Anvar, Int. J. Environ. Sci. Technol., 2017, 14, 2149–2154 CrossRef CAS.
- J. G. Bruno, J. Microbiol. Methods, 2022, 193, 106406 CrossRef CAS PubMed.
- Z. Guan, Y. Sun, C. Ma, J. J. Lee, S. Zhang, X. Zhang, Z. Guo and Y. Du, Biosens. Bioelectron., 2023, 236, 115438 CrossRef CAS PubMed.
- J. Zhao, M. Han, A. Ma, F. Jiang, R. Chen, Y. Dong, X. Wang, S. Ruan and Y. Chen, J. Hazard. Mater., 2024, 466, 133648 CrossRef CAS PubMed.
- H. Li, H. Xu, S. Yao, S. Wei, X. Shi, C. Zhao, J. Li and J. Wang, Talanta, 2024, 270, 125505 CrossRef CAS PubMed.
- F. Zhao, H. Yan, Y. Zheng, Y. Zu, S. Yang, H. Hu, S. Shi, H. Liang and X. Niu, Food Chem., 2023, 426, 136581 CrossRef CAS PubMed.
- L. Li, Q. Li, Z. Liao, Y. Sun, Q. Cheng, Y. Song, E. Song and W. Tan, Anal. Chem., 2018, 90, 9621–9628 CrossRef CAS PubMed.
- J. Ren, G. Liang, Y. Man, A. Li, X. Jin, Q. Liu and L. Pan, PLoS One, 2019, 14, e0218325 CrossRef CAS PubMed.
- D. Vijian, S. V. Chinni, L. S. Yin, B. Lertanantawong and W. Surareungchai, Biosens. Bioelectron., 2016, 77, 805–811 CrossRef CAS PubMed.
- P. Murasova, A. Kovarova, J. Kasparova, I. Brozkova, A. Hamiot, J. Pekarkova, B. Dupuy, J. Drbohlavova, Z. Bilkova and L. Korecka, J. Electroanal. Chem., 2020, 863, 114051 CrossRef CAS.
- Y. Zhang, F. Ren, G. Wang, T. Liao, Y. Hao and H. Zhang, Sens. Actuators, B, 2021, 329, 129273 CrossRef CAS.
- S. Tu, M. Golden, W. F. Fett, A. Gehring and P. Irwin, J. Food Saf., 2003, 23, 75–89 CrossRef.
- B. Meermann and V. Nischwitz, J. Anal. At. Spectrom., 2018, 33, 1432–1468 RSC.
- X. Wang, W. Chen, H. Yang, X. Zhang, M. Deng, X. Zhou, K. Huang, P. Chen and B. Ying, Microchim. Acta, 2020, 187, 453 CrossRef CAS PubMed.
- X. Huang, Z. Zhang, L. Chen, Y. Lin, R. Zeng, J. Xu, S. Chen, J. Zhang, H. Cai, H. Zhou and P. Sun, Biosens. Bioelectron., 2022, 212, 114414 CrossRef CAS PubMed.
- S. Yao, B. Pang, Y. Fu, X. Song, K. Xu, J. Li, J. Wang and C. Zhao, Sens. Actuators, B, 2022, 359, 131581 CrossRef CAS.
- X. Xu, J. Chen, B. Li, L. Tang and J. Jiang, Analyst, 2019, 144, 1725–1730 RSC.
- Y. Lin and A. T. Hamme II, J. Mater. Chem. B, 2015, 3, 3573–3582 RSC.
- W. Xiuji, J. Lanlan, G. Wei, H. Liuqin and S. H. And, At. Spectrosc., 2017, 38, 44–50 CrossRef.
- S. Crestani, A. Leitolis, L. F. O. Lima, M. A. Krieger and L. Foti, J. Immunol. Methods, 2016, 435, 17–26 CrossRef CAS PubMed.
- N. Sun, Y. Ding, Z. Tao, H. You, X. Hua and M. Wang, Food Chem., 2018, 257, 289–294 CrossRef CAS PubMed.
- K. Kang and H. Jang, Bull. Korean Chem. Soc., 2016, 37, 1391–1392 CrossRef CAS.
- T. Chen, Q. Chu, M. Li, G. Han and X. Li, J. Nanobiotechnol., 2021, 19, 206 CrossRef CAS PubMed.
- Q. Xu, X. Guo, L. Xu, Y. Ying, Y. Wu, Y. Wen and H. Yang, Sens. Actuators, B, 2017, 241, 1008–1013 CrossRef CAS.
- S. Xuan, F. Wang, Y. J. Wang, J. C. Yu and K. C. Leung, J. Mater. Chem., 2010, 20, 5086–5094 RSC.
- M. Esmaeilpour, A. R. Sardarian and J. Javidi, Appl. Catal., 2012, 445–446, 359–367 CrossRef CAS.
- N. Xu, H. Yan, X. Jiao, L. Jiang, R. Zhang, J. Wang, Z. Liu, Z. Liu, Y. Gu, F. Gang, X. Wang, L. Zhao and X. Sun, J. Cryst. Growth, 2020, 547, 125780 CrossRef CAS.
- T. Jesionowski, F. Ciesielczyk and A. Krysztafkiewicz, Mater. Chem. Phys., 2010, 119, 65–74 CrossRef CAS.
- S. Xuan, L. Hao, W. Jiang, X. Gong, Y. Hu and Z. Chen, J. Magn. Magn. Mater., 2007, 308, 210–213 CrossRef CAS.
- Y. Zhang, S. Wang, S. Shen and B. Zhao, Chem. Eng. J., 2013, 233, 258–264 CrossRef CAS.
- D. Li, W. Y. Teoh, J. J. Gooding, C. Selomulya and R. Amal, Adv. Funct. Mater., 2010, 20, 1767–1777 CrossRef CAS.
- Y. Liu, W. Luo, Q. Fan, H. Ma, Y. Yin and J. Guan, Adv. Funct. Mater., 2023, 33, 2303470 CrossRef CAS.
- D. Wan, G. Wang, W. Li and X. Wei, Appl. Surf. Sci., 2017, 413, 398–407 CrossRef CAS.
- Y. Dang, G. Wang, G. Su, Z. Lu, Y. Wang, T. Liu, X. Pu, X. Wang, C. Wu, C. Song, Q. Zhao, H. Rao and M. Sun, ACS Nano, 2022, 16, 4536–4550 CrossRef CAS PubMed.
- R. De Palma, S. Peeters, M. J. Van Bael, H. Van den Rul, K. Bonroy, W. Laureyn, J. Mullens, G. Borghs and G. Maes, Chem. Mater., 2007, 19, 1821–1831 CrossRef CAS.
- M. C. Stuparu and A. Khan, J. Polym. Sci., Part A: Polym. Chem., 2016, 54, 3057–3070 CrossRef CAS.
- L. Wu, S. Zhou, Y. Yun, L. Zhu, B. Li and W. Zhang, Sens. Actuators, B, 2022, 356, 131124 CrossRef CAS.
- Q. Sun, X. Ma, Y. Lu, S. Wang and H. Zhong, Powder Technol., 2021, 380, 421–429 CrossRef CAS.
- X. Liu, S. Zhang, Z. Cheng, X. Wei, T. Yang, Y. Yu, M. Chen and J. Wang, Anal. Chem., 2018, 90, 12116–12122 CrossRef CAS PubMed.
- Y. Zhang, T. Wu, Q. Cui, Z. Qu, Y. Zhang, H. Ma and Q. Wei, Biosens. Bioelectron., 2023, 222, 114992 CrossRef CAS PubMed.
- S. A. Sufyan, B. van Devener, P. Perez and M. M. Nigra, ACS Appl. Mater. Interfaces, 2023, 15, 1210–1218 CrossRef CAS PubMed.
- Y. Xing, J. Han, X. Wu, D. T. Pierce and J. X. Zhao, Microchim. Acta, 2019, 187, 56 CrossRef PubMed.
- Y. Negishi, K. Nobusada and T. Tsukuda, J. Am. Chem. Soc., 2005, 127, 5261–5270 CrossRef CAS PubMed.
- J. Xing, Z. Xu and B. Deng, Polymers, 2018, 10, 83 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4tb00291a |
| This journal is © The Royal Society of Chemistry 2024 |