Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Acidic polymers reversibly deactivate phages due to pH changes†
Huba L. Martona,
Antonia P. Sagona*c,
Peter Kilbrided and
Matthew I. Gibson *abef
*abef
aDepartment of Chemistry, University of Warwick, Coventry, CV4 7AL, UK. E-mail: matt.gibson@manchester.ac.uk; Fax: +44 247 652 4112
bWarwick Medical School, University of Warwick, Coventry, CV4 7AL, UK
cSchool of Life Sciences, University of Warwick, Coventry, CV4 7AL, UK. E-mail: A.Sagona@warwick.ac.uk
dAsymptote, Cytiva, Chivers Way, Cambridge, CB24 9BZ, UK
eDepartment of Chemistry, University of Manchester, Oxford Road, Manchester, M13 9PL, UK
fManchester Institute of Biotechnology, University of Manchester, 131 Princess Street, Manchester, M1 7DN, UK
First published on 23rd August 2024
Abstract
Bacteriophages are promising as therapeutics and biotechnological tools, but they also present a problem for routine and commercial bacterial cultures, where contamination must be avoided. Poly(carboxylic acids) have been reported to inhibit phages’ ability to infect their bacterial hosts and hence offer an exciting route to discover additives to prevent infection. Their mechanism and limitations have not been explored. Here, we report the role of pH in inactivating phages to determine if the polymers are unique or simply acidic. It is shown that lower pH (=3) triggered by either acidic polymers or similar changes in pH using HCl lead to inhibition. There is no inhibitory activity at higher pHs (in growth media). This was shown across a panel of phages and different molecular weights of commercial and controlled-radical polymerization-derived poly(acrylic acid)s. It is shown that poly(acrylic acid) leads to reversible deactivation of phage, but when the pH is adjusted using HCl alone the phage is irreversibly deactivated. Further experiments using metal binders ruled out ion depletion as the mode of action. These results show that polymeric phage inhibitors may work by unique mechanisms of action and that pH alone cannot explain the observed effects whilst also placing constraints on the practical utility of poly(acrylic acid).
Introduction
Biosynthetic pathway editing is used in bacteria to produce high-value chemicals and natural products.1 The broad uses of bacteria in food production range from dairy products, including cheese and yoghurt, to pickled products, such as sauerkraut, gherkins and olives, using lactic acid bacteria.1 Hence, for the use of bacteria in any application area, it is essential to exclude bacteriophages (phages – bacteria selective viruses), a common cause of infection leading to financial and scientific losses. Bacteriophages are amongst the most abundant organisms on earth and are present wherever their hosts are.2 Phages have potential as alternatives to antibiotics,3–5 for food safety,6 veterinary settings,7 and biotechnology for ligand selection.8–10Despite their wide biotechnological use, phage contamination in bacterial cultures leads to complete loss of the culture. It requires starting re-culturing from uninfected stocks, carrying significant cost implications for academic and industrial settings. For example, removing phages from raw materials in the food industry is almost impossible, often leading to process collapse.11–13 Present-day mitigation tools include good microbiology practices, working under aseptic conditions and vigorous cleaning or autoclaving. However, mitigation is not always successful as phages are robust and can survive in almost every condition.14 An alternative option is to engineer bacterial strains intrinsically resistant to phages using gene editing technology, but this is not trivial and may not suit all bacterial hosts.15 Re-engineering strains optimised for a particular bio-refinery or changing the refinery's processes is not always easy. Hence, a practical solution would be an anti-phage additive, comparable to how antibiotics are routinely used in mammalian cell cultures to prevent bacterial infection.16 Several studies explored the use of phages in treatment7,17,18 and ligand screening,9,10,19 but very few tools to inhibit them. In contrast, investigations of mammalian viruses led to the discovery of viral inhibitors20,21 and the successful re-purposing of existing inhibitors.22 The evolutionary phage prevention/reducing system bacteria acquired evolving with their phage predators, relying on protein components, restriction-modification, and CRISPR defences,23–25 are not easy to re-purpose as an anti-phage additive for biotechnological processes. Some studies reported phage-inhibiting molecules discovered in Streptomyces26 and some aminoglycoside antibiotics.27 Due to antimicrobial resistance (AMR) concerns, the latter is not desirable for large-scale biotechnological applications.
Sulphated polymers, which mimic heparin sulphate anchors on cell membranes and some modified cyclodextrins, have been recently shown to be broad-spectrum virucides against various human pathogenic viruses.28,29 Poly(carboxylic acids) have also been reported to inhibit human viruses.30–32 It is well established that polymers can also be deployed as anti-bacterial agents, mimicking cationic host-defence peptides.33–36 Mild acidic conditions have also been reported to inhibit mammalian viruses.37 Apart from the carboxylic acids, fatty acids and phenolic acids have been reported as mammalian virus inhibitors.38,39 We have previously reported that poly(carboxylic acids) can inhibit phage replication.40 However, the mode of action is still being determined, and these polymers’ limits and practical utility have yet to be explored.
Here, we report the further investigation of poly(acrylic acid) as a phage inhibitor, focusing on the role of pH. The phage is irreversibly inactivated after exposure to low pH (3) using HCl. In contrast the inactivation is fully reversible when using poly(acrylic acid) to obtain the same pH. This supports the hypothesis that the pH is crucial to inactivation but that the polymers may play a unique role. Depletion of divalent metal ions under the tested conditions was ruled out as a contributor using metal binding macrocycles. This supports the exploration of biomaterials to control phage infection but also highlights that the pH is (currently) the most significant contributor to function.
Results and discussion
Poly(acrylic acid) was previously reported to inhibit the infectivity of Escherichia coli targeting bacteriophage through a virustatic (reversible) mechanism without impacting host biotechnological processes.40 To further elucidate the inhibition mechanism and to compare in-house synthesised (using RAFT polymerisation) (full details in ESI†) versus commercially available PAA on inhibition, commercial poly(acrylic acid) (5000 g mol−1) was tested. The rationale is that other users might buy commercial polymers but these are supplied in both acid and basic forms: a distinction we show here to be crucial. Commercial PAA was purchased as a sodium salt form (Fig. 1), so upon direct dissolution in water, it gives a higher pH value than the same experiment using the acid form (which was synthesised in-house). In addition to the observations of the polyacids, we wanted to rule in/out a mechanism of inhibition for bacteriophages involving the sequestration of divalent ions, such as calcium (Ca2+), which are essential for the proliferation of bacteriophages.41–45 Calcium ions can increase the rate at which bacteriophages bind to their host,46 which may be replaced by other divalent ions such as magnesium (Mg2+) or manganese (Mn2+),45 due to phage adaptability. We hypothesised that the PAA (and poly(methacrylic acid)) could sequester one or more of these divalent ions, leading to the observed inhibitory activity. To address this, beta-cyclodextrins47–49 and crown ethers (18-crown-6 and 15-crown-5 ether)50,51 were included in our initial screening (Fig. 1) as model metal ion binders. Briefly, each compound was incubated with K1F-GFP and T4 bacteriophages in SM-II buffer with an additive concentration of 10, 15 or 20 mg mL−1 overnight to allow interaction between the two. The incubated solution was then added to a culture of E. coli EV36 or E. coli K-12 (K1F-GFP and T4 phage hosts) grown for 4 hours at 37 °C after seeding. Bacteriophage-infected liquid cultures were grown for 20 hours at 37 °C (24 hours total). Bacterial growth was measured by monitoring the increase in optical density at 600 nm (OD600). An increase in OD indicates bacterial growth and hence no phage activity, whilst viable (infectious) phages would inhibit the bacterial growth initially (at t = 5 hours) before rebounding during extended culture. The initial low-throughput assay (Fig. S1†) showed that the only bacteriophage inhibitors were the positive controls used (synthetic PAA 32 and PAA 372, numbers corresponding to degrees of polymerisation, full details in Table S1†). The metal chelators had no activity, and neither did the commercial PAA deployed as the sodium salt. Direct dissolution of commercial (Na salt) and home-made (acid form) PAA showed a pH of ∼8 and ∼3, respectively. This observation suggested that pH contributes to phage dissolution and was further explored.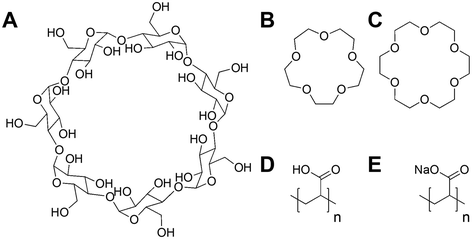 | ||
| Fig. 1 Compounds screened for phage inhibition. (A) β-Cyclodextrin; (B) 15-crown-5 ether; (C) 18-crown-6 ether; (D) RAFT synthesised poly(acrylic acid) acid form (full synthesis details in ESI†); (E) commercial poly(acrylic acid) sodium salt form. | ||
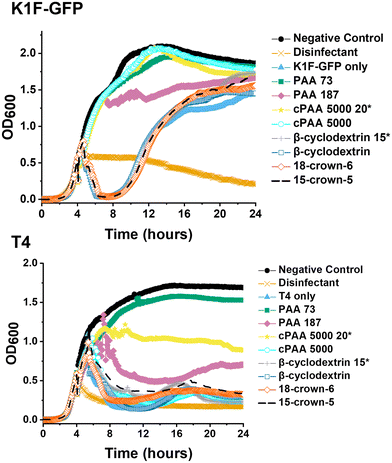
Commercial PAA aliquots were acidified with dilute HCl to pH 3. The final commercial PAA concentration of 10 mg mL−1 and 20 mg mL−1 matched the previously reported MIC of synthetic PAA for K1F-GFP and T4, respectively.40 Post-acidification, the same screening phage inhibition assay as in a previous report was used,40 including all tested compounds for comparative purposes (Fig. 2).
The data in Fig. 2 demonstrated that acidification of the commercial (sodium salt) PAA to pH 3 re-activated its phage inhibitory ability at both 20 mg mL−1 and 10 mg mL−1 for K1F-GFP with a notably increased potency at 20 mg mL−1 for T4, matching the performance of the in-house synthesised (acid form) PAA. This result confirms the hypothesis that poly(acids) phage-inhibitory activity is linked to pH, and when used directly as the sodium salt (as in the commercial sample), no activity is seen.
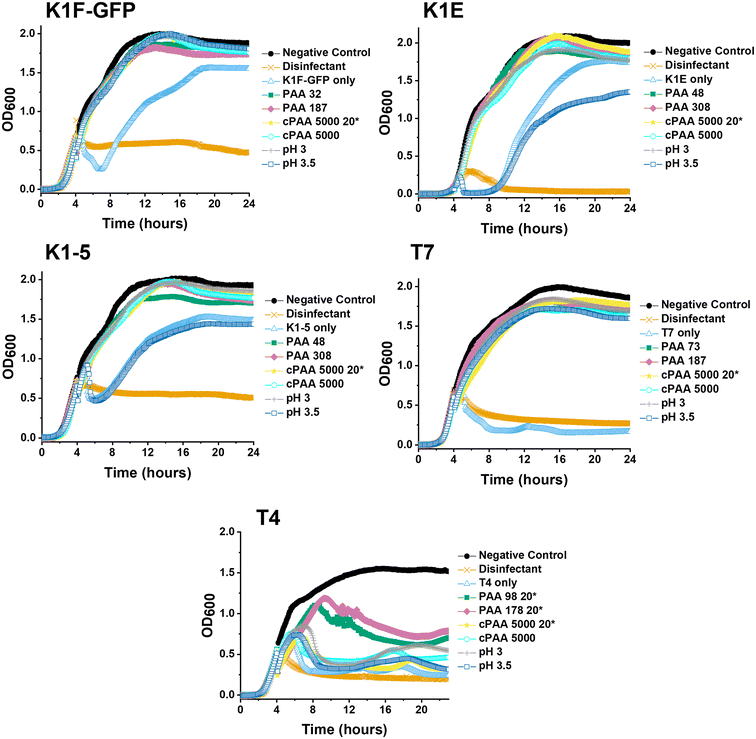
Guided by the above, a higher-throughput assay was carried out for five different E. coli targeting bacteriophages (K1F-GFP, K1E, K1-5, T7, and T4) to investigate our initial observations further. Commercial PAA sodium salt 5000 g mol−1 (cPAA) synthetic PAA 98, and PAA 178 (number represents the degree of polymerisation) were used at 10 mg mL−1 for all phages, except T4, where 20 mg mL−1 is needed to reach the MIC value and to avoid false negatives. The commercial PAAs were acidified to pH 3 before testing, and controls of acidified SM-II buffers to pH 3 and 3.5 were prepared to test the effects of low pH without PAA (Fig. 3). Spot tests (solid phase growth assay) for phage were also performed to quantify the impact further, using 1–6 10-fold dilution segmented plates of the corresponding E. coli host lawn (Fig. S2†). The high-throughput assay revealed phage infectivity inhibition for all the (pH = 3) polymer samples and the acidified SM-II buffer. These results were confirmed using the spot test data by the absence of phage plaques on the plates. This critical observation suggested low pH was vital to the phage inhibition mechanism.
The variable pH inhibition was investigated after confirming that acidity (low pH) assisted phage inhibition for the five bacteriophages. Before the growth curve assay, three separate series of dose–response conditions were performed: acidification of the SM-II buffer without PAA to pH 3, commercial PAA to pH 3 and NaOH basification of the synthetic PAA to pH 5.5. This dose–response showed that acidification of the commercial PAA (decreasing pH) leads to an ‘activation’ of the phage inhibition (Fig. S3†). The reverse of this trend also held, with basification (increasing pH) leading to the ‘deactivation’ of the otherwise potent 10 mg mL−1 synthetic PAA, which enabled bacterial growth, i.e. lack of phage inhibition (Fig. S3†). The minimum inhibitory pH values from the dose–response curves are summarised in Table 1. In all three conditions, a pH value below 3 inhibited all three tested phages (K1F-GFP, K1-5, and T7), regardless of the starting pH, before increasing or decreasing this.
The data above showed that an acidic environment was a vital aspect of the E. coli targeting phage inhibition mechanism with or without the presence of poly(acrylic acid). Low pH-assisted inhibition of mammalian viruses has been shown, with Janiga et al. reporting mild acidic pH inhibition of Herpes simplex virus (HSV).37 Damonte reported the inhibition of Junin virus (JUNV), an arenavirus, by fatty acids, including capric, lauric, myristic, palmitic, and stearic acids.38 Mechanistic studies concluded that lauric acid inhibited a late maturation stage in the replication cycle of JUNV rather than the inactivation of virions. In contrast, varied chain-length fatty acids had negligible anti-JUNV activity.38 Silva Jr. reported the in vitro inhibition of canine distemper virus (CDV) by phenolic acids (cis-cinnamic acid, trans-cinnamic acid, and ferulic acid).39 Elrod reported the inhibitory activity of medium-chain fatty acids, including caprylic, capric, and lauric acids, on the African swine fever virus (ASFv).52
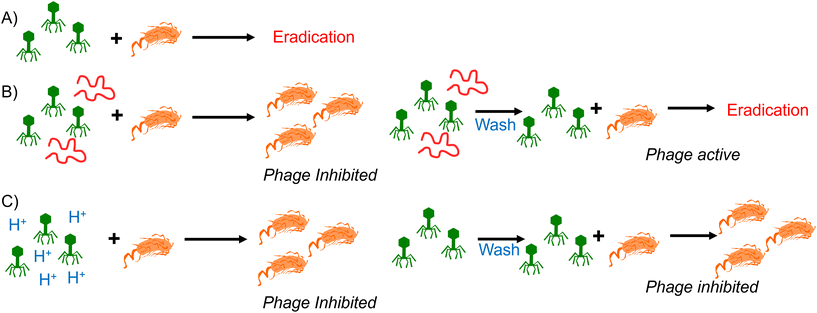
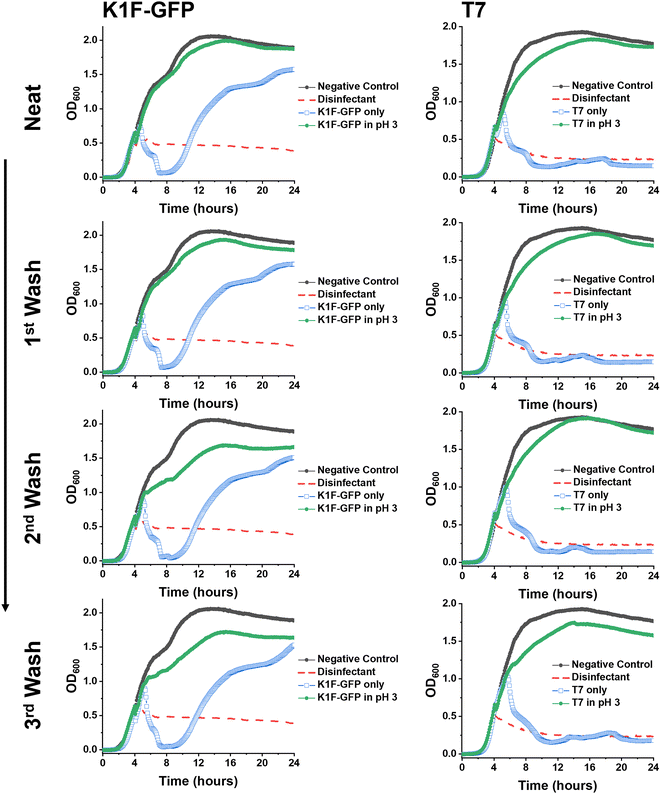
Synthetic PAA was previously reported to inhibit phage activity through a virustatic (reversible) mechanism.40 The low pH high-throughput assay and the pH dose–response data both suggested that below pH 3, the bacteriophages were inhibited, with and without the presence of PAA. Based on this, the next question was whether the mechanism of inhibition was simply due to a low pH or whether acidic PAA inhibited the phages differently from acidity alone. Hence, an experiment was devised to determine if the acidic (low pH) conditions permanently (i.e., virucidal) or transiently (i.e., virustatic) inhibit the phages, so that the data was comparable to washing out the polymers. Briefly, K1F-GFP and T7 phages were incubated with acidified SM-II to pH 3 (minimum inhibitory pH, so no subsequent infection would be expected) for 24 hours. After incubation, each acidified phage solution was diluted (washed out), which increased the pH above 3 (the inhibition threshold) before being added to the E. coli host. Up to three 10-fold dilutions, the pH 3 exposed bacteriophages were still unable to eradicate the bacteria, which confirmed a virucidal (irreversible) mechanism of inhibition at low pH conditions (Fig. 4). This was confirmed by the phage infectivity in the untreated phage controls applied at equal PFU per mL to account for the dilution. These contrast when PAA is used against the phages using the same experimental set up, which shows fully reversibly inhibitory activity.40
Taking into account the impact of pH on the inhibition of bacteriophages, two pH tracking assays were devised to measure the changes in pH at each stage of the growth curve assay and pH wash assay from incubated aliquot to addition to LB media. The difference between synthetic PAA and synthetic poly(methacrylic acid) (PMA) was compared in the first pH tracking assay, as PMA was reported as a weak phage inhibitor.40 Briefly, the pH was measured where a corresponding phage-free SM-II buffer replaced the phage-containing volume to mimic the former. The tracking assay showed that this led to an increase in the pH of PMA across dilutions (Table S1†) as would be expected. At 10 mg mL−1, the difference in pH between PMA/PAA was ∼0.3, which may explain the difference in inhibition potency due to the pKa differences between PAA and PMA. Diluting the PAA in LB media led to similar pHs compared to the pH solution alone, confirming that the final bacterial exposure conditions were identical (Table S2†).
This data confirms that the decrease in pH is essential for the anti-phage activity of poly(carboxylic acids) and that simply lowering pH is an effective tool to neutralise phage contamination. However, it also showed that phages that had been neutralised by poly(carboxylic acids) recovered their activity when the pH was returned to neutral. The reasons for this remain unclear, but an electrostatic interaction between the polymer and the phage surface could lead to a protective effect against the drop in pH and hence the poly(acrylic acid) serve both as source of low pH but mitigate some of the impact. Fig. 5 summarizes our observations of polymers at the same pH, reversibly inactivating phage, but the same pH without polymer is irreversible deactivation. It shows that polymers have distinct mechanisms and might give an opportunity to discover materials that are selective and more active for phage eradication.
Conclusions
Here, we report a comparison of the inhibition of bacteriophages by polymeric acids to the inhibition caused by simple modulation of pH. Poly(acrylic acid) in its acid form was first shown to be more active than its sodium salt, consistent with the pH playing a pivotal role. Acidification of commercial poly(acrylic acid) sodium salt to pH 3 re-activated the phage inhibitory mechanism. It was observed that if a phage solution was acidified to pH 3 (using HCl), phage was irreversibly deactivated, such that when applied to a bacterial culture (with the pH recovered back to that of the growth media), the bacteria could grow as expected. In contrast, when the poly(acrylic acid)s were cycled from pH3 back to the growth media, the phage activity was recovered (i.e. bacteria growth inhibited), confirming that poly(carboxylic acid)s have a fully reversible mode of action that is linked to pH, but still distinct. As a potential mode of action, the sequestration of divalent metal ions by the poly(acids) was ruled out through testing of metal binding macrocycles, which showed no impact on phage function under the conditions tested. These results show that polymeric phage inhibitors are potent but function by a mechanism that includes lowering the pH and hence cannot simply be integrated into bacterial cultures unless the bacteria tolerate low pH. However, the reversible nature of the inactivation suggests a unique mechanism that might be exploitable in next-generation polymeric phage inhibitors and will form the basis of future studies.Data access
Background data is available in the ESI.†Conflicts of interest
MIG, APD and HLM are named inventors on a patent application relating to this work.Acknowledgements
MIG thanks the ERC for a Consolidator Grant (866056) and the Royal Society for an Industry Fellowship (191037) joint with Cytiva. The Biotechnology and Biological Sciences Research Council (BBSRC) Midlands Integrative Biosciences Training Partnership (MIBTP) [BB/M01116X/1] and Cytiva fund HLM Studentship. The Engineering and Physical Sciences Research Council (EPSRC) [EP/S001255/1] is thanked for the support of A. P. S. A. P. S. would like to acknowledge Dr Dean Scholl, AvidBiotics Corporation, for providing us with the phage K1F, K1E, and K1-5 and Drs Eric R. Vimr and Susan M. Steenbergen, thank you for giving us the E. coli EV36 strain. For open access, the author has applied a Creative Commons 18 Attribution (CC BY) license to any author-accepted manuscript version arising from this submission.References
- J. Zhang, L. G. Hansen, O. Gudich, K. Viehrig, L. M. M. Lassen, L. Schrübbers, K. B. Adhikari, P. Rubaszka, E. Carrasquer-Alvarez, L. Chen, V. D’Ambrosio, B. Lehka, A. K. Haidar, S. Nallapareddy, K. Giannakou, M. Laloux, D. Arsovska, M. A. K. Jørgensen, L. J. G. Chan, M. Kristensen, H. B. Christensen, S. Sudarsan, E. A. Stander, E. Baidoo, C. J. Petzold, T. Wulff, S. E. O’Connor, V. Courdavault, M. K. Jensen and J. D. Keasling, A Microbial Supply Chain for Production of the Anti-Cancer Drug Vinblastine, Nature, 2022, 609, 341–347 CrossRef CAS PubMed.
- M. R. R. J. Clokie, A. D. Millard, A. V. Letarov and S. Heaphy, Phages in Nature, Bacteriophage, 2011, 1(1), 31–45 CrossRef PubMed.
- Y. Yang, W. Shen, Q. Zhong, Q. Chen, X. He, J. L. Baker, K. Xiong, X. Jin, J. Wang, F. Hu and S. Le, Development of a Bacteriophage Cocktail to Constrain the Emergence of Phage-Resistant Pseudomonas Aeruginosa, Front. Microbiol., 2020, 11, 327 CrossRef PubMed.
- X. Li, Y. He, Z. Wang, J. Wei, T. Hu, J. Si and G. Tao, A Combination Therapy of Phages and Antibiotics : Two Is Better than One, Int. J. Biol. Sci., 2021, 17, 3573–3582 CrossRef CAS PubMed.
- R. Międzybrodzki, J. Borysowski, B. Weber-Dąbrowska, W. Fortuna, S. Letkiewicz, K. Szufnarowski, Z. Pawełczyk, P. Rogóż, M. Kłak, E. Wojtasik and A. Górski, Clinical Aspects of Phage Therapy, Adv. Virus Res., 2012, 73–121 Search PubMed.
- R. Lewis and C. Hill, Overcoming Barriers to Phage Application in Food and Feed, Curr. Opin. Biotechnol., 2020, 61, 38–44 CrossRef CAS PubMed.
- M. A. S. Khan and S. R. Rahman, Use of Phages to Treat Antimicrobial-Resistant Salmonella Infections in Poultry, Vet. Sci., 2022, 9, 438 Search PubMed.
- N. K. Petty, T. J. Evans, P. C. Fineran and G. P. C. Salmond, Biotechnological Exploitation of Bacteriophage Research, Trends Biotechnol., 2007, 25(1), 7–15 CrossRef CAS PubMed.
- C. A. Stevens, F. Bachtiger, X. D. Kong, L. A. Abriata, G. C. Sosso, M. I. Gibson and H. A. Klok, A Minimalistic Cyclic Ice-Binding Peptide from Phage Display, Nat. Commun., 2021, 12(1), 2675 CrossRef CAS PubMed.
- J. Pande, M. M. Szewczyk and A. K. Grover, Phage Display: Concept, Innovations, Applications and Future, Biotechnol. Adv., 2010, 28(6), 849–858 CrossRef CAS PubMed.
- J. Mahony, J. Murphy, V. Sinderen and D. Lactococcal, 936-Type Phages and Dairy Fermentation Problems: From Detection to Evolution and Prevention, Front. Microbiol., 2012, 3, 335 Search PubMed.
- W. D. Sing and T. R. Klaenhammer, A Strategy for Rotation of Different Bacteriophage Defenses in a Lactococcal Single-Strain Starter Culture System, Appl. Environ. Microbiol., 1993, 59(2), 365–372 CrossRef CAS PubMed.
- J. E. Garneau and S. Moineau, Bacteriophages of Lactic Acid Bacteria and Their Impact on Milk Fermentations, Microb. Cell Fact., 2011, 10(SUPPL. 1), S20 CrossRef PubMed.
- L. Marcin, Strategies of Phage Contamination Prevention in Industry, Open J. Bacteriol., 2020, 4(1), 020–023 CrossRef.
- R. H. Baltz, Bacteriophage-Resistant Industrial Fermentation Strains: From the Cradle to CRISPR/Cas9, J. Ind. Microbiol. Biotechnol., 2018, 45(11), 1003–1006 CrossRef CAS PubMed.
- D. Perlman, [7] Use of Antibiotics in Cell Culture Media, Methods Enzymol., 1979, 58(C), 110–116 CAS.
- A. Kakasis and G. Panitsa, Bacteriophage Therapy as an Alternative Treatment for Human Infections. A Comprehensive Review, Int. J. Antimicrob. Agents, 2019, 53(1), 16–21 CrossRef CAS PubMed.
- D. M. Lin, B. Koskella and H. C. Lin, Phage Therapy: An Alternative to Antibiotics in the Age of Multi-Drug Resistance, World J. Gastrointest. Pharmacol. Ther., 2017, 8(3), 162 CrossRef PubMed.
- M. Sojitra, S. Sarkar, J. Maghera, E. Rodrigues, E. J. Carpenter, S. Seth, D. Ferrer Vinals, N. J. Bennett, R. Reddy, A. Khalil, X. Xue, M. R. Bell, R. B. Zheng, P. Zhang, C. Nycholat, J. J. Bailey, C. C. Ling, T. L. Lowary, J. C. Paulson, M. S. Macauley and R. Derda, Genetically Encoded Multivalent Liquid Glycan Array Displayed on M13 Bacteriophage, Nat. Chem. Biol., 2021, 17(7), 806–816 CrossRef CAS PubMed.
- E. De Clercq and E. Strategies, The Design of Antiviral Drugs, Nat. Rev. Drug Discov., 2002, 1(1), 13–25 CrossRef PubMed.
- E. De Clercq, Antivirals and Antiviral Strategies, Nat. Rev. Microbiol., 2004, 704–720 CrossRef CAS PubMed.
- L. Riva, S. Yuan, X. Yin, L. Martin-Sancho, N. Matsunaga, L. Pache, S. Burgstaller-Muehlbacher, P. D. De Jesus, P. Teriete, M. V. Hull, M. W. Chang, J. F. W. Chan, J. Cao, V. K. M. Poon, K. M. Herbert, K. Cheng, T. T. H. Nguyen, A. Rubanov, Y. Pu, C. Nguyen, A. Choi, R. Rathnasinghe, M. Schotsaert, L. Miorin, M. Dejosez, T. P. Zwaka, K. Y. Sit, L. Martinez-Sobrido, W. C. Liu, K. M. White, M. E. Chapman, E. K. Lendy, R. J. Glynne, R. Albrecht, E. Ruppin, A. D. Mesecar, J. R. Johnson, C. Benner, R. Sun, P. G. Schultz, A. I. Su, A. García-Sastre, A. K. Chatterjee, K. Y. Yuen and S. K. Chanda, Discovery of SARS-CoV-2 Antiviral Drugs through Large-Scale Compound Repurposing, Nature, 2020, 586(7827), 113–119 CrossRef CAS PubMed.
- K. D. Seed, Battling Phages: How Bacteria Defend against Viral Attack, PLoS Pathog., 2015, 11(6), e1004847 CrossRef PubMed.
- G. Ofir, S. Melamed, H. Sberro, Z. Mukamel, S. Silverman, G. Yaakov, S. Doron and R. Sorek, DISARM Is a Widespread Bacterial Defence System with Broad Anti-Phage Activities, Nat. Microbiol., 2018, 3(1), 90–98 CrossRef CAS PubMed.
- S. Doron, S. Melamed, G. Ofir, A. Leavitt, A. Lopatina, M. Keren, G. Amitai and R. Sorek, Systematic Discovery of Antiphage Defense Systems in the Microbial Pangenome, Science, 2018, 359(6379), eaar4120 CrossRef PubMed.
- S. Kronheim, M. Daniel-Ivad, Z. Duan, S. Hwang, A. I. Wong, I. Mantel, J. R. Nodwell and K. L. Maxwell, A Chemical Defence against Phage Infection, Nature, 2018, 564(7735), 283–286 CrossRef CAS PubMed.
- L. Kever, A. Hardy, T. Luthe, M. Hünnefeld, C. Gätgens, L. Milke, J. Wiechert, J. Wittmann, C. Moraru, J. Marienhagen and J. Frunzke, Aminoglycoside Antibiotics Inhibit Phage Infection by Blocking an Early Step of the Infection Cycle, mBio, 2022, 13(3), e0078322 CrossRef PubMed.
- S. T. Jones, V. Cagno, M. Janeček, D. Ortiz, N. Gasilova, J. Piret, M. Gasbarri, D. A. Constant, Y. Han, L. Vuković, P. Král, L. Kaiser, S. Huang, S. Constant, K. Kirkegaard, G. Boivin, F. Stellacci and C. Tapparel, Modified Cyclodextrins as Broad-Spectrum Antivirals, Sci. Adv., 2020, 6(5), eaax9318 CrossRef CAS PubMed.
- V. Cagno, E. D. Tseligka, S. T. Jones and C. Tapparel, Heparan Sulfate Proteoglycans and Viral Attachment: True Receptors or Adaptation Bias?, Viruses, 2019, 11(7), 596 CrossRef CAS PubMed.
- P. De Somer, E. De Clercq, A. Billiau, E. Schonne and M. Claesen, Antiviral Activity of Polyacrylic and Polymethacrylic Acids, J. Virol., 1968, 2(9), 878–885 CrossRef CAS PubMed.
- R. H. Bianculli, J. D. Mase and M. D. Schulz, Antiviral Polymers: Past Approaches and Future Possibilities, Macromolecules, 2020, 53(21), 9158–9186 CrossRef CAS.
- F. Schandock, C. F. Riber, A. Röcker, J. A. Müller, M. Harms, P. Gajda, K. Zuwala, A. H. F. Andersen, K. B. Løvschall, M. Tolstrup, F. Kreppel, J. Münch and A. N. Zelikin, Macromolecular Antiviral Agents against Zika, Ebola, SARS, and Other Pathogenic Viruses, Adv. Healthcare Mater., 2017, 6(23), 1700748 CrossRef PubMed.
- T. Ikeda, H. Yamaguchi and S. Tazuke, New Polymeric Biocides: Synthesis and Antibacterial Activities of Polycations with Pendant Biguanide Groups, Antimicrob. Agents Chemother., 1984, 26(2), 139–144 CrossRef CAS PubMed.
- G. N. Tew, D. Liu, B. Chen, R. J. Doerksen, J. Kaplan, P. J. Carroll, M. L. Klein and W. F. DeGrado, De Novo Design of Biomimetic Antimicrobial Polymers, Proc. Natl. Acad. Sci. U. S. A., 2002, 99(8), 5110–5114 CrossRef CAS PubMed.
- D. J. Phillips, J. Harrison, S.-J. J. Richards, D. E. Mitchell, E. Tichauer, A. T. M. M. Hubbard, C. Guy, I. Hands-Portman, E. Fullam and M. I. Gibson, Evaluation of the Antimicrobial Activity of Cationic Polymers against Mycobacteria: Toward Antitubercular Macromolecules, Biomacromolecules, 2017, 18(5), 1592–1599 CrossRef CAS PubMed.
- R. E. W. Hancock and H.-G. Sahl, Antimicrobial and Host-Defense Peptides as New Anti-Infective Therapeutic Strategies, Nat. Biotechnol., 2006, 24(12), 1551–1557 CrossRef CAS PubMed.
- K. S. Rosenthal, J. Killius, C. M. Hodnichak, T. M. Venetta, L. Gyurgyik and K. Janiga, Mild Acidic PH Inhibition of the Major Pathway of Herpes Simplex Virus Entry into HEp-2 Cells, J. Gen. Virol., 1989, 70(4), 857–867 CrossRef PubMed.
- S. Bartolotta, C. C. García, N. A. Candurra and E. B. Damonte, Effect of Fatty Acids on Arenavirus Replication: Inhibition of Virus Production by Lauric Acid, Arch. Virol., 2001, 146(4), 777–790 CrossRef CAS PubMed.
- O. V. Carvalho, C. V. Botelho, C. G. T. Ferreira, H. C. C. Ferreira, M. R. Santos, M. A. N. Diaz, T. T. Oliveira, J. A. P. Soares-Martins, M. R. Almeida and A. Silva Jr, In Vitro Inhibition of Canine Distemper Virus by Flavonoids and Phenolic Acids: Implications of Structural Differences for Antiviral Design, Res. Vet. Sci., 2013, 95(2), 717–724 CrossRef CAS PubMed.
- H. L. Marton, P. Kilbride, A. Ahmad, A. P. Sagona and M. I. Gibson, Anionic Synthetic Polymers Prevent Bacteriophage Infection, J. Am. Chem. Soc., 2023, 145(16), 8794–8799 CrossRef CAS PubMed.
- H. Y. Steensma and J. Blok, Effect of Calcium Ions on the Infection of Bacillus Subtilis by Bacteriophage SF 6, J. Gen. Virol., 1979, 42(2), 305–314 CrossRef CAS PubMed.
- F. Shafia and T. L. Thompson, Calcium ion requirement for proliferation of bacteriophage Φμ-4, J. Bacteriol., 1964, 88(2), 293–296 CrossRef CAS PubMed.
- E. F. Landry and R. M. Zsigray, Effects of Calcium on the Lytic Cycle of Bacillus Subtilis Phage 41c, J. Gen. Virol., 1980, 51(1), 125–135 CrossRef CAS PubMed.
- V. Cvirkaitė-Krupovič, M. Krupovič, R. Daugelavičius and D. H. Bamford, Calcium Ion-Dependent Entry of the Membrane-Containing Bacteriophage PM2 into Its Pseudoalteromonas Host, Virology, 2010, 405(1), 120–128 CrossRef PubMed.
- J. Mahony, D. M. Tremblay, S. J. Labrie, S. Moineau and D. van Sinderen, Investigating the Requirement for Calcium during Lactococcal Phage Infection, Int. J. Food Microbiol., 2015, 201, 47–51 CrossRef CAS PubMed.
- S. Chhibber, T. Kaur and S. Kaur, Essential Role of Calcium in the Infection Process of Broad–spectrum Methicillin–resistant Staphylococcus Aureus Bacteriophage, J. Basic Microbiol., 2014, 54(8), 775–780 CrossRef CAS PubMed.
- P. Chakraborty, M. Neumaier, P. Weis and M. M. Kappes, Exploring Isomerism in Isolated Cyclodextrin Oligomers through Trapped Ion Mobility Mass Spectrometry, J. Am. Soc. Mass Spectrom., 2023, 34(4), 676–684 CrossRef CAS PubMed.
- H. Dossmann, L. Fontaine, T. Weisgerber, V. Bonnet, E. Monflier, A. Ponchel and C. Przybylski, First Steps to Rationalize Host–Guest Interaction between α-, β-, and γ-Cyclodextrin and Divalent First-Row Transition and Post-Transition Metals (Subgroups VIIB, VIIIB, and IIB), Inorg. Chem., 2021, 60(2), 930–943 CrossRef CAS PubMed.
- E. C. Silva, C. G. Gomes, J. Pina, R. F. P. Pereira, D. Murtinho, A. R. Fajardo and A. J. M. Valente, Carbon Quantum Dots-Containing Poly(β-Cyclodextrin) for Simultaneous Removal and Detection of Metal Ions from Water, Carbohydr. Polym., 2024, 323, 121464 CrossRef CAS PubMed.
- J. S. Bradshaw and R. M. Izatt, Crown Ethers: The Search for Selective Ion Ligating Agents, Acc. Chem. Res., 1997, 30(8), 338–345 CrossRef CAS.
- S. D. Alexandratos and C. L. Stine, Synthesis of Ion-Selective Polymer-Supported Crown Ethers: A Review, React. Funct. Polym., 2004, 60, 3–16 CrossRef CAS.
- J. A. Jackman, A. Hakobyan, H. Zakaryan and C. C. Elrod, Inhibition of African Swine Fever Virus in Liquid and Feed by Medium-Chain Fatty Acids and Glycerol Monolaurate, J. Anim. Sci. Biotechnol., 2020, 11(1), 1–10 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4lp00202d |
| This journal is © The Royal Society of Chemistry 2024 |

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
: