Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Zinc(II) complexes of 3-bromo-5-chloro-salicylaldehyde: characterization and biological activity†
Ariadni
Zianna
 *a,
Ellie
Vradi
b,
Antonios G.
Hatzidimitriou
a,
Stavros
Kalogiannis
b and
George
Psomas
*a,
Ellie
Vradi
b,
Antonios G.
Hatzidimitriou
a,
Stavros
Kalogiannis
b and
George
Psomas
 *a
*a
aLaboratory of Inorganic Chemistry, Department of Chemistry, Aristotle University of Thessaloniki, GR-54124 Thessaloniki, Greece. E-mail: ariadnezianna@gmail.com; gepsomas@chem.auth.gr
bDepartment of Nutritional Sciences and Dietetics, International Hellenic University, Sindos, GR-57400 Thessaloniki, Greece
First published on 7th November 2022
Abstract
Five neutral zinc(II) complexes of 3-bromo-5-chloro-salicylaldehyde (3-Br-5-Cl-saloH) were synthesized in the absence or presence of the nitrogen-donor co-ligands 2,2′-bipyridine (bipy), 1,10-phenanthroline (phen), 2,9-dimethyl-1,10-phenanthroline (neoc), or 2,2′-bipyridylamine (bipyam) and were characterized by various techniques. The obtained complexes were [Zn(3-Br-5-Cl-salo)2(H2O)2] (1), [Zn(3-Br-5-Cl-salo)2(bipy)] (2), [Zn(3-Br-5-Cl-salo)2(phen)] (3), [Zn(3-Br-5-Cl-salo)2(neoc)] (4) and [Zn(3-Br-5-Cl-salo)2(bipyam)] (5). The crystal structures of complexes 1 and 3 were determined by single-crystal X-ray crystallography. The interaction of the compounds with calf-thymus DNA takes place via intercalation. The compounds may moderately cleave pBR322 plasmid DNA at a concentration of 500 μM. The compounds may bind tightly and reversibly to serum albumins. The antioxidant activity of the compounds was examined towards 1,1-diphenyl-picrylhydrazyl and 2,2′-azinobis(3-ethylbenzothiazoline-6-sulfonic acid) radicals and H2O2. The antimicrobial potency of the compounds was investigated against Staphylococcus aureus ATCC 6538, Bacillus subtilis ATCC 6633, Escherichia coli NCTC 29212 and Xanthomonas campestris ATCC 1395.
1. Introduction
In recent years, there has been increasing interest in the design of new complexes suitable as drug candidates. In the human body, a drug interacts with DNA, which is a storage and carrier molecule of genetic information, RNA and proteins.1–4 Until recently, most compounds used as medicines were almost exclusively purely organic. At the beginning of the 20th century, the first arsenic organometallic complex for the treatment of syphilis (salvarsan) was applied. Since then, many other metal complexes were discovered, such as the Au complex agent Auranofin and the Pt-based anticancer drugs cisplatin, oxaliplatin and carboplatin. Despite their high activity in such diseases, little attention has been paid to their application as antimicrobial compounds.5Salicylaldehyde is a natural product with oily pale-yellow color and bitter almond odor and is an ingredient of a defensive secretion of some leaf beetle species.6 A number of substituted salicylaldehydes have been found to present significant antimicrobial properties against bacteria and yeasts, while some complexes could be used as potential therapeutic agents against Candida infections. In order to present high antimicrobial activity, substituents on the benzene ring are required, and halogenation often gives highly active compounds; however, their antimicrobial potency is not easily predictable and the mechanism of action of these agents still remains unknown. One possibility is that their antimicrobial activity is due to the formation of Schiff bases with amino groups of the microbial cells.7–9
Coordination of salicylaldehydes to metal ions may enhance their antimicrobial properties.1,10–12 Apart from their antimicrobial properties, salicylaldehydes and their metal complexes have the ability to interact with important biomolecules, such as DNA and RNA. For example, mixed-ligand Cu(II) complexes of 5-nitro-salicylaldehyde and 2-hydroxynaphthaldehyde act as artificial nucleases,1 a Cu(II) complex with 2-hydroxynaphthaldehyde presented interesting results regarding its DNA/RNA binding, cleavage and cytotoxicity13 and a series of mixed-ligand Cu(II) complexes of substituted salicylaldehydes and α-diimines exhibited antiproliferative properties.14
Among the plethora of metal ions, zinc is the second most common trace element in the human body and it is vital to the structure and function of circa 2800 macromolecules and 300 enzymes and is a component of 10% of human proteins. Zinc metallothioneins are metal-binding proteins that present high affinity to divalent trace metals. Metallothioneins can scavenge hydroxyl radicals 300 times higher than glutathione, so they protect biological structures and DNA from oxidative damage. Zinc is also a co-factor of the cytosolic and extracellular Zn/Cu SOD enzyme, which acts as an ROS scavenger by catalyzing the dismutation of O2 radicals into the less harmful O2 and H2O2. Zinc also plays an important role in innate and adaptive immunity and is very important for maintaining the membrane barrier structure and function.15 Furthermore, zinc complexes with biological activity are found in the literature. Zinc oxide and baby zinc are used for skin injuries and infections, and deadly diarrhea in Asian and African countries, respectively.16 Other zinc complexes have anticancer,17 antidiabetic,18 anticonvulsant,19 anti-inflammatory,20 antimicrobial21 and antioxidant22 properties.
Our research is focused on the synthesis, characterization and investigation of the biological profile of metal complexes with substituted salicylaldehydes. Our previous works concerned metal complexes of mono- (X-salo) and bis-substituted (3,5-diX-salo)2 salicylaldehydes.23–28 Continuing the search in this field, we have chosen 3-bromo-5-chloro-salicylaldehyde (3-Br-5-Cl-saloH, Fig. 1(A)), a ligand with two different substituents on the benzene ring (3-bromo and 5-chloro). A thorough search in the literature may reveal the existence of a copper(II)14 and two cobalt(II) complexes29,30 with 3-bromo-5-chloro-salicylaldehyde.
We have synthesized five novel Zn(II) complexes of 3-Br-5-Cl-saloH in the absence or presence of the α-diimines 2,2′-bipyridine (bipy), 1,10-phenanthroline (phen), 2,9-dimethyl-1,10-phenanthroline (neoc), or 2,2′-bipyridylamine (bipyam) (Fig. 1), formulated as [Zn(3-Br-5-Cl-salo)2(H2O)2] (1), [Zn(3-Br-5-Cl-salo)2(bipy)] (2), [Zn(3-Br-5-Cl-salo)2(phen)] (3), [Zn(3-Br-5-Cl-salo)2(neoc)] (4) and [Zn(3-Br-5-Cl-salo)2(bipyam)]. All complexes were characterized by physicochemical and spectroscopic (IR, UV–vis and 1H NMR) techniques and the crystal structures of complexes 1 and 3 were determined by single-crystal X-ray crystallography.
The interaction of the compounds with calf-thymus (CT) DNA was investigated by UV–vis spectroscopy, by viscosity measurements, and via their ability to displace ethidium bromide (EB) from the DNA–EB conjugate. The potential nuclease-like activity of the compounds was evaluated via the cleavage of supercoiled circular pBR322 plasmid DNA (pDNA) examined by agarose gel electrophoresis. The in vitro affinity of the complexes to bind to human serum albumin (HSA) and bovine serum albumin (BSA) was evaluated by fluorescence emission spectroscopy and the corresponding binding constants were determined. The antimicrobial activity of the compounds was examined against two Gram-positive microorganisms (Staphylococcus aureus ATCC 6538 (S. aureus) and Bacillus subtilis ATCC 6633 (B. subtilis)) and two Gram-negative microorganisms (Escherichia coli NCTC 29212 (E. coli) and Xanthomonas campestris ATCC 1395 (X. campestris)), and the minimum inhibitory concentration (MIC) was determined. The potential antioxidant activity of the compounds was evaluated by determining their ability to scavenge 1,1-diphenyl-picrylhydrazyl (DPPH) and 2,2′-azinobis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) free radicals and to reduce H2O2.
2 Experimental
2.1 Materials – instrumentation – physical measurements
All chemicals and solvents were reagent grade and were used as purchased from commercial sources: 3-Br-5-Cl-saloH, bipy, phen, neoc, bipyam, Zn(NO3)2·4H2O, CH3ONa, trisodium citrate, NaCl, BSA, HSA, CT DNA, EB, ABTS, K2S2O8, NaH2PO4, nordihydroguaiaretic acid (NDGA) and butylated hydroxytoluene (BHT) were purchased from Sigma-Aldrich Co.; 6-hydroxy-2,5,7,8-tetramethylchromane-2-carboxylic acid (Trolox) was purchased from J&K; DPPH was bought from TCI; L-ascorbic acid and all solvents were purchased from Chemlab.Infrared (IR) spectra (400–4000 cm−1) were recorded on a Nicolet FT-IR 6700 spectrometer with samples prepared as KBr pellets (abbreviations used: s = strong, m = medium, sm = strong-medium, w = weak). UV–visible (UV–vis) spectra were recorded as nujol mulls and in DMSO solutions at concentrations in the range 10−4– 5 × 10−3 M on a Hitachi U-2001 dual-beam spectrophotometer. C, H and N elemental analyses were performed on a PerkinElmer 240B elemental microanalyzer. Molecular conductivity measurements of 1 mM DMSO solution of the complexes were carried out with a Crison Basic 30 conductometer. Fluorescence spectra were recorded in solution on a Hitachi F-7000 fluorescence spectrophotometer. Viscosity experiments were carried out using an ALPHA L Fungilab rotational viscometer equipped with an 18 mL LCP spindle and the measurements were performed at 100 rpm. 1H NMR spectra were recorded on an Agilent 500/54 (500 MHz for 1H) spectrometer using DMSO-d6 as a solvent (abbreviations used: s = singlet, d = doublet, dd = double-doublet, br = broad, m = multiplet).
DNA stock solution was prepared by dilution of CT DNA with buffer (containing 150 mM NaCl and 15 mM trisodium citrate at pH 7.0) followed by stirring for 1 h, and kept at 4 °C for no longer than a week. The stock solution of CT DNA gave a ratio of UV absorbance at 260 and 280 nm (A260/A280) of ∼1.88, indicating that the DNA was sufficiently free of protein contamination.31 The DNA concentration per nucleotide was determined by UV absorbance at 260 nm after 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 dilution using ε = 6600 M−1 cm−1.32
20 dilution using ε = 6600 M−1 cm−1.32
2.2 Synthesis of the complexes
Complexes 1–5 were synthesized according to the published procedure.25 Complex 1 was prepared by the addition of a methanolic solution of 3-Br-5-Cl-saloH (1 mmol, 235 mg) deprotonated by CH3ONa (1 mmol, 54 mg) to a methanolic solution of Zn(NO3)2·4H2O (0.5 mmol, 130 mg) at room temperature. The reaction mixture was stirred for 1 h, filtered and left for slow evaporation at room temperature. For the mixed-ligand complexes 2–5, a methanolic solution of the corresponding α-diimine (0.5 mmol) was added to the above mixture and the reaction mixture was stirred further for 1 h, filtered and left for slow evaporation at room temperature. After a few days, yellow single-crystals of complexes 1 and 3 suitable for X-ray structure determination were collected and air-dried.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1308(m), v(C–O → Zn). UV–vis: as nujol mulls, λ (nm): 339, 421; in DMSO, λ (nm) (ε, M−1 cm−1): 338 (2600), 423 (6800). 1H NMR (DMSO-d6), δ (ppm): 9.45 (2H, s, H7 3-Br-5-Cl-salo), 7.71 (2H, d, H6 3-Br-5-Cl-salo), 7.52 (2H, dd, H4 3-Br-5-Cl-salo). ΛM (in 1 mM DMSO solution) = 6 mho cm2 mol−1.
O); 1308(m), v(C–O → Zn). UV–vis: as nujol mulls, λ (nm): 339, 421; in DMSO, λ (nm) (ε, M−1 cm−1): 338 (2600), 423 (6800). 1H NMR (DMSO-d6), δ (ppm): 9.45 (2H, s, H7 3-Br-5-Cl-salo), 7.71 (2H, d, H6 3-Br-5-Cl-salo), 7.52 (2H, dd, H4 3-Br-5-Cl-salo). ΛM (in 1 mM DMSO solution) = 6 mho cm2 mol−1.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1318(m) ν(C–O → Zn), 767(m) ρ(C–H)bipy; UV–vis: as nujol mulls, λ (nm): 310(sh), 422; in DMSO, λ (nm) (ε, M−1 cm−1): 309 (sh), 422 (5500). 1H NMR (DMSO-d6), δ (ppm): 9.55 (2H, s, H7 3-Br-5-Cl-salo), 8.72 (2H, H3- and H3′-bipy), 8.62 (2H, br, H4- and H4′-bipy), 8.25 (2H, m, H5- and H5′-bipy), 7.74 (2H, d, H6- and H6′-bipy), 7.62 (2H, d, H6 3-Br-5-Cl-salo), 7.38 (2H, dd, H4 3-Br-5-Cl-salo). ΛM (in 1 mM DMSO solution) = 12 mho cm2 mol−1.
O), 1318(m) ν(C–O → Zn), 767(m) ρ(C–H)bipy; UV–vis: as nujol mulls, λ (nm): 310(sh), 422; in DMSO, λ (nm) (ε, M−1 cm−1): 309 (sh), 422 (5500). 1H NMR (DMSO-d6), δ (ppm): 9.55 (2H, s, H7 3-Br-5-Cl-salo), 8.72 (2H, H3- and H3′-bipy), 8.62 (2H, br, H4- and H4′-bipy), 8.25 (2H, m, H5- and H5′-bipy), 7.74 (2H, d, H6- and H6′-bipy), 7.62 (2H, d, H6 3-Br-5-Cl-salo), 7.38 (2H, dd, H4 3-Br-5-Cl-salo). ΛM (in 1 mM DMSO solution) = 12 mho cm2 mol−1.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1318(m), ν(C–O → Zn); 724(m), ρ(C–H)phen. UV–vis: as nujol mulls, λ (nm): 325 (sh), 422; in DMSO, λ (nm) (ε, M−1 cm−1): 425 (5000). 1H NMR (DMSO-d6), δ (ppm): 9.58 (2H, s, H7 3-Br-5-Cl-salo), 9.04 (2H, br, H2- and H9-phen), 8.91 (2H, H4- and H7-phen), 8.28 (2H, s, H5- and H6-phen), 8.11 (2H, m, H3- and H8-phen), 7.61 (2H, br, H6 3-Br-5-Cl-salo), 7.34 (2H, m, H4 3-Br-5-Cl-salo). ΛM (in 1 mM DMSO solution) = 15 mho cm2 mol−1.
O); 1318(m), ν(C–O → Zn); 724(m), ρ(C–H)phen. UV–vis: as nujol mulls, λ (nm): 325 (sh), 422; in DMSO, λ (nm) (ε, M−1 cm−1): 425 (5000). 1H NMR (DMSO-d6), δ (ppm): 9.58 (2H, s, H7 3-Br-5-Cl-salo), 9.04 (2H, br, H2- and H9-phen), 8.91 (2H, H4- and H7-phen), 8.28 (2H, s, H5- and H6-phen), 8.11 (2H, m, H3- and H8-phen), 7.61 (2H, br, H6 3-Br-5-Cl-salo), 7.34 (2H, m, H4 3-Br-5-Cl-salo). ΛM (in 1 mM DMSO solution) = 15 mho cm2 mol−1.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1324(m), ν(C–O → Zn), 729 (sm), ρ(C–H)neoc. UV–vis: as nujol mulls, λ (nm): 329(sh), 418; in DMSO, λ (nm) (ε, M−1 cm−1): 329 (sh), 418 (11400). 1H NMR (DMSO-d6), δ (ppm): 9.46 (2H, s, H7 3-Br-5-Cl-salo), 8.77 (2H, d, H4- and H7-neoc), 8.32 (2H, s, H5- and H6-neoc), 7.68 (2H, br, H3- and H8-neoc), 7.60 (2H, br, H6 3-Br-5-Cl-salo), 7.44 (2H, br, H4 3-Br-5-Cl-salo), 3.00 (6H, CH3-neoc). ΛM (in 1 mM DMSO solution) = 10 mho cm2 mol−1.
O); 1324(m), ν(C–O → Zn), 729 (sm), ρ(C–H)neoc. UV–vis: as nujol mulls, λ (nm): 329(sh), 418; in DMSO, λ (nm) (ε, M−1 cm−1): 329 (sh), 418 (11400). 1H NMR (DMSO-d6), δ (ppm): 9.46 (2H, s, H7 3-Br-5-Cl-salo), 8.77 (2H, d, H4- and H7-neoc), 8.32 (2H, s, H5- and H6-neoc), 7.68 (2H, br, H3- and H8-neoc), 7.60 (2H, br, H6 3-Br-5-Cl-salo), 7.44 (2H, br, H4 3-Br-5-Cl-salo), 3.00 (6H, CH3-neoc). ΛM (in 1 mM DMSO solution) = 10 mho cm2 mol−1.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1596(s), ν(C
O); 1596(s), ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1315(s), ν(C–O → Zn); 767(m), ρ(C–H)bipyam. UV–vis: as nujol mulls, λ (nm): 309 (sh), 335, 424; in DMSO, λ (nm) (ε, M−1 cm−1): 309 (sh), 337 (7000), 425 (1250). 1H NMR (DMSO-d6), δ (ppm): 9.98 (1H, br s, N–H bipyam), 9.55 (2H, s, H7 3,5-diBr-salo), 8.35 (2H, d, H3- and H3′-bipyam), 7.74 (4H, m, H5-, H5′-, H6- and H6′-bipyam), 7.62 (2H, d, H6 3-Br-5-Cl-salo), 7.51 (2H, dd, H4 3-Br-5-Cl-salo), 6.96 (2H, br m, H4- and H4′-bipyam). ΛM (in 1 mM DMSO solution) = 10 mho cm2 mol−1.
N); 1315(s), ν(C–O → Zn); 767(m), ρ(C–H)bipyam. UV–vis: as nujol mulls, λ (nm): 309 (sh), 335, 424; in DMSO, λ (nm) (ε, M−1 cm−1): 309 (sh), 337 (7000), 425 (1250). 1H NMR (DMSO-d6), δ (ppm): 9.98 (1H, br s, N–H bipyam), 9.55 (2H, s, H7 3,5-diBr-salo), 8.35 (2H, d, H3- and H3′-bipyam), 7.74 (4H, m, H5-, H5′-, H6- and H6′-bipyam), 7.62 (2H, d, H6 3-Br-5-Cl-salo), 7.51 (2H, dd, H4 3-Br-5-Cl-salo), 6.96 (2H, br m, H4- and H4′-bipyam). ΛM (in 1 mM DMSO solution) = 10 mho cm2 mol−1.
2.3 X-ray crystal structure determination
Single-crystals of complexes 1 and 3 suitable for crystal structure analysis were obtained by slow evaporation of their mother liquids at RT. They were mounted at room temperature on a Bruker Kappa APEX2 diffractometer equipped with a Triumph monochromator using Mo Kα radiation. Unit cell dimensions were determined and refined from the angular settings of at least 163 high intensity reflections in the range 10 < θ < 20°. Intensity data were recorded using φ and ω-scans. Both crystals presented no decay during the data collection. The frames collected for each crystal were integrated with the Bruker SAINT Software package,33 using a narrow-frame algorithm. Data were corrected for absorption using a numerical method (SADABS) based on crystal dimensions.34 The structures were solved using the SUPERFLIP package,35 incorporated in Crystals. Data refinement (full-matrix least-squares methods on F2) and all subsequent calculations were carried out using the Crystals version 14.61 build 6236 program package.36 All non-hydrogen atoms were refined anisotropically. Hydrogen atoms were located by difference maps at their expected positions and refined using soft constraints. By the end of the refinement, they were positioned geometrically using riding constraints to bonded atoms. Crystallographic data for complexes 1 and 3 are presented in Table S1.†2.4 Study of the biological profile of the compounds
The biological activity (interaction with DNA and albumins and antioxidant and antimicrobial activities) of the compounds was evaluated in vitro after the compounds were dissolved in DMSO (1 mM), due to their low solubility in water. The studies were conducted in the presence of aqueous buffer solutions, where mixing of each solution never exceeded 5% DMSO (v/v) in the final solution. Control experiments were undertaken to assess the effect of DMSO on the data. Minimum or no changes were observed in the spectra of the SAs or CT DNA and appropriate corrections were performed, when needed.The interaction of the compounds with CT DNA was examined thoroughly by UV–vis spectroscopy and viscosity measurements, and via competitive studies with EB by fluorescence emission spectroscopy. The ability of the compounds to cleave pBR322 plasmid DNA was studied by agarose gel electrophoresis. The serum albumin-(BSA or HSA)-binding was studied through tryptophan fluorescence quenching experiments. The antioxidant activity of the compounds was evaluated by determining their ability to scavenge the ABTS and DPPH radicals and to reduce H2O2. Each experiment was performed in triplicate and the standard deviation of absorbance was <10% of the mean. The antimicrobial activity of the compounds was evaluated by MIC values towards two Gram-(−) and two Gram-(+) bacterial species. All the specific protocols and relevant equations involved in the in vitro study of the biological activity of the compounds are presented in the ESI (sections S1–S5†).
3 Results and discussion
3.1 Synthesis, general aspects and characterization of the complexes
Complex 1 was prepared in methanol from the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 ratio reaction of Zn(NO3)2·4H2O with 3-Br-5-Cl-salo− (deprotonated by CH3ONa) according to a previously published procedure.25 Furthermore, the reaction of methanolic solutions of Zn(NO3)2·4H2O with 3-Br-5-Cl-salo− in the presence of the α-diimines bipy, phen, neoc or bipyam in a 1
2 ratio reaction of Zn(NO3)2·4H2O with 3-Br-5-Cl-salo− (deprotonated by CH3ONa) according to a previously published procedure.25 Furthermore, the reaction of methanolic solutions of Zn(NO3)2·4H2O with 3-Br-5-Cl-salo− in the presence of the α-diimines bipy, phen, neoc or bipyam in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Zn2+)
1 (Zn2+)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) (3-Br-5-Cl-salo−)
(3-Br-5-Cl-salo−)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) (α-diimine) ratio led to the formation of complexes 2–5.
(α-diimine) ratio led to the formation of complexes 2–5.
All complexes were found soluble in DMF and DMSO but insoluble in most organic solvents, H2O and Et2O. They are non-electrolytes in DMSO solution, since the values of the ΛM of the complexes in 1 mM DMSO solution were found in the range 6–15 mho cm2 mol−1,37 and elemental analysis data showed a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 Zn(II)
2 Zn(II)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) (3-Br-5-Cl-salo−) composition. The crystal structures of complexes 1 and 3 were further determined by single-crystal X-ray diffraction analysis.
(3-Br-5-Cl-salo−) composition. The crystal structures of complexes 1 and 3 were further determined by single-crystal X-ray diffraction analysis.
The findings of the IR spectroscopy confirmed the deprotonation and suggested the bidentate binding mode of the 3-Br-5-Cl-salo− ligand and the co-existence of the α-diimines. Free 3-Br-5-Cl-saloH presents in the IR spectrum two peaks attributed to the stretching and bending vibrations of the phenolic OH, at ∼3200 cm−1 and 1400 cm−1, respectively. In the IR spectra of complexes 1–5, these two peaks disappear, due to the deprotonation and the coordination of the phenolato group of the ligand to the Zn ion. The peak of the aldehyde bond ν(HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) at ∼1653 cm−1 of free 3-Br-5-Cl-saloH is shifted towards 1625–1640 cm−1 in the IR spectra of complexes 1–5, indicating the coordination through the carbonyl oxygen. The bands due to the C–O stretching vibrations at 1280 cm−1 of free 3-Br-5-Cl-saloH are found in the IR spectra of complexes 1–5 at 1308–1324 cm−1, because of the coordination of the phenolato oxygen of the ligand. Finally, the presence of the α-diimines was confirmed by the characteristic bands assigned to the corresponding out-of-plane ρ(C–H) vibrations at 724–767 cm−1.38 The findings from the IR spectroscopy are in agreement with the determined X-ray crystal structures.
O) at ∼1653 cm−1 of free 3-Br-5-Cl-saloH is shifted towards 1625–1640 cm−1 in the IR spectra of complexes 1–5, indicating the coordination through the carbonyl oxygen. The bands due to the C–O stretching vibrations at 1280 cm−1 of free 3-Br-5-Cl-saloH are found in the IR spectra of complexes 1–5 at 1308–1324 cm−1, because of the coordination of the phenolato oxygen of the ligand. Finally, the presence of the α-diimines was confirmed by the characteristic bands assigned to the corresponding out-of-plane ρ(C–H) vibrations at 724–767 cm−1.38 The findings from the IR spectroscopy are in agreement with the determined X-ray crystal structures.
The UV–vis spectra of the complexes were recorded in the solid state as nujol mulls and in DMSO and buffer solutions (150 mM NaCl and 15 mM trisodium citrate at pH values 6–8, regulated by HCl solution). No significant changes (shift of the λmax or new peaks) were observed in the spectra in the solid and liquid states, which is evidence that the complexes retain their structures in the pH range 6–8.25
1H NMR spectroscopy has also been used to confirm the binding of the 3-Br-5-Cl-salo ligand and α-diimines and the stability of the complexes in solution. The deprotonation of the phenol hydrogen is inferred by the absence of the –OH signal, which appears as a broad peak at δ = 12 ppm in the 1H NMR spectrum of the free ligand. The 1H NMR spectra of the complexes give signals attributable to the aldehyde proton at δ ∼9.50 ppm. All sets of signals related to the presence of the ligands in the corresponding compounds are present; three for the 3-Br-5-Cl-salo− and four for the α-diimine ligands. All signals are slightly shifted downfield as expected upon binding to the zinc ion. Therefore, the 1H NMR spectra are consistent with the obtained structures of 1–5.
The 1H NMR spectra were also recorded for different time intervals (0, 24 and 72 h) and remained unchanged (representatively shown for complex 2 in Fig. S1†). The absence of an additional set of signals related to the dissociated ligands suggests that all complexes remain intact in solution. This fact in combination with the stability of UV–vis spectra in solution, the non-dissociation of the DMSO solutions and the formation of stable chelate rings upon coordination of the 3-Br-5-Cl-salo− ligands and the α-diimine co-ligands with zinc(II) may reveal that the complexes remain intact in solution during the time necessary for the reported biological studies.
3.2 Structures of the complexes
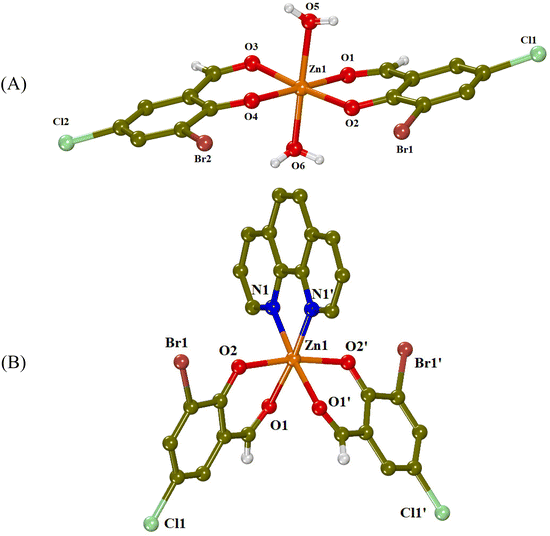 | ||
| Fig. 2 Crystal structures of (A) complex 1 and (B) complex 3 (symmetry code: (′) −x + 1, y, −z + 3/2). Aromatic hydrogen atoms are omitted for clarity. | ||
| Bond | Distance (Å) | Bond | Distance (Å) |
|---|---|---|---|
| Zn1–O1 | 2.095(3) | Zn1–O4 | 2.042(3) |
| Zn1–O2 | 2.019(3) | Zn1–O5 | 2.106(3) |
| Zn1–O3 | 2.067(3) | Zn1–O6 | 2.099(3) |
| Bonds | Angle (°) | Bonds | Angle (°) |
|---|---|---|---|
| O1–Zn1–O2 | 87.14(13) | O2–Zn1–O3 | 175.46(13) |
| O1–Zn1–O3 | 88.81(13) | O2–Zn1–O4 | 96.23(13) |
| O1–Zn1–O4 | 175.69(12) | O2–Zn1–O5 | 94.99(13) |
| O1–Zn1–O5 | 85.75(13) | O2–Zn1–O6 | 93.77(14) |
| O1–Zn1–O6 | 87.20(13) | O4–Zn1–O5 | 91.26(13) |
| O3–Zn1–O4 | 87.92(13) | O4–Zn1–O6 | 95.24(13) |
| O3–Zn1–O5 | 86.75(13) | O5–Zn1–O6 | 168.47(13) |
| O3–Zn1–O6 | 83.98(13) |
| Bond | Distance (Å) | Bond | Distance (Å) |
|---|---|---|---|
| Zn1–O1 | 2.153(3) | Zn1–N1 | 2.115(4) |
| Zn1–O2 | 2.036(3) |
| Bonds | Angle (°) | Bonds | Angle (°) |
|---|---|---|---|
| Symmetry code: (i) −x + 1, y, −z + 3/2. | |||
| O1–Zn1–O1i | 87.97(18) | O2–Zn1–O2i | 169.58(15) |
| O1–Zn1–O2 | 84.47(12) | O2–Zn1–N1 | 95.52(12) |
| O1–Zn1–O2i | 88.03(12) | O2–Zn1–N1i | 92.50(12) |
| O1–Zn1–N1 | 96.39(13) | N1–Zn1–N1i | 79.4(2) |
| O1–Zn1–N1i | 174.61(15) | ||
Complex 1 is a mononuclear zinc(II) complex (Fig. 2(A)), which consists of two deprotonated 3-Br-5-Cl-salo− ligands lying at the equatorial sites and two aqua ligands occupying the apical sites, completing thus the distorted octahedron around the Zn(II) ion (ZnO6 coordination sphere). The two 3-Br-5-Cl-salo− ligands are coordinated to zinc(II) in a bidentate chelating mode through their carbonyl oxygen (O1, O3) and their phenolato (O2, O4) oxygen atoms with the Zn–Ocarbonyl distances being slightly longer than the Zn–Ophenolato distances (Table 1) as expected and in accordance with the literature.25
The crystal structure of complex 1 is further stabilized by intermolecular hydrogen bonds between the aqua hydrogen atoms and phenolato oxygen atoms O2 and O4 of neighboring molecules (Table S2†).
In complex 3 (Fig. 2(B)), the Zn(II) ion has a ZnN2O4 coordination sphere, which consists of two deprotonated 3-Br-5-Cl-salo− ligands, coordinated in a bidentate chelating mode through their carbonyl (O1) and phenolato (O2) oxygen atoms, and a chelating phen ligand bound through its nitrogen atoms. The phenolato oxygen atoms are in trans positions to each other, with an O2i–Zn1–O2 angle of 169.58(15)° and the carbonyl oxygens are at cis positions, with an O1i–Zn1–O1 angle of 87.97(18)° (Table 2). This kind of arrangement is often found in [M(X–salo)2(α-diimine)] complexes.24
3.3 Study of the biological profile of the complexes
A number of different studies have been performed in order to screen the biological profile of 3-Br-5-Cl-saloH and its complexes 1–5. These studies may give insight into how the complexes interact with DNA and serum albumins and help investigate their antioxidant and antimicrobial properties.The study of the interaction of coordination complexes with DNA is important, since DNA is often a key target for the development of anticancer drugs and antibacterial agents, which owe their properties to the interaction with DNA and/or the induction of DNA cleavage.39 When a compound can interact with DNA, it should also be able to get transported to reach its potential target. Serum albumins usually play the role of transporting drugs and/or bioactive compounds towards the environment of the biological targets; therefore, it is of vital importance to study the interactions of bioactive compounds with such proteins. The investigation of the binding strength and the mechanism of interaction of small molecules with serum albumins is important for the clarification of the pharmacodynamics and pharmacokinetics, as they play an important role in drug absorption, distribution, metabolism and excretion. BSA is usually selected as a relevant model, because of its structural similarity to human serum albumin (76%), its low cost and easy availability.40 However, the report on plasma protein binding is now an FDA requirement in screening potential therapeutic agents, so the study of the interaction with HSA is also important.41
Coordination compounds may also serve as an alternative to traditional synthetic antioxidants, because of their variety in coordination number, different geometries and oxidation states, which facilitates and favors the redox processes associated with their antioxidant action.42 But this variety also makes coordination compounds very useful in accessing another highly underexplored chemical space for drug development, the design of new antimicrobial agents. Metal complexes have exhibited noteworthy antimicrobial activity as a result of synergism with antimicrobial ligands.7 Metal complexes could lead to exchange or release of ligands, redox activation, catalytic generation of reactive oxygen species (ROS) and depletion of essential substrates making them able to stop enzymatic activities, inhibit membrane function or induce DNA damage in microbes.5
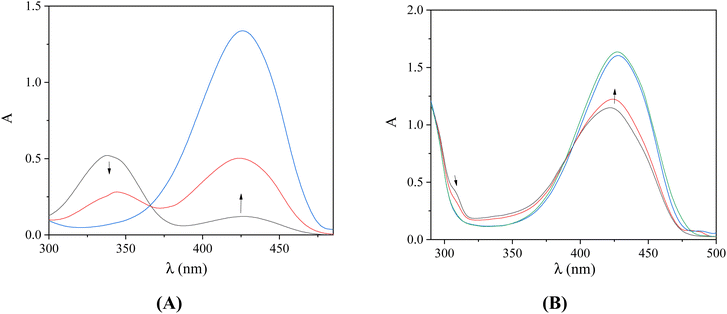
UV–vis spectroscopy is used initially to study the interaction between a compound and CT DNA and to estimate its binding strength, since any interaction between DNA and the compound may perturb the band(s) observed in the spectra.45 For this reason, the UV–vis spectra of the compounds (10−4 M) were recorded in the presence of increasing amounts of CT DNA (Fig. 3 and S2†) and the changes of the λmax were recorded. In the UV–vis spectra of the compounds, two bands are observed: band I located at around 309–340 nm and band II at 413–427 nm. Upon addition of CT DNA, medium-to-intense hypochromism is observed for band I accompanied by a red-shift, while band II exhibits only hyperchromism (Table 3). These changes may suggest the binding of the complexes to CT DNA,46 although sufficient evidence of the interaction mode with DNA cannot be obtained. For this purpose, DNA viscosity measurements and EB-competitive studies were conducted.
| Compound | Band (ΔA/A0 (%)a, Δλ (nm)b) | K b (M−1) |
|---|---|---|
| a “+” denotes hyperchromism and “−” denotes hypochromism. b “+” denotes red-shift and “−” denotes blue-shift. c “elim” denotes elimination of the band. | ||
| 3-Br-5-Cl-saloH | 338(<−50, elimc); 427(>+50, 0) | 1.13(±0.08) × 104 |
| Complex 1 | 338(−27, elim); 413(>+50, +10) | 1.10(±0.21) × 106 |
| Complex 2 | 309(<−50, elim); 422(+39, +5) | 6.74(±0.08) × 105 |
| Complex 3 | 325(<−50, elim); 422(+45, +5) | 7.16(±0.08) × 105 |
| Complex 4 | 329(<−20, 0); 418(+47, +10) | 1.18(±0.08) × 106 |
| Complex 5 | 337(−48, +13); 425(>+50, 0) | 4.31(±0.08) × 106 |
In order to estimate the binding strength of the compounds to CT-DNA, the DNA-binding constants (Kb) of the compounds were calculated with the Wolfe–Shimer equation (eqn (S1)†)47 and the plots [DNA]/(εA − εf) versus [DNA] (Fig. S3†). All complexes 1–5 are tighter DNA-binders than free 3-Br-5-Cl-saloH with complex 5 presenting the highest Kb constant (Table 3). The Kb constants of complexes 1–5 (Table 3) are relatively high (of the order 105–106 M−1) and higher than that of the classical intercalator EB (= 1.23(±0.07) × 105 M−1).48 The Kb constants are similar to other zinc(II)-salicylaldehyde complexes found in the literature.11,24,25
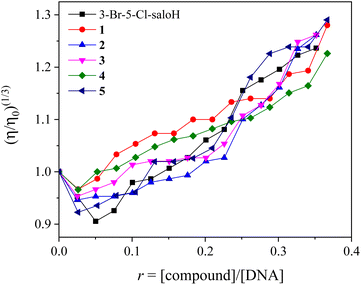
In order to further investigate the interaction of the compounds with CT DNA, the changes of the viscosity of a DNA solution in the presence of the compounds were monitored, since the viscosity of DNA is usually related to its length changes, induced by the interaction with a compound. An increase in DNA-viscosity may be interpreted as intercalation, since the distance between the DNA–base pairs gets longer, which leads to an increase in the relative DNA-length. In the case of interaction on the DNA surface (i.e., non-classical intercalation), the relative DNA-length does not change significantly and a negligible decrease in the DNA-viscosity may be observed.49
The DNA-viscosity measurements were carried out on CT DNA solutions (0.1 mM) by adding increasing amounts of the compounds (up to the value of r = 0.36) at room temperature. The relative viscosity of DNA exhibited an increase upon addition of the compounds (Fig. 4). In particular, all complexes exhibited a higher initial increase in their relative DNA-viscosity than the free 3-Br-5-Cl-saloH. The observed behavior of the DNA-viscosity may be considered evidence of the existence of an intercalative interaction mode with DNA clarifying thus the UV–vis spectroscopic features.
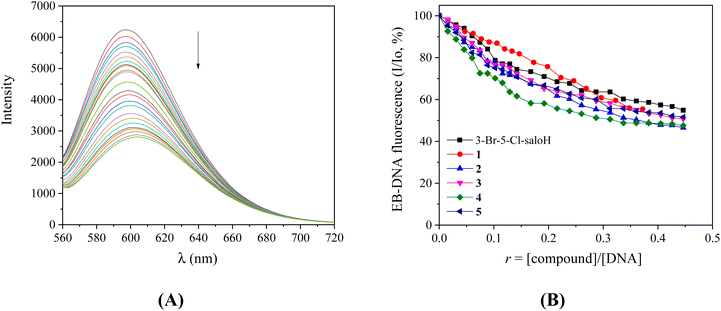
In order to further investigate the interaction of the compounds with CT DNA, competitive studies with the typical intercalator EB were carried out. A solution containing the EB–DNA conjugate shows an intense fluorescence emission band at 592 nm (with λexcitation = 540 nm) as a result of the intercalation of the planar EB-phenanthridine ring in-between adjacent DNA–base pairs. For this reason, EB is considered a typical indicator of DNA-intercalation.50 If a compound that intercalates to DNA equally or stronger than EB is added into this solution, significant quenching of the EB–DNA fluorescence emission may be observed. The compounds do not show any fluorescence emission bands at room temperature in solution or in the presence of CT DNA or EB under the same experimental conditions (λex = 540 nm).
The fluorescence emission spectra of a pre-treated solution containing the EB–DNA adduct ([EB] = 20 μM, [DNA] = 26 μM) were recorded in the presence of increasing amounts of each compound up to the value of r = 0.45 (representatively shown for complex 3 in Fig. 5(A)). The addition of the compounds resulted in a notable quenching of the fluorescence emission band at 592 nm (which was up to 53.4% for complex 2 of the initial EB–DNA fluorescence, Fig. 5(B) and Table 4). Therefore, the induced quenching may indicate the ability of the compounds to displace EB from the EB–DNA adduct, revealing indirectly the interaction with CT DNA by the intercalative mode.50,51
| Compound | ΔI/I0 (%) | K SV (M−1) | k q (M1 s−1) |
|---|---|---|---|
| 3-Br-5-Cl-saloH | 45.1 | 2.86(±0.07) × 104 | 1.24(±0.03) × 1013 |
| Complex 1 | 44.6 | 3.40(±0.10) × 104 | 1.48(±0.04) × 1012 |
| Complex 2 | 53.4 | 4.16(±0.06) × 104 | 1.81(±0.02) × 1012 |
| Complex 3 | 49.2 | 3.40(±0.06) × 104 | 1.43(±0.03) × 1012 |
| Complex 4 | 52.4 | 5.36(±0.18) × 104 | 2.33(±0.08) × 1012 |
| Complex 5 | 48.4 | 3.33(±0.08) × 104 | 1.45(±0.04) × 1012 |
The KSV constants (Table 4) of the compounds were calculated with the Stern–Volmer equation (eqn (S2)†)50 and the Stern–Volmer plots (Fig. S4†). The values of KSV were found relatively high, which is an indication of a tight binding of the complexes to DNA. Complex 4 exhibits the highest KSV constant among the complexes. The reported constants are of the same magnitude as those found for zinc complexes of salicylaldehydes.11,24,25 The EB–DNA quenching constants of the compounds (kq) were calculated with eqn (S3)† (considering τ0 = 23 ns as the fluorescence lifetime of the EB–DNA system).52 A piece of evidence that the quenching of the EB–DNA fluorescence takes place via a static mechanism can be deduced from the fact that the values of the kq constant of the compounds (Table 4) were found higher than 1010 M−1 s−1.51
From all the above experiments (UV-Vis titration studies, viscosity measurements and competitive studies with EB), it can be concluded that 3-Br-5-Cl-saloH and its complexes 1–5 interact with CT DNA via an intercalative mode and present relatively strong binding ability.
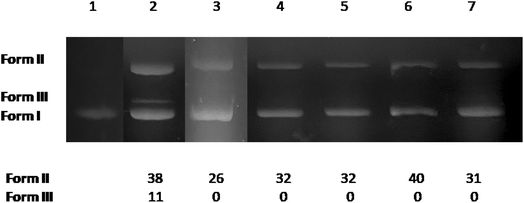
The potential DNA-cleaving activity by complexes 1–5 (Fig. 6, lanes 3–7) was revealed as single-stranded (ss) nicks in the supercoiled DNA forming relaxed circular DNA (form II), while an additional double-stranded (ds) nick was revealed for 3-Br-5-Cl-saloH (Fig. 6, lane 2) leading to the formation of linear DNA (form III). The percentages of ss and ds shown in Fig. 6 were calculated with eqn (S4) and (S5).† At a high concentration of 500 μM, the compounds present moderate DNA-cleaving activity up to 40% (Fig. 6, lane 6, for complex 4) which is slightly higher than their Zn(II) analogues with 3,5-dibromo-salicylaldehyde and 3,5-dichloro-salicylaldehyde.11,24
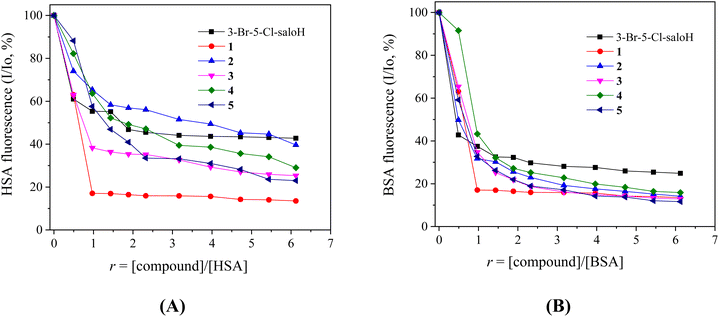
When the compounds (10−4 M) were added incrementally into a SA solution (3 μM), a significant quenching of the fluorescence emission bands of the albumins at λem = 340–345 nm was observed and the quenching induced by the compounds was less pronounced in the case of HSA (Fig. 7). The observed quenching can be attributed to the changes in the tryptophan environment of the albumins, due to the possible denaturation of their secondary structure, resulting from the binding of the compounds to SA.56
The SA-quenching constants (kq) of the compounds (Table 5) (calculated from the corresponding Stern–Volmer plots (Fig. S5 and S6†) and the Stern–Volmer quenching equations (eqn (S2) and (S3)†)) are much higher than 1010 M−1 s−1, which is an indication of a static quenching mechanism39 and verifies the interaction of the compounds with the albumins, with complexes 3 and 1 presenting the highest kq constants for BSA and HSA, respectively. The kq constants of complexes 1–5 are similar to those reported for similar Zn(II) with substituted salicylaldehydes as ligands.11,24,25
| Compound | k q(BSA) (M−1 s−1) | K (BSA) (M−1) | k q(HSA) (M−1 s−1) | K (HSA) (M−1) |
|---|---|---|---|---|
| 3-Br-5-Cl-saloH | 1.36(±0.10) × 1013 | 1.44(±0.07) × 106 | 1.83(±0.11) × 1013 | 1.26(±0.04) × 106 |
| Complex 1 | 3.97(±0.12) × 1013 | 3.49(±0.25) × 106 | 4.38(±0.15) × 1013 | 3.45(±0.17) × 106 |
| Complex 2 | 2.58(±0.11) × 1013 | 8.21(±0.18) × 105 | 6.55(±0.28) × 1012 | 4.62(±0.17) × 105 |
| Complex 3 | 5.85(±0.34) × 1013 | 1.02(±0.04) × 106 | 1.80(±0.10) × 1013 | 5.55(±0.13) × 105 |
| Complex 4 | 2.24(±0.08) × 1013 | 6.89(±0.20) × 105 | 1.21(±0.06) × 1013 | 2.87(±0.05) × 105 |
| Complex 5 | 3.54(±0.15) × 1013 | 8.64(±0.22) × 105 | 1.83(±0.09) × 1013 | 3.23(±0.13) × 105 |
The SA-binding constants (K) of the compounds (calculated from the corresponding Scatchard plots (Fig. S7 and S8†) and the Scatchard equation (eqn (S7)†)) (Table 5) are relatively high. This implies a tight interaction of the compounds with the albumins, which allows them to get transported towards the environment of their potential biological targets. The K values were found significantly lower than the constant of avidin (which is of the order 1015 M−1 and is regarded as the limit between the reversible and irreversible interactions), so a reversible interaction of the complexes with serum albumins may be assumed, and the complexes may get released when they approach close to the environment of their targets.57 Complex 1 exhibited the highest K constants for both albumins among the complexes.
The DPPH-radical assay was developed in the 1950s and has often been used to assess the antioxidant capacity of several metal complexes.42 It correlates to ageing prevention and anti-cancer and anti-inflammation activities.59 The ability to scavenge the cationic ABTS radicals (ABTS˙+) has often been used as a tool to measure the total antioxidant activity of a complex.48 Finally, H2O2, which is generated in vivo by several oxidase enzymes and activated by phagocytes, helps in killing several bacterial and fungal strains and may serve as a messenger molecule in the synthesis and activation of several inflammatory mediators. When a scavenger is incubated with H2O2, using a peroxidase assay system, the loss of H2O2 can be measured.60 For the above reasons, compounds able to scavenge free radicals or to inhibit their formation, may be found helpful in the treatment of inflammations.
Within this context, the antioxidant activity of the compounds was examined regarding the scavenging of DPPH and ABTS radicals and reduction of H2O2 and was compared with that of the well-known antioxidant agents NDGA, BHT, Trolox and L-ascorbic acid, which are used as reference antioxidant agents.61,62 The results are summarized in Table 6.
| Compound | DPPH%, 30 min | DPPH%, 60 min | ABTS% | H2O2% |
|---|---|---|---|---|
| 3-Br-5-Cl-saloH | 52.79 ± 1.13 | 53.64 ± 0.83 | 14.52 ± 0.46 | 80.86 ± 0.77 |
| Complex 1 | 55.25 ± 1.14 | 56.68 ± 1.77 | 27.50 ± 1.05 | 99.52 ± 0.43 |
| Complex 2 | 59.98 ± 0.16 | 61.76 ± 0.13 | 46.20 ± 0.12 | 85.06 ± 1.49 |
| Complex 3 | 53.83 ± 0.98 | 55.15 ± 1.34 | 72.58 ± 0.23 | 99.63 ± 0.10 |
| Complex 4 | 52.38 ± 0.65 | 52.77 ± 1.06 | 52.19 ± 1.04 | 95.09 ± 0.64 |
| Complex 5 | 54.99 ± 1.10 | 56.13 ± 1.30 | 59.06 ± 1.33 | 98.02 ± 0.25 |
| NDGA | 87.08 ± 0.12 | 87.47 ± 0.12 | Not tested | Not tested |
| BHT | 61.30 ± 1.16 | 79.78 ± 1.12 | Not tested | Not tested |
| Trolox | Not tested | Not tested | 98.10 ± 0.48 | Not tested |
| L-Ascorbic acid | Not tested | Not tested | Not tested | 60.80 ± 0.20 |
The DPPH-scavenging ability of the compounds was rather high and rather time-independent. The reported zinc(II) complexes were in most cases more active than 3-Br-5-Cl-saloH with complex 2 presenting the highest DPPH-scavenging activity, although lower than that of the reference compounds BHT and NDGA. All complexes were found to be more active ABTS-scavengers than free 3-Br-5-Cl-saloH, with complex 3 being the best ABTS-scavenger among the compounds. Concerning the reduction of H2O2, the complexes are more efficient than the free ligand and more than the reference compound L-ascorbic acid. Complexes 1 and 3 presented the highest reducing ability of H2O2.
In general, complexes 1–5 presented better antioxidant activity than free 3-Br-5-Cl-saloH, indicating that the coordination to zinc(II) ion improved the antioxidant ability of the free ligand. Compared with the analogous zinc(II) complexes of 3,5-diCl-saloH and 3,5-diBr-saloH, complexes 1–5 exhibited higher ability to scavenge DPPH and ABTS radicals and were found more active in reducing H2O2.11,24,25
In an effort to further clarify the nature of halogen-substitution on the benzene ring, the in vitro antimicrobial activity of the compounds was examined against two Gram-positive microorganisms (S. aureus and B. subtilis) and two Gram-negative microorganisms (E. coli and X. campestris), and the corresponding MIC values were determined. The results are found in Table 7.
| Compound | X. campestris | E. coli | B. subtilis | S. aureus |
|---|---|---|---|---|
| 3-Br-5-Cl-saloH | 100 (425) | 100 (425) | 100 (425) | 100 (425) |
| Complex 1 | 200 (351) | >200 (>351) | 50 (88) | 100 (175) |
| Complex 2 | 200 (290) | >200 (>290) | 50 (72) | 100 (144) |
| Complex 3 | 100 (140) | 50 (70) | 50 (70) | 50 (70) |
| Complex 4 | 200 (269) | 50 (67) | 25 (34) | 50 (67) |
| Complex 5 | 200 (284) | 100 (142) | 25 (35) | 50 (71) |
3-Br-5-Cl-saloH presented equal antimicrobial activity against all tested microorganisms (MIC = 100 μg mL−1) but lower than 3,5-dibromo-salicylaldehyde (MIC = 25–50 μg mL−1) and 3,5-dichloro-salicylaldehyde (MIC = 50 μg mL−1)11,24 against the same microorganisms suggesting that the combination of two different halogens leads to lower activity.
Complexes 1–5 presented better results against Gram(+) microorganisms and complexes 4 and 5 presented the best activity with regard to B. subtilis (MIC = 25 μg mL−1). Compared with their analogous zinc(II) complexes with 3,5-diCl-saloH or 3,5-diBr-saloH, complexes 1–5 presented lower antimicrobial activity in accordance with the corresponding saloH ligands,11,24 so as a premature conclusion, we could say that the use of two different halogens in the benzene ring did not seem to improve the antimicrobial properties of the complexes, but further studies need to be performed.
4. Conclusions
Five zinc(II) complexes of 3-Br-5-Cl-salicylaldehyde were prepared and characterized by various techniques. The crystal structures of complexes 1 and 3 were determined by single-crystal X-ray crystallography, revealing the bidentate coordination of the ligand to the metal ion. The complexes may interact with CT DNA via intercalation and the highest DNA-binding constant was found for complex 5 [Zn(3-Br-5-Cl-salo)2(bipyam)] (Kb = 4.31(±0.08) × 106 M−1). The complexes can moderately cleave pBR322 plasmid DNA to relaxed circular DNA (up to 40%) at a concentration of 500 μM. The complexes may bind tightly and reversibly to bovine and human serum albumin. Concerning the antioxidant activity of the compounds, all complexes 1–5 exhibited noteworthy scavenging ability of DPPH radicals (DPPH% = 52.38–61.76%), and significant ABTS-scavenging activity up to 72.58%. Almost all complexes 1–5 exhibited very high ability to reduce H2O2 (up to 99.63%), which constitutes a promising conclusion regarding their potential applications in the future. The antimicrobial activity of complexes 1–5 against two Gram-negative (X. campestris and E. coli) and two Gram-positive (B. subtilis and S. aureus) bacterial strains was tested. The activity of the complexes against the Gram(−) bacteria tested was low and the complexes were found to be more active against Gram(+) microorganisms; in particular, the best MIC values were determined for complexes 4 and 5 against B. subtilis.A plethora of different studies were employed to investigate the biological profile of 3-Br-5-Cl-saloH and its complexes 1–5. The tight intercalative binding of the complexes with CT DNA and the potential cleavage of plasmid DNA induced by the complexes in combination with the noteworthy antioxidant potency (scavenging of free radicals and reduction of hydrogen peroxide) may serve as the first indicative step towards anticancer activity. The reported antimicrobial activity may also be interconnected with DNA-binding and the DNA-cleavage properties of the complexes, as there are numerous commonly used antibacterial drugs (e.g. quinolones) that target bacterial DNA and/or the inhibition of enzymes related to the replication of DNA (e.g. topoisomerases and DNA-gyrases). In all cases, the role of serum albumins should also be taken into consideration for biological studies mainly due to their role as carriers of drugs and/or bioactive compounds towards the environment of their biological targets, although in some cases it is difficult for the albumins to approach too close to such targets (e.g. in the case of nuclear DNA).
The combination of different techniques may provide not only preliminary information of the biological potency of the compounds but also important evidence of how the complexes interact with DNA and serum albumins and of their antioxidant and antimicrobial properties, increasing future prospects for further studies of their biological properties with the employment of more elaborate experiments regarding their potential use as pharmaceutical agents.
Abbreviations
| 3-Br-5-Cl-saloH | 3-bromo-5-chloro-salicylaldehyde |
| ABTS | 2,2′-Azinobis(3-ethylbenzothiazoline-6-sulfonic acid |
| B. subtilis | Bacillus subtilis ATCC 6633 |
| bipy | 2,2′-Bipyridine |
| bipyam | 2,2′-Bipyridylamine |
| BSA | Bovine serum albumin |
| CT | Calf-thymus |
| DPPH | 1,1-Diphenyl-picrylhydrazyl |
| ds | Double-stranded |
| E. coli | Escherichia coli NCTC 29212 |
| EB | Ethidium bromide, 3,8-diamino-5-ethyl-6-phenyl-phenanthridinium bromide |
| HSA | Human serum albumin |
| K | SA-binding constant |
| K b | DNA-binding constant |
| k q | Quenching constant |
| K SV | Stern–Volmer constant |
| MIC | Minimum inhibitory concentration |
| NDGA | Nordihydroguaiaretic acid |
| Neoc | 2,9-Dimethyl-1,10-phenanthroline, neocuproine |
| pDNA | Supercoiled circular pBR322 plasmid DNA |
| phen | 1,10-Phenanthroline |
| r | [Compound]/[DNA] ratio or [compound]/[albumin] ratio |
| S. aureus | Staphylococcus aureus ATCC 6538 |
| SA | Serum albumin |
| ss | Single-stranded |
| Trolox | 6-Hydroxy-2,5,7,8-tetramethylchromane-2-carboxylic acid |
| X. campestris | Xanthomonas campestris ATCC 1395 |
Conflicts of interest
There are no conflicts to declare.Acknowledgements
AZ has received a postdoctoral scholarship for this research which is co-financed by Greece and the European Union (European Social Fund – ESF) through the Operational Programme “Human Resources Development, Education and Lifelong Learning” in the context of the project “Reinforcement of Postdoctoral Researchers – 2nd Cycle” (MIS–5033021), implemented by the State Scholarships Foundation (IKY).References
- A. Jayamani, R. Bellam, G. Gopu, S. O. Ojwach and N. Sengottuvelan, Polyhedron, 2018, 156, 138–149 CrossRef CAS.
- J. A. Lee, M. T. Uhlik, C. M. Moxham, D. Tomandl and D. J. Sall, J. Med. Chem., 2012, 55, 4527–4538 CrossRef CAS PubMed.
- C. H. Ng, K. C. Kong, S. T. Von, P. Balraj, P. Jensen, E. Thirthagiri, H. Hamada and M. Chikira, Dalton Trans., 2008, 4, 447–454 RSC.
- A. I. Matesanz, C. Hernandez, A. Rodriguez and P. Souza, Dalton Trans., 2011, 40, 5738–5745 RSC.
- M. Claudel, J. V. Schwarte and K. M. Fromm, Chemistry, 2020, 2, 849–899 CrossRef CAS.
- G. Pauls, T. Becker, P. Rahfeld, R. R. Gretscher, C. Paetz, J. Pasteels, S. H. von Reuss, A. Burse and W. Boland, J. Chem. Ecol., 2016, 42, 240–248 CrossRef CAS PubMed.
- E. Pelttari, E. Karhumaki, J. Langshaw, H. Perakyla and H. Elo, Z. Naturforsch., C: J. Biosci., 2007, 62, 487–497 CrossRef CAS.
- E. Pelttari, M. Lehtinen and H. Elo, Z. Naturforsch., C: J. Biosci., 2011, 66, 571–580 CrossRef CAS.
- H. Elo, M. Kuure and E. Pelttari, Eur. J. Med. Chem., 2015, 92, 750–753 CrossRef CAS.
- S. Ntanatsidis, S. Perontsis, S. Konstantopoulou, S. Kalogiannis, A. G. Hatzidimitriou, A. N. Papadopoulos and G. Psomas, J. Inorg. Biochem., 2022, 227, 111693 CrossRef CAS.
- A. Zianna, E. Geromichalou, G. Geromichalos, A.-M. Fiotaki, A. G. Hatzidimitriou, S. Kalogiannis and G. Psomas, J. Inorg. Biochem., 2022, 226, 111659 CrossRef CAS PubMed.
- A. Zianna, G. Geromichalos, A.-M. Fiotaki, A. G. Hatzidimitriou, S. Kalogiannis and G. Psomas, Pharmaceuticals, 2022, 15, 886 CrossRef CAS PubMed.
- S. Parveen, K. Fatima, S. Zehra and F. Arjmand, J. Biomol. Struct. Dyn., 2021, 39, 6070–6083 CrossRef CAS PubMed.
- N. Kordestani, H. A. Rudbari, A. R. Fernandes, L. R. Raposo, P. V. Baptista, D. Ferreira, G. Bruno, G. Bella, R. Scopelliti, J. D. Braun, D. E. Herbert and O. Blacque, ACS Comb. Sci., 2020, 22, 89–99 CrossRef CAS PubMed.
- M. Jarosz, M. Olbert, G. Wyszogrodzka, K. Młyniec and T. Librowski, Inflammopharmacology, 2017, 25, 11–24 CrossRef CAS PubMed.
- C. P. Larson, U. R. Saha and H. Nazrul, PLoS Med., 2009, 6, e1000175 CrossRef PubMed.
- M. Porchia, M. Pellei, F. Del Bello and C. Santini, Molecules, 2020, 25, 5814 CrossRef CAS.
- C. I. Chukwuma, S. S. Mashele, K. C. Eze, G. R. Matowane, S. Md. Islam, S. L. Bonnet, A. E. M. Noreljaleel and L. M. Ramorobi, Pharmacol. Res., 2020, 155, 104744 CrossRef CAS PubMed.
- J. d'Angelo, G. Morgant, N. E. Ghermani, D. Desmaele, B. Fraisse, F. Bonhomme, E. Dichi, M. Sghaier, Y. Li, Y. Journaux and J. R. J. Sorenson, Polyhedron, 2008, 27, 537–546 CrossRef.
- Q. Zhou, T. W. Hambley, B. J. Kennedy, P. A. Lay, P. Turner, B. Warwick, J. R. Biffin and H. L. Regtop, Inorg. Chem., 2000, 39, 3742–3748 CrossRef CAS PubMed.
- H. Ali, S. Omar, M. Darawsheh and H. Fares, J. Coord. Chem., 2016, 69, 1110–1122 CrossRef.
- C. Kakoulidou, S. Kalogiannis, P. Angaridis and G. Psomas, Polyhedron, 2019, 166, 98–108 CrossRef CAS.
- M. Sumar Ristovic, A. Zianna, G. Psomas, A. Hatzidimitriou, E. Coutouli-Argyropoulou and M. Lalia-Kantouri, Mater. Sci. Eng., C, 2016, 61, 579–590 CrossRef.
- A. Zianna, G. Geromichalos, E. Psoma, S. Kalogiannis, A. G. Hatzidimitriou and G. Psomas, J. Inorg. Biochem., 2022, 229, 111727 CrossRef CAS PubMed.
- A. Zianna, G. Psomas, A. Hatzidimitriou, E. Coutouli-Argyropoulou and M. Lalia-Kantouri, J. Inorg. Biochem., 2013, 127, 116–126 CrossRef CAS PubMed.
- A. Zianna, G. Psomas, A. Hatzidimitriou and M. Lalia-Kantouri, RSC Adv., 2015, 5, 37495–37511 RSC.
- A. Zianna, G. Psomas, A. Hatzidimitriou and M. Lalia-Kantouri, J. Inorg. Biochem., 2016, 163, 131–142 CrossRef CAS PubMed.
- A. Zianna, G. D. Geromichalos, A. G. Hatzidimitriou, E. Coutouli-Argyropoulou, M. Lalia-Kantouri and G. Psomas, J. Inorg. Biochem., 2019, 194, 85–96 CrossRef CAS PubMed.
- Y. J. Han, L. Wang, Q.-B. Li and L. W. Xue, Acta Chim. Slov., 2017, 64, 179–185 CrossRef CAS PubMed.
- Q. Wu, Y. Tang, L. Xu, Y. Lei, T. Liu, F. Jiang, Q. Zi and Y. Qiao, Z. Kristallogr. - New Cryst. Struct., 2018, 233, 967–968 CrossRef CAS.
- J. Marmur, J. Mol. Biol., 1961, 3, 208–211 CrossRef CAS.
- M. F. Reichmann, S. A. Rice, C. A. Thomas and P. Doty, J. Am. Chem. Soc., 1954, 76, 3047–3053 CrossRef CAS.
- Bruker Analytical X–ray Systems, Inc. Apex2, Version 2 User Manual, M86-E01078, Madison, WI, 2006 Search PubMed.
- Siemens Industrial Automation, Inc., SADABS: Area–Detector Absorption Correction, Madison, WI, 1996 Search PubMed.
- L. Palatinus and G. Chapuis, J. Appl. Crystallogr., 2007, 40, 786–790 CrossRef CAS.
- P. W. Betteridge, J. R. Carruthers, R. I. Cooper, K. Prout and D. J. Watkin, J. Appl. Crystallogr., 2003, 36, 1487 CrossRef CAS.
- W. J. Geary, Coord. Chem. Rev., 1971, 7, 81–122 CrossRef CAS.
- K. Nakamoto, Infrared and Raman Spectra of Inorganic and Coordination Compounds, Part B: Applications in Coordination, Organometallic, and Bioinorganic Chemistry, Wiley, New Jersey, sixth Ed., 2009 Search PubMed.
- N. Raman and S. Sobha, Spectrochim. Acta, Part A, 2012, 93, 250–259 CrossRef CAS PubMed.
- T. Topala, A. Bodoki, L. Oprean and R. Oprean, Clujul Med., 2014, 87, 215–219 Search PubMed.
- N. Shahabadi, S. M. Fili and S. Kashanian, J. Coord. Chem., 2018, 71, 329–341 CrossRef CAS.
- R. C. Marchi, I. A. S. Campos, V. T. Santana and R. M. Carlos, Coord. Chem. Rev., 2022, 451, 2142752 CrossRef.
- B. M. Zeglis, V. C. Pierre and J. K. Barton, Chem. Commun., 2007, 4565–4576 RSC.
- Q. Zhang, J. Liu, H. Chao, G. Xue and L. Ji, J. Inorg. Biochem., 2001, 83, 49–55 CrossRef CAS.
- G. Pratviel, J. Bernadou and B. Meunier, Adv. Inorg. Chem., 1998, 45, 251–262 CrossRef CAS.
- A. M. Pyle, J. P. Rehmann, R. Meshoyrer, C. V. Kumar, N. J. Turro and J. K. Barton, J. Am. Chem. Soc., 1989, 111, 3053–3063 Search PubMed.
- A. Wolfe, G. Shimer and T. Meehan, Biochemistry, 1987, 26, 6392–6396 CrossRef CAS PubMed.
- A. Dimitrakopoulou, C. Dendrinou-Samara, A. A. Pantazaki, M. Alexiou, E. Nordlander and D. P. Kessissoglou, J. Inorg. Biochem., 2008, 102, 618–628 CrossRef CAS PubMed.
- J. L. Garcia-Gimenez, M. Gonzalez-Alvarez, M. Liu-Gonzalez, B. Macias, J. Borras and G. Alzuet, J. Inorg. Biochem., 2009, 103, 923–934 CrossRef PubMed.
- J. R. Lakowicz, Principles of Fluorescence Spectroscopy, Plenum Press, NY, 3rd edn, 2006 Search PubMed.
- G. Zhao, H. Lin, S. Zhu, H. Sun and Y. Chen, J. Inorg. Biochem., 1998, 70, 219–226 CrossRef CAS PubMed.
- D. P. Heller and C. L. Greenstock, Biophys. Chem., 1994, 50, 305–312 CrossRef CAS.
- X. M. He and D. C. Carter, Nature, 1992, 358, 209–215 CrossRef CAS PubMed.
- C. Tan, J. Liu, H. Li, W. Zheng, S. Shi, L. Chen and L. Ji, J. Inorg. Biochem., 2008, 102, 347–358 CrossRef CAS PubMed.
- L. Stella, A. L. Capodilupo and M. Bietti, Chem. Commun., 2008, 4744–4746 RSC.
- V. Rajendiran, R. Karthik, M. Palaniandavar, H. Stoeckli-Evans, V. S. Periasamy, M. A. Akbarsha, B. S. Srinag and H. Krishnamurthy, Inorg. Chem., 2007, 46, 8208–8221 CrossRef CAS PubMed.
- O. H. Laitinen, V. P. Hytonen, H. R. Nordlund and M. S. Kulomaa, Cell. Mol. Life Sci., 2006, 63, 2992–3017 CrossRef CAS.
- R. Cini, G. Giorgi, A. Cinquantini, C. Rossi and M. Sabat, Inorg. Chem., 1990, 29, 5197–5200 CrossRef CAS.
- C. Kontogiorgis and D. Hadjipavlou-Litina, J. Enzyme Inhib. Med. Chem., 2003, 180, 63–69 CrossRef PubMed.
- G. K. Jayaprakasha, L. Rao and K. Sakariah, Bioorg. Med. Chem., 2004, 12, 5141–5146 CrossRef CAS PubMed.
- S. Dairi, M. Carbonneau, T. Galeano-Diaz, H. Remini, F. Dahmoune, O. Aoun, A. Belbahi, C. Lauret, J. Cristol and K. Madani, Food Chem., 2017, 237, 297–304 CrossRef CAS PubMed.
- C. Kontogiorgis, M. Ntella, L. Mpompou, F. Karallaki, A. Papadopoulos, D. Hadjipavlou-Litina and D. Lazari, J. Enzyme Inhib. Med. Chem., 2016, 31, 154–159 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC 2191080 and 2191081. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d2dt02404g |
| This journal is © The Royal Society of Chemistry 2022 |