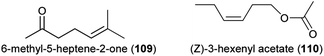Open Access Article
Open Access ArticleChemistry, biosynthesis and biology of floral volatiles: roles in pollination and other functions
Stefan
Dötterl
 *a and
Jonathan
Gershenzon
*a and
Jonathan
Gershenzon
 *b
*b
aDepartment of Environment & Biodiversity, Paris Lodron University Salzburg, Hellbrunnerstr 34, 5020 Salzburg, Austria. E-mail: stefan.doetterl@plus.ac.at
bDepartment of Biochemistry, Max Planck Institute for Chemical Ecology, Hans-Knöll-Straße 8, 07745 Jena, Germany. E-mail: gershenzon@ice.mpg.de
First published on 4th September 2023
Abstract
Covering: 2010 to 2023
Floral volatiles are a chemically diverse group of plant metabolites that serve multiple functions. Their composition is shaped by environmental, ecological and evolutionary factors. This review will summarize recent advances in floral scent research from chemical, molecular and ecological perspectives. It will focus on the major chemical classes of floral volatiles, on notable new structures, and on recent discoveries regarding the biosynthesis and the regulation of volatile emission. Special attention will be devoted to the various functions of floral volatiles, not only as attractants for different types of pollinators, but also as defenses of flowers against enemies. We will also summarize recent findings on how floral volatiles are affected by abiotic stressors, such as increased temperatures and drought, and by other organisms, such as herbivores and flower-dwelling microbes. Finally, this review will indicate current research gaps, such as the very limited knowledge of the isomeric pattern of chiral compounds and its importance in interspecific interactions.
1 Introduction
Floral volatiles are key mediators of plant–pollinator interactions.1–6 Considering that flowering plants (angiosperms) are by far the most diverse group of terrestrial plants and that 85% of these plants are pollinated by animals,7 among them various crops,8 pollinator attraction by olfactory means is a highly important process in terrestrial ecosystems and for crop production. About 1700 floral volatiles had already been described more than a decade ago,4 but many new compounds have been discovered since then,e.g.,9–12 making floral scent a high dimensional trait with many possible ways to vary among species. Indeed, floral scents are highly variable among species, either due to differences in the compounds emitted or due to differences in the absolute or relative amounts of single scent compounds. Thus, it is not surprising that floral scents are involved in shaping plant–pollinator networks.13–15 Quite recently, it has been demonstrated that floral volatiles serve additional functions, such as defending flowers against enemies (e.g., pathogens)16 and attracting predators of plant pests.17 It was also shown that they are attractive to florivores and their occurrence is influenced by biotic and abiotic stressors at the organ, plant and population levels. As our knowledge of floral volatile biosynthesis has increased, especially on the regulation of biosynthesis, we have learned more about the mechanisms underlying these patterns. This review summarizes advances in floral scent research since 2010 from chemical, molecular and ecological perspectives, excluding recent findings on the olfactory communication between orchids and their pollinators, given that this topic is considered by a recent review.18 It will also indicate current research gaps, such as the very limited knowledge of the occurrence and ratio of stereoisomers of chiral compounds and its importance in interspecific interactions.2 Reports of notable new floral volatiles
Floral volatiles have long been classified into a few basic groups according to their chemistry and probable biosynthetic origin: fatty acid derivatives, terpenoids, phenylpropanoids/benzenoids and other amino acid derivatives. During the period of this review, specific compounds from each of these groups were identified as floral volatiles for the first time. Representatives of these compounds are summarized in Table 1 and Fig. 1–5, and the biological roles in pollination of several of these compounds are discussed later in the chapter. Compounds isolated from orchids are also covered by Perkins et al.,18 but are included here for completeness.| Compound number and name | Plant species | Family | Reference |
|---|---|---|---|
| Fatty acid derivatives | |||
| (1) (6Z,9Z)-1,6,9-tricosatriene | Pterostylis orbiculata | Orchidaceae | 21 |
| (2) (E)-2-octen-1-yl acetate | Ceropegia sandersonii | Apocynaceae | 22 |
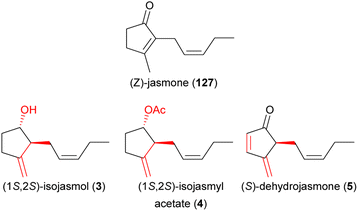
| (3) isojasmol | Thaumatophyllum mello-baretoanum, Xanthosoma hylaeae, Ludovia lancifolia | Araceae, Cyclanthaceae | 11, 12 and 23 |
| (4) isojasmyl acetate | Xanthosoma hylaeae | Araceae | 11 |
| (5) dehydrojasmone | Thaumatophyllum mello-baretoanum | Araceae | 11 |
| (6) trans-3-methyldodecano-4-lactone | Passiflora chocoensis, Kefersteinia aurorae, Scuticaria salesiana, Lycaste brevispatha | Passifloraceae, Orchidaceae | 24 |
| (7) trans-(Z)-3-methyldodec-7-eno-4-lactone | Kefersteinia, Scuticaria salesiana, Lycaste brevispatha, Chaubardiella dalessandroi, Mormodes romanii | Orchidaceae | 24 |
| (8) trans-(Z)-3-methyldodec-6-eno-4-lactone | Kefersteinia pellita, Chaubardiella dalessandroi, Mormodes romanii | Orchidaceae | 24 |
| (9) cis-(Z)-3-methyldodec-6-eno-4-lactone | Kefersteinia pellita, Chaubardiella dalessandroi, Mormodes romanii | Orchidaceae | 24 |
| (10) (Z)-deca-7,9-dieno-5-lactone | Gardenia brighamii | Rubiaceae | 24 |
| (11) (16S,9Z)-16-ethylhexadec-9-enolide | Disa forficaria | Orchidaceae | 25 |
| (12) (Z)-3-isopropylpent-3-en-1-ol | Syngonium hastiferum | Araceae | 26 |
| (13) (S)-2-(tetrahydrofuran-2-yl)acetic acid | Cryptostylis ovata | Orchidaceae | 27 |
| (14) methyl-(S)-2-(tetrahydrofuran-2-yl)acetate | Cryptostylis ovata | Orchidaceae | 27 |
| (15) ethyl-(S)-2-(tetrahydrofuran-2-yl)acetate | Cryptostylis ovata | Orchidaceae | 27 |
| (16) 2-ethyl-5-propylcyclohexan-1,3-dione (chiloglottone 1) | Chiloglottis | Orchidaceae | 28 |
| (17) 2-ethyl-5-pentylcyclohexan-1,3-dione (chiloglottone 2) | Chiloglottis | Orchidaceae | 28 |
| (18) 2-butyl-5-methylcyclohexan-1,3-dione (chiloglottone 3) | Chiloglottis | Orchidaceae | 28 |
| (19) 5-allyl-2-ethylcyclohexan-1,3-dione (chiloglottone 4) | Chiloglottis | Orchidaceae | 28 |
| (20) 2-butyl-5-propylcyclohexan-1,3-dione (chiloglottone 5) | Chiloglottis | Orchidaceae | 28 |
| (21) 2-hexyl-5-methylcyclohexan-1,3-dione (chiloglottone 6) | Chiloglottis | Orchidaceae | 28 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Terpenoids | |||
| (22) (Z)-2-allylidene-6-methylhept-5-en-1-ol: (Z)-filamentol | Yucca | Asparagaceae | 29 |
| (23) (Z)-2-allylidene-6-methylhept-5-en-1-al: (Z)-filamental | Yucca | Asparagaceae | 29 |
| (24) (Z,R)-3-allylidene-5-(2-methylprop-1-enyl) dihydro(3H)furan-2-one: (Z)-filamentolide | Yucca | Asparagaceae | 29 |
| (25) (E)-2-allylidene-6-methylhept-5-en-1-ol: (E)-filamentol | Yucca | Asparagaceae | 29 |
| (26) (E)-2-allylidene-6-methylhept-5-en-1-al: (E)-filamental | Yucca | Asparagaceae | 29 |
| (27) (E,R)-3-allylidene-5-(2-methylprop-1-enyl) dihydro(3H)furan-2-one: (E)-filamentolide | Yucca | Asparagaceae | 29 |
| (28) 2-(4-methylpent-3-en-1-yl)cyclopent-3-enone: filamentone | Yucca | Asparagaceae | 29 |
| (29) (E)-4,8-dimethylnona-1,3,7-trien-5-yl acetate | Philodendron squamiferum | Araceae | 11 |
| (30) 4-hydroxy-3-methyl-6S-(pentan-2S-yl)-5,6-dihydro-2H-pyran-2-one (drakolide) | Drakaea micrantha | Orchidaceae | 9 |
| (31) (+)-copalol | Cryptanthus burle-marxii | Bromeliaceae | 20 |
| (32) (β)-safranol | Anthurium salvadorense | Araceae | 24 |
| (33) safranyl acetate | Anthurium salvadorense | Araceae | 24 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Phenylpropanoids/benzenoids | |||
| (34) 2-methoxy-4-vinyl phenol | Dichaea pendula | Orchidaceae | 30 |
| (35) 2-methoxy-6-methylacetophenone | Colocasia gigantea, Cyphomandra divaricata, Pterygodium | Araceae, Solanaceae, Orchidaceae | 24 |
| (36) (Z)-hept-4-en-2-yl salicylate | Saraca asoca | Fabaceae | 24 |
| (37) 3-acetyloxy-4-phenylbutan-2-one | Ceropegia stenantha | Apocynaceae | 31 |
| (38) 3-acetyloxy-1-phenylbutan-2-one | Ceropegia stenantha | Apocynaceae | 31 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Other amino acid derivatives | |||
| (39) 2-hydroxymethyl-3,5,6-trimethylpyrazine | Drakaea livida | Orchidaceae | 32 |
| (40) 2-hydroxymethyl-3,5-dimethyl-6-ethylpyrazine | Drakaea micrantha | Orchidaceae | 9 |
| (41) 2-hydroxymethyl-3-(3-methylbutyl)-5-methylpyrazine | Drakaea livida | Orchidaceae | 33 |
| (42) (3,5,6-trimethylpyrazin-2-yl) methyl 3-methylbutanoate | Drakaea livida | Orchidaceae | 32 |
| (43) (3,6-dimethylpyrazin-2-yl) methyl 3-methylbutanoate | Drakaea livida | Orchidaceae | 32 |
| (44) (3,5,6-trimethylpyrazin-2-yl) methyl (2S)-methylbutanoate | Drakaea livida | Orchidaceae | 32 |
| (45) 2-(3-methylbutyl)-3,5,6-trimethylpyrazine | Drakaea livida | Orchidaceae | 32 |
| (46) (E)-N-(2-methylbutyl)-1-(pyridin-3-yl)methanimine | Pyrus communis | Rosaceae | 10 |
| (47) (E)-N-(3-methylbutyl)-1-(pyridin-3-yl)methanimine | Pyrus communis | Rosaceae | 10 |
| (48) 4-methyl-5-vinylthiazole | Annona, Caladium bicolor | Annonaceae, Araceae | 34 |
| (49) 2-(methylthio)-4-(hydroxy)phenol | Caladenia crebra | Orchidaceae | 35 |
| (50) 2-(methylthio)-4-(hydroxymethyl)phenol | Caladenia crebra | Orchidaceae | 35 |
| (51) 2-(methylthio)-4-(formyl)phenol | Caladenia crebra | Orchidaceae | 35 |
| (52) 2,3-dihydroxypropyl isovalerate | Luisia teres | Orchidaceae | 36 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Miscellaneous | |||
| (53) 1,2-diacetin | Multiple | Multiple | 19 |
| (54) 1,3-diacetin | Multiple | Multiple | 19 |
| (55) 2,3-butanediol acetate | Arum palaestinum | Araceae | 37 |
| (56) acetoin acetate | Arum palaestinum | Araceae | 37 |
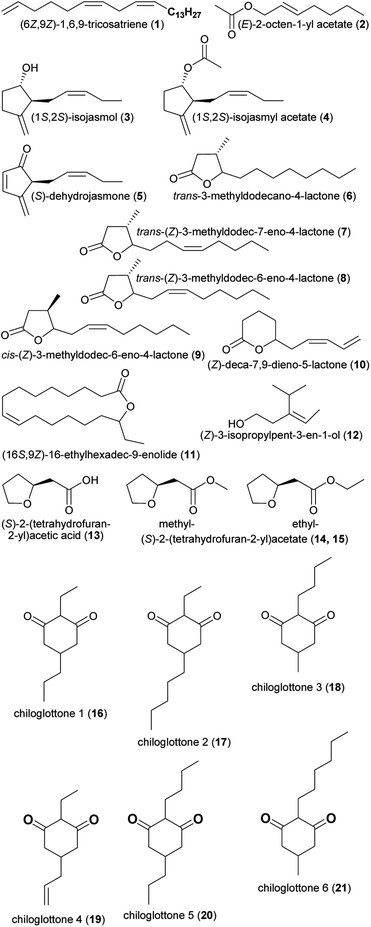 | ||
| Fig. 1 Fatty acid derivatives listed in Table 1. | ||
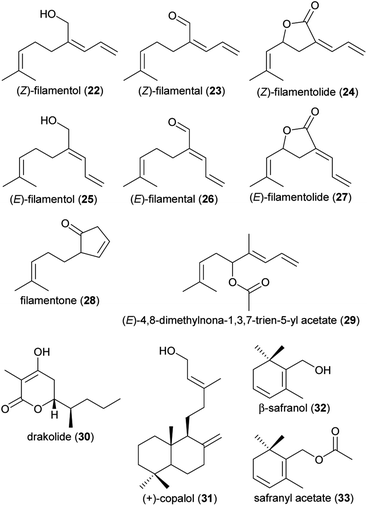 | ||
| Fig. 2 Terpenoids listed in Table 1. | ||
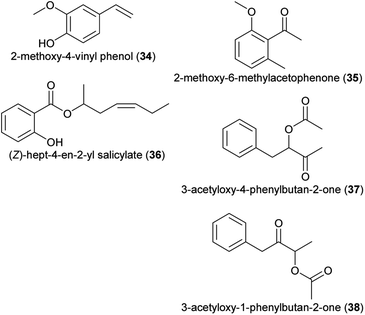 | ||
| Fig. 3 Phenylpropanoids/benzenoids listed in Table 1. | ||
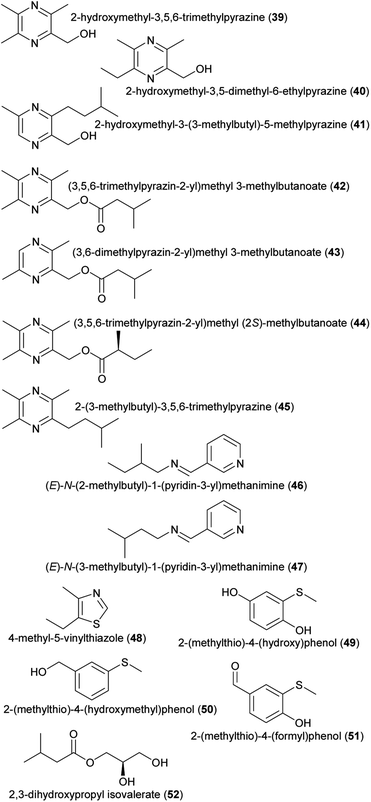 | ||
| Fig. 4 Other amino acid derivatives listed in Table 1. | ||
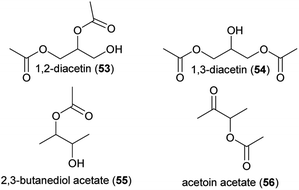 | ||
| Fig. 5 Miscellaneous compounds listed in Table 1. | ||
The length of this list of new floral volatiles suggests that many more substances still remain to be discovered in the blends emitted from flowers. This is not simply because of advances in analytical instrumentation that provide ever increasing sensitivity. Some of the new volatiles are compounds that had been missed in earlier surveys that relied only on headspace trapping methods, rather than also carrying out solvent extraction from floral tissues.19 Floral volatiles of low volatility, such as the C20 diterpene alcohol, copalol,20 are especially difficult to detect by headspace analysis. However, most of the new volatiles reported are constituents of floral blends of species that had not been previously examined.
The discovery of new floral volatiles in new species is consistent with the concept that plant scents have been generally selected to be unique to attract specific pollinators and ensure a high degree of intraspecific gene flow during pollen transfer. The release of unique compounds has been described as creating a private channel between the plant and pollinator species,5,19 with volatiles detected by the intended receiver, but not by other potential flower visitors. On the other hand, specificity can also be achieved by the emission of blends of widespread compounds in unique proportions, and new floral blends dominated by previously characterized widespread compounds are continually reported.
3 Biosynthesis of floral volatiles
Recent progress in understanding the biosynthesis of floral volatiles has been dominated by reports of new genes and enzymes participating in volatile formation. Advances in DNA sequencing technology and the availability of genome and transcriptome data for many plant species have now made it possible to readily identify genes in floral volatile formation. Differential gene expression studies based on RNA-Seq allow researchers to focus on genes expressed specifically in floral organs and at the precise time of volatile emission. In addition, co-expression surveys can help to identify biosynthetic genes as long as one gene of a pathway is already known, since the genes of a single pathway are usually co-regulated. Meanwhile, increasing knowledge of the enzymes participating in floral scent formation has provided more target gene families to search for. Once candidate genes are chosen, advances in heterologous expression of the encoded enzymes both in model plants and in microbial systems for in vivo characterization have made it much easier to determine their biochemical function.3.1 Fatty acid derivatives
The methylation of carboxylic acids is a common step in the biosynthesis of floral volatiles. These reactions are catalyzed by SABATH methyltransferases,38 which carry out the O-methylation of small carboxylic acids to form common floral volatiles, such as the benzenoids methyl salicylate and methyl benzoate. The methyl group itself is derived from S-adenosyl methionine (SAM). Recently, a SABATH O-methyltransferase was characterized that forms methyl hexanoate (57), the major constituent of floral scent of the water lily Victoria cruziana (Fig. 6).39 In addition, a jasmonic acid O-methyltransferase was identified from Cymbidium faberi that produces methyl jasmonate (58), the major floral scent component of this orchid.40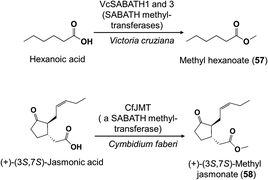 | ||
| Fig. 6 Methylation of carboxylic acids in biosynthesis of volatile fatty acid derivatives in flowers. | ||
3.2 Terpenoids
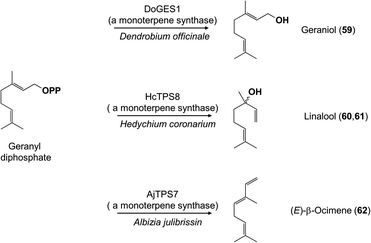
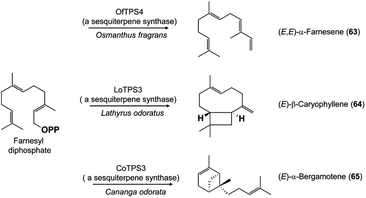
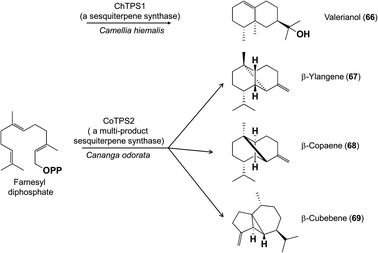
The multitude of terpene carbon skeletons found in floral scents can nearly all be attributed to catalysis by terpene synthases. This large enzyme family converts acyclic prenyl diphosphates via carbocation-dependent reactions into an enormous variety of cyclic and acyclic products that are often released directly as floral volatiles or otherwise metabolized further. The great diversity of floral terpenes arises because each plant typically has a sizable group of different terpene synthases,41 plus some terpene synthases produce multiple products from a single substrate.42 Many new terpene synthases involved in floral scent formation have been characterized in the period under review, especially in horticulturally or economically important plants such as sweet osmanthus (Osmanthus fragrans, Oleaceae),43 and species of the orchid genera Dendrobium44,45 and Phalaenopsis (moth orchids).46 Although too numerous to mention individually, most of these terpene synthases make widespread floral volatiles for which terpene synthases are already known, like the monoterpenes geraniol (59),46,47 linalool (60 and/or 61),43,46,48–50 and (E)-β-ocimene (62)43,49 (Fig. 7), or the sesquiterpenes (E,E)-α-farnesene (63),43 (E)-β-caryophyllene (64)44 and (E)-α-bergamotene (65)51 (Fig. 8). These enzymes share their basic properties with other terpene synthases making the same products, but often have divergent sequences. Terpene synthases making products such as geraniol, linalool and (E)-β-ocimene have originated independently many times over the course of plant evolution.52 However, other new terpene synthases make more unusual products, such as the sesquiterpene valerianol (66)53 or unusual blends of products, such as the sesquiterpenes β-ylangene (67), β-copaene (68) and β-cubebene (69)51 (Fig. 9) or others.48–50 One group of monoterpene synthases from the flowers of Nicotiana species all make a similar mixture of seven products dominated by 1,8-cineole (70) with some α-terpineol (71) and lesser amounts of α-pinene (72), β-pinene (73), sabinene (74), limonene (75) and β-myrcene (76) (Fig. 10).54 Interestingly, terpene synthase differences are at least in part responsible for the divergent pollination syndromes of two Mimulus (Phrymaceae) species: the hummingbird-pollinated M. cardinalis and the bumblebee-pollinated M. lewisii. The latter makes several monoterpenes, (E)-β-ocimene, limonene and myrcene, not produced by the former. These differences can be ascribed to two terpene synthases, an ocimene synthase that is functional in M. lewisii, but not in M. cardinalis, and a limonene–myrcene synthase that is expressed at higher levels in M. lewisii than in M. cardinalis.55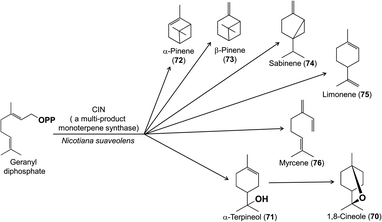 | ||
| Fig. 10 A multi-product monoterpene synthase that synthesizes a mixture known as the “cineole cassette”. 1,8-Cineole is formed via an enzyme-bound α-terpineol intermediate. | ||
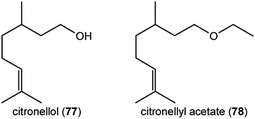
One of the most exciting advances of recent research on floral volatile biosynthesis is the discovery of a new pathway for the formation of the common monoterpene alcohol geraniol (59) in rose (Rosa x hybrida) flowers.56 Geraniol in flowers has been previously shown to be made via terpene synthases,46,47 whose mechanisms involve simple cleavage of the diphosphate moiety from the precursor geranyl diphosphate with the resulting carbocation being quenched by water to form geraniol. Rose flowers were also assumed to have a geraniol-forming terpene synthase, but geranyl diphosphate was instead found to be hydrolyzed in a completely unexpected way: a diphosphohydrolase from the Nudix family cleaves one phosphate moiety to give geranyl monophosphate, which is then further hydrolyzed to geraniol by a second phosphatase (Fig. 11). The role of this Nudix hydrolase was first suggested by QTL (Quantitative Trait Loci) mapping in crosses between rose genotypes with different scents, and confirmed by RNA interference of the corresponding gene, which reduced emission of geraniol.56 More recently, a Nudix hydrolase from another rose species, Rosa rugosa, was identified, and expression of the encoding gene was shown to correlate with the formation of geraniol and derivatives, such as citronellol (77) and citronellyl acetate (78) as in R. x hybrida.57
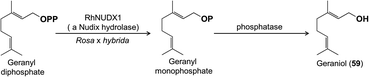 | ||
| Fig. 11 Formation of geraniol in roses via a new pathway involving a Nudix hydrolase instead of a terpene synthase. | ||
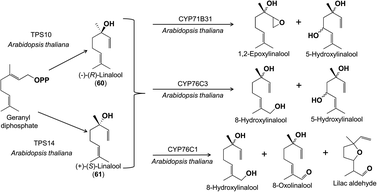
When terpene synthase products in flowers are further metabolized, the reactions involved are frequently oxidations carried out by cytochrome P450s, the largest family of enzymes in plant metabolism. A series of fascinating studies has described the role of cytochrome P450s in the oxidation of terpenes in Arabidopsis thaliana flowers. Although this species is largely self-pollinated, the flowers still emit a diverse blend of volatiles, especially terpenes. Their biosynthesis starts from geranyl diphosphate with flower-localized terpene synthases (TPS10 and 14) that produce the alcohols (−)- and (+)-linalool (60, 61), respectively. Next, these are converted by two flower-localized P450s (CYP71B31 and CYP76C3) to products with additional hydroxyl groups or epoxide and aldehyde functions that accumulate in the flowers (Fig. 12).58 A third floral P450 in this species (CYP76C1) converts the linalools into an even larger range of products with alcohol, aldehyde, carboxylic acid and cyclic ether functions.59 These extensively oxidized products act as herbivore deterrents, and thus the P450s can be considered as enzymes that transform linalool, frequently reported as a pollinator attractant, to herbivore defense compounds (but see below) in A. thaliana, a self-compatible plant that is not dependent on pollination for seed set. A parallel process involves sesquiterpenes in A. thaliana flowers with the olefins produced by terpene synthase TPS11 being oxidized to alcohols and ketones by yet another P450 enzyme (CYP706A3) (Fig. 13).16 In this case, the oxidized products again accumulate in flowers and serve as anti-herbivore defenses, in contrast to the more volatile sesquiterpene olefin substrates, which are emitted as attractants.
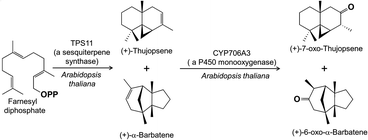 | ||
| Fig. 13 Biosynthesis of oxidized sesquiterpene volatiles in Arabidopsis thaliana flowers that function as herbivore deterrents. | ||
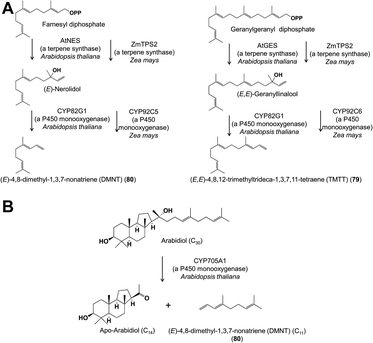
Cytochrome P450 enzymes have also been implicated in the formation of two irregular terpenes, the C16 (E,E)-4,8,12-trimethyltrideca-1,3,7,11-tetraene (TMTT) (79) and the C11 (E)-4,8-dimethyl-1,3,7-nonatriene (DMNT) (80), which are frequent constituents of floral scent, especially in night-blooming white flowers. TMTT and DMNT are also formed in the vegetative parts of plants after herbivore damage. Biosynthetic studies on these two compounds carried out with damaged A. thaliana and maize foliage demonstrated that the C20 alcohol geranyllinalool, a terpene synthase product, was cleaved to form TMTT, and the C15 nerolidol cleaved to form DMNT, respectively (Fig. 14A).60,61 The enzymes involved in both plants are cytochrome P450s (CYP82G1 in A. thaliana, CYP92C5 and C6 in maize), but are from different subfamilies suggesting independent evolution of the pathway in different plant lineages. Given that the A. thaliana CYP82G1 is expressed after herbivore damage in leaves as well as constitutively in flowers, TMTT and DMNT seem very likely to be formed by similar pathways in flowers. Arabidopsis thaliana also possess a second completely different DMNT pathway in roots involving the cleavage of a triterpene (C30) precursor by yet another cytochrome P450 (CYP705A1) to form the C11 product (Fig. 14B).62
Another group of floral terpenes formed by cleavage reactions are those derived from carotenoids by catalysis of carotenoid cleavage dioxygenases (CCDs) instead of P450s. Here there are reports of the floral volatiles α- and β-ionone (81, 82) being produced from α- and β-carotene in Osmanthus fragrans by a CCD1 enzyme,63 while β-ionone is produced by a CCD4 enzyme from β-carotene and lutein in Crocus sativus, the source of saffron64 (Fig. 15).
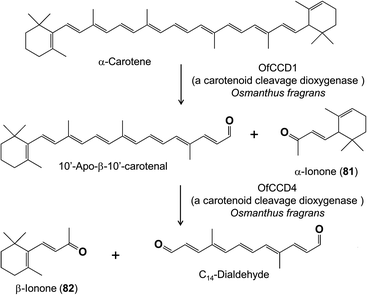 | ||
| Fig. 15 Cleavage of a carotenoid to volatile apocarotenoids by carotenoid cleavage dioxygenase enzymes. | ||
3.3 Phenylpropanoids and benzenoids
The biosynthesis of most phenylpropanoid and benzenoid floral volatiles from intermediates of primary metabolism typically requires longer pathways than those to terpenoid floral volatiles. One of the exceptions is the pathway leading from phenylalanine to the widely-encountered phenylacetaldehyde (83). This is a two-step decarboxylation-amine oxidation reaction catalyzed by phenylacetaldehyde synthase in a pyridoxal 5′-phosphate-dependent manner (Fig. 16). Described originally from petunia flowers,65 this enzyme of the aromatic amino acid decarboxylase family has also been reported from the flowers of A. thaliana and rose (R. x hybrida).66,67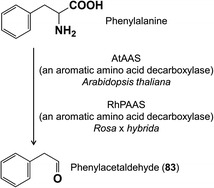 | ||
| Fig. 16 Biosynthesis of phenylacetaldehyde from phenylalanine occurs via a two-step reaction catalyzed by phenylacetaldehyde synthase, a member of the aromatic amino acid decarboxylase family. | ||
Phenylacetaldehyde is next subject to reduction to form 2-phenylethanol (84), a well-known component of rose scent. The responsible enzyme, a member of the short chain dehydrogenase/reductase family previously characterized from R. x hybrida ‘Hoh-Jun’ flowers68 and tomato fruit, was more recently characterized in detail from R. x damascena flowers (Fig. 17).69 A QTL study employing a R. x hybrida mapping population segregating for 2-phenylethanol emission implicated an allele of phenylacetaldehyde synthase as being critical for 2-phenylethanol formation.70 Curiously, expression of the encoding gene was found to be active already in the flower bud stage causing 2-phenylethanol to accumulate in the petals as a glycoside. A UDP-glucosyltransferase responsible for glucosylation of 2-phenylethanol (and benzyl alcohol) was recently identified in petunia.71
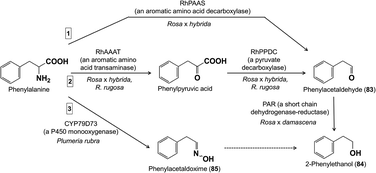 | ||
| Fig. 17 2-Phenylethanol is formed by three different biosynthetic routes, and more than one may operate in a single plant species depending on the environmental conditions. The steps between phenylacetaldoxime and 2-phenylethanol are not known. Other floral volatiles are also formed by different pathways in different species or by similar pathways employing different enzymes.52 | ||
A second pathway to 2-phenylethanol in roses (R. x hybrida) also starts from phenylalanine. This aromatic amino acid is first deaminated to phenylpyruvate by an aminotransferase, then decarboxylated to phenylacetaldehyde by a phenylpyruvate decarboxylase, and finally reduced as above to 2-phenylethanol (Fig. 17).72 Studies with isotopically-labeled phenylalanine demonstrated that the route via phenylpyruvate operates especially in summer due to higher temperatures. The authors suggest that the greater efficiency of the pathway via phenylpyruvate helps maintain 2-phenylethanol formation in summer despite reduced petal growth. Both pathways also seem to operate in R. rugosa flowers since the respective genes were expressed and overexpression of either the phenylacetaldehyde synthase gene or the phenylalanine aminotransferase gene increased 2-phenylethanol formation.73
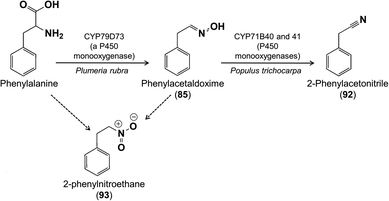
And, there is even a third pathway for 2-phenylethanol biosynthesis in flowers! In Plumeria rubra (frangipani, Apocynaceae), phenylalanine is oxidized by a cytochrome P450 of the CYP79 family (CYP79D73) to phenylacetaldoxime (85), which was shown to be an intermediate in 2-phenylethanol formation by in vivo feeding (Fig. 17).74 Expression of the CYP79D73 gene in Nicotiana benthamiana also resulted in 2-phenylethanol emission, but the biosynthetic steps from phenylacetaldoxime to 2-phenylethanol are not yet known. The evolution of three different pathways to 2-phenylethanol in flowers might suggest that it is easy to evolve a pathway to this compound or that it has a strong innate attractiveness for pollinating insects (see below). Multiple pathways for 2-phenylethanol formation are also known in poplar leaves.75
Along with 2-phenylethanol, the volatile 1-phenylethanol (86) also occurs in flowers, such as those of Camellia sinensis (tea). Isotopic tracer experiments demonstrated that 1-phenylethanol is derived from phenylalanine via the intermediate acetophenone (Fig. 18).76 Several genes and enzymes involved in the conversion of acetophenone to 1-phenylethanol have been identified in tea from the short chain dehydrogenase/reductase family.77–79 Interestingly, 1-phenylethanol is present as both (R)- and (S)-enantiomers and separate enzymes are responsible for making each enantiomer.
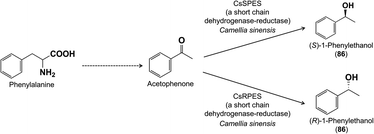 | ||
| Fig. 18 The formation of 1-phenylethanol. Only the last step of the pathway has been elucidated in plants. | ||
Another well-known group of phenylpropanoid volatiles are the phenylpropenes, including eugenol, chavicol and estragole, found in flowers and the foliage of certain herbs. Their biosynthesis leads from phenylalanine to the phenylpropanoids, which are then acetylated to serve as substrates for an NADPH-dependent reductase that forms the final phenylpropene by acetate elimination and reduction (Fig. 19).80 Since this original report, genes participating in other steps in phenylpropene formation have been identified, such as in O-methylation of caffeoyl-CoA to feruloyl-CoA in petunia,81 the acetylation of coniferyl alcohol in Prunus mume,82 and the conversion of coniferyl acetate to eugenol (87) in roses.83
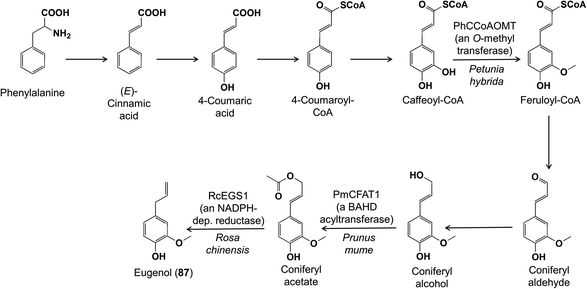 | ||
| Fig. 19 The biosynthesis of the phenylpropene volatile eugenol (87) has been previously elucidated. Shown are new reports of enzymes involved in the biosynthetic pathway. | ||
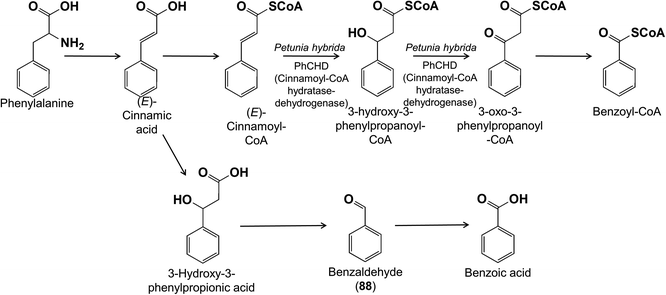
The formation of benzenoid volatiles requires the shortening of the C3 side chain of their phenylpropanoid precursors. Our understanding of this biosynthetic process has increased significantly in recent years thanks largely to work in petunia flowers as well as A. thaliana, but curiously several pathways may co-occur (reviewed in84). Evidence is strongest for the operation of a β-oxidation pathway from cinnamic to benzoic acid that removes a C2 fragment from the C3 side chain in a manner analogous to the β-oxidation of fatty acids. The reaction sequence includes hydration of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bond of the side chain, oxidation of the resulting alcohol to give a β-keto function, and a reverse aldol cleavage to release the C2 fragment as acetyl-CoA (Fig. 20). These steps proceed via CoA-bound intermediates in the peroxisome. The most recently characterized enzyme was the cinnamoyl-CoA hydratase-dehydrogenase, which catalyzes the consecutive hydration and oxidation steps, thereby filling the last gap in this pathway.85
C bond of the side chain, oxidation of the resulting alcohol to give a β-keto function, and a reverse aldol cleavage to release the C2 fragment as acetyl-CoA (Fig. 20). These steps proceed via CoA-bound intermediates in the peroxisome. The most recently characterized enzyme was the cinnamoyl-CoA hydratase-dehydrogenase, which catalyzes the consecutive hydration and oxidation steps, thereby filling the last gap in this pathway.85
A non-oxidative pathway for benzenoid formation is also known that follows a similar chemical logic, but the reverse aldol cleavage leads to benzaldehyde (88) rather than benzoyl-CoA, which is then oxidized to benzoic acid (Fig. 20). The non-oxidative sequence occurs in the cytosol with free or CoA-bound intermediates. According to flux data from petunia flowers, both the β-oxidative and non-oxidative pathways participate in the formation of benzenoid volatiles with their contributions depending on light conditions and time of day.84
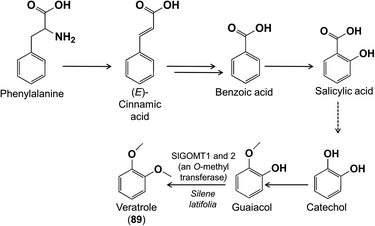
A benzenoid with two O-methyl groups is veratrole (1,2-dimethoxybenzene) (89) found in the flowers of Silene latifolia (Caryophyllaceae). Feeding of potential precursors established that this benzenoid is derived from phenylalanine via (E)-cinnamic acid, benzoic acid and salicylic acid.86 Decarboxylation results in catechol,87 which is then O-methylated sequentially to give 1,2-dimethoxybenzene (Fig. 21).86,87 The activities of the two O-methylation steps have been demonstrated,86,87 and one of the genes identified.86,88
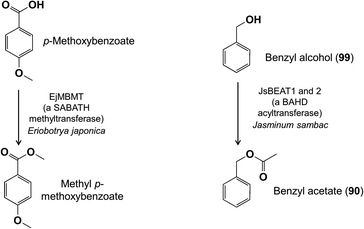
An O-methyltransferase has also been identified from the flowers of loquat (Eriobotrya japonica, Rosaceae) that reacts preferentially with phenylpropanoids and benzenoids containing para-OH and meta-OCH3 functions in vitro and methylates the free OH to add a para-OCH3 group to this ring system.89 Loquat flowers also possess a carboxylate-methylating activity from the SABATH family that converts p-methoxybenzoic acid to its corresponding methyl ester (Fig. 22).90 The enzyme has broad specificity and also methylates benzoic acid and jasmonic acid. Meanwhile, a BAHD acyltransferase was identified in Jasminum sambac flowers that reacts with benzyl alcohol and acetyl-CoA to form benzyl acetate (90) (Fig. 22).91
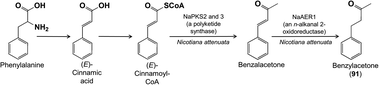
A more elaborate benzenoid is benzylacetone (91) from the flowers of Nicotiana attenuata. Transcript analysis of RNA isolated from single corolla cells and gene co-expression studies helped suggest a biosynthetic pathway that could be confirmed by silencing candidate genes.92 Based in part on previous studies on benzylacetone93 and other phenylbutanoids,94 the pathway was found to proceed from phenylalanine via (E)-cinnamic acid and (E)-cinnamoyl-CoA. Next comes a single carbon extension: a condensation reaction of (E)-cinnamoyl-CoA and malonyl-CoA catalyzed by a polyketide synthase with an additional loss of CO2 (besides the typical loss of one molecule of CO2 from malonate in polyketide synthase catalysis). This is followed by a final reduction to give benzylacetone (Fig. 23).
3.4 Other amino acid derivatives
Relative to other classes of floral volatiles, few reports have appeared on the biosynthesis of amino acid derivatives. Most of these feature aldoximes, well known from the floral scent of species of Oenothera (Onagraceae)95 and Angraecum sesquipedale (Orchidaceae),96 and as herbivore-induced volatiles from vegetative plant parts and biosynthetic intermediates in the formation of anti-herbivore defenses such as cyanogenic glycosides. Aldoximes are all derived from amino acids under the catalysis of the CYP79 family of cytochrome P450 enzymes.97,98 The most common substrates are the branched chain amino acids, valine, leucine and isoleucine, as well as phenylalanine and tyrosine, while both (E)- and (Z)-configurations are present in the products. Once formed, the isomers of phenylacetaldoxime (85) are potential precursors of other nitrogen-containing floral volatiles, including 2-phenylacetonitrile (benzyl nitrile, benzyl cyanide) (92) and 2-phenylnitroethane (93), based on feeding experiments in Plumeria rubra flowers (Fig. 24).74 The conversion of phenylacetaldoxime to 2-phenylacetonitrile is catalyzed by another family of P450 enzymes, the CYP71s, though this has only been demonstrated in connection with vegetative volatiles99 and cyanogenic glycoside formation,98 and not yet in flowers. Floral phenylacetaldoximes are also precursors of 2-phenylethanol and other benzenoid compounds in flowers, as described above in 3.3, but the reactions involved have not yet been elucidated.96Intriguing reports describe an amino acid-derived floral volatile that also has a role in the biosynthesis of a floral pigment. The volatile indole (94), derived from the tryptophan biosynthetic pathway, is released directly from the flowers of certain poppy species,100 and also appears to combine with flavonoids to form the unique pigments called nudicaulins101via reactions that still need investigation.
4 Regulation of floral volatile emission
In the period under review, nearly as many new findings have been published on the regulation of floral volatile biosynthesis as on the biosynthetic pathways themselves. Knowledge about regulation has given new insights into how floral volatile pathways operate in the plant as well as providing additional tools to manipulate volatile emission.4.1 Transcription factors
The majority of the regulatory studies have focused on transcription factors, which have been found to be critical in controlling the time and place of volatile biosynthesis and hence emission in flowers. Transcription factors limit synthesis to particular floral organs, stages of development and times of day. The first transcription factor identified as controlling floral volatile emission was ODORANT1 (ODO1) from petunia (Petunia x hybrida), which stimulates the formation of the phenylpropanoid and benzenoid compounds that dominate petunia scent.102 First discovered by comparing the transcripts of scented and non-scented petunia lines, this R2R3-MYB family transcription factor was also identified as a regulatory element in a quantitative genetics study involving crosses between two petunia species with big difference in their floral volatile spectra.103 After ODO1 was reported, a second MYB transcription factor, EMISSION OF BENZENOIDS II (EOBII), was observed to stimulate volatile biosynthesis in petunia and its expression was also correlated with the peak of emission.104 Research revealed that EOBII stimulates ODO1 expression by binding to the ODO1 promoter.105 Further complexity comes from a third MYB transcription factor in petunia flowers, EOB1, which is upstream of EOBII and ODO1 and activates both of them, while ODO1 inhibits EOBI expression.106 These transcription factors activate genes of phenylpropanoid and benzenoid biosynthesis as well as earlier metabolic steps, such as the shikimate pathway, which produces phenylalanine, the precursor to nearly all petunia flower volatiles.104,106,107 Still other MYB transcription factors seem to be involved in stimulating volatile formation in petunia flowers that target genes other than biosynthetic genes.108 Petunia transcription factors have also been discovered that negatively regulate floral emission. A member of the ethylene response factor (ERF) family binds to EOBI and so inhibits its binding to promoters of floral biosynthetic genes.109 Hence ethylene downregulates volatile formation in petunia. Other hormones that suppress volatile formation include gibberellins and auxin. Gibberellin signaling is blocked by a DELLA transcription factor that activates EOBII,110 while auxin suppresses volatile emission by decreasing phenylalanine levels.111MYB transcription factors regulate floral volatile formation in other plant species as well. A MYB called PAP1, known to activate phenylpropanoid biosynthesis in A. thaliana, increases phenylpropanoid volatile formation in roses.112 Terpenoid volatiles are also influenced by MYB transcription factors in plants, such as roses,112Freesia x hybrida and A. thaliana.113 For example, several different MYB factors regulate the formation of terpene synthase products in F. x hybrida and A. thaliana by binding directly to the promoters of terpene synthases. An antagonistic effect on volatile formation is caused by the basic helix-loop-helix (bHLH) transcription factor MYC2, which blocks MYB induction of monoterpene synthases, but stimulates expression of sesquiterpene synthases.113 The expression of MYC2 in turn is induced by both jasmonic acid and gibberellin signaling pathways.114 In the orchid Dendrobium officinale, a jasmonate-responsive bHLH transcription factor was also reported to regulate the expression of a monoterpene synthase gene, but here the bHLH transcription factor promoted the expression of this gene, which encodes linalool (60 and 61) formation, instead of blocking it.115
An increasingly popular species for floral volatile studies is the white ginger lily (Hedychium coronarium), which produces a range of terpenoid and benzenoid volatiles. MYB transcription factors also play a role in regulating volatile emission here.116 For example, H. coronarium MYB factors were identified that bind to the promoter regions of floral biosynthetic genes after their expression is activated by auxin.117 Another H. coronarium transcription factor binds to the promoter region of an (E)-β-ocimene synthase,118 which produces a major floral volatile of H. coronarium.
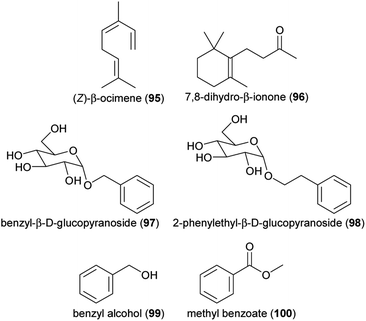
Terpenoids, such as linalool (60 and 61), (Z)-β-ocimene (95) and β-ionone (82), are dominant floral volatiles in another species well-investigated for floral volatiles, sweet osmanthus (Osmanthus fragrans). Here, genomic and transcriptomic studies suggested that R2R3-MYB and WRKY transcription factors play a role in controlling volatile emission.119,120 MYB transcription factors were reported to be involved in regulating terpene emission in a cultivar of lily (Lilium sp.) as well, with one MYB demonstrated to bind to the promoter region of an ocimene synthase.121 Lilies also contain a MYB similar to petunia ODO1 that upregulates genes involved in volatile phenylpropanoid and benzenoid biosynthesis and the shikimate pathway.122
4.2 Other factors regulating biosynthesis
The pattern of gene expression is usually assumed to exert a dominant influence on the quantity and composition of floral volatiles formed. Support for this assertion comes not only from the many transcript studies and the impact of transcription factors, but also from the results of artificial selection experiments on the floral volatiles of the fast-cycling Brassica rapa plants.123 After only three generations of selection for increased emission, elevated amounts of phenylpropanoid and benzenoid volatiles were evident and these were shown to arise from elevated expression of genes of the phenylpropanoid pathway and earlier stages in biosynthesis, such as the shikimate pathway, which supplies the precursor phenylalanine. This amino acid is often a limiting factor in phenylpropanoid and benzenoid volatile formation as emphasized by construction of transgenic petunia lines with increased phenylalanine supply due to overexpression of a bacterial shikimate pathway enzyme that is insensitive to feedback inhibition from phenylalanine.124 These lines not only accumulated higher amounts of phenylalanine but also emitted higher amounts of volatiles than wild-type lines. Moreover, direct feeding of phenylalanine to cut flowers also increased the emission of phenylalanine-derived volatiles.125 The importance of primary metabolite precursors in regulating flower volatile formation was also highlighted by research on a Lilium cultivar where glucose and fructose application increased the expression of genes for terpenoid volatile formation and terpenoid emission.126 There was a regulatory context in this study too, since sugar kinases found to be highly expressed in flowers increased volatile emission when overexpressed and decreased emission when silenced.Gene expression is also controlled by epigenetic influences, including DNA methylation, histone modification and chromatin remodeling. Using the petunia model system, research has found that histone acetylation during flower anthesis facilitated the activation of specific genes for the biosynthesis of the phenylpropanoid and benzenoid volatiles of petunia flowers.127 Included in this list are genes encoding the main steps of the phenylpropanoid pathway plus those for the shikimate pathway. At the same time, genes encoding enzymes in pathways that compete for phenylalanine, such as those involved in the formation of flavonoids and lignin, were repressed. Long, non-coding RNAs may also influence gene expression at transcriptional and post-transcriptional stages. A recent investigation on roses implicated several long, non-coding RNAs in controlling floral volatile release from Rosa x hybrida.128 Silencing of one such sequence increased the emission of the apocarotenoid, 7,8-dihydro-β-ionone (96).
4.3 Factors regulating storage, transport and emission
Once synthesized, floral volatiles have been generally assumed to be emitted from floral tissues into the atmosphere by simple diffusion through cell membranes. Thanks to recent breakthroughs in our knowledge, however, it is now clear that storage and transport can also regulate emission, while simple diffusion plays only a minor role. Based on the chemical properties of typical floral volatiles, Natalia Dudareva and coworkers129 calculated that simple physical diffusion would require such high concentrations to build up in volatile-producing cells that these would become toxic, suggesting emission must occur instead via biological carriers. The Dudareva laboratory did indeed discover such a carrier, identifying a transport protein located in the plasma membranes of petunia flower cells that exports volatiles.130 As a member of the ATP-binding cassette (ABC) transporter family closely related to previously described wax transporters, this protein was found to be specific for benzenoids. It was present almost exclusively in the cells of open flowers, and expression of the corresponding gene was controlled like so many other volatile biosynthetic genes in petunia by the transcription factor ODORANT1. Interestingly, silencing of the transporter not only reduced emission and increased accumulation of volatiles in flowers, but also caused signs of cell damage due to the build-up of volatiles to toxic levels in living cells.After volatiles traverse the plasma membrane, the cuticle of floral organs also acts as a barrier to emission. The cuticle, however, has an unexpectedly complex role in the emission process serving also as a sink for volatiles prior to emission that protects cells from toxicity. Curiously, when cuticle thickness in petunia was reduced by silencing a petal wax transporter, the resulting flowers had lower rather than greater volatile emission rates since the reduction in sink size exerted negative feedback on volatile biosynthesis.131 Cuticular resistance was greater for petunia volatiles of lower volatility, such as benzyl alcohol (99) and eugenol (87), which have large storage pools relative to their emission rates. In contrast, higher volatility compounds, such as methyl benzoate (100) and benzaldehyde (88), have smaller storage pools relative to emission rate and emission more closely matches their rate of biosynthesis. The existence of additional control points between floral volatile synthesis and emission was suspected for many years based on the isolation of glycosylated volatiles from various plant organs. From petunia flowers, for example, glycosylated phenylpropanoids and benzenoids, including benzyl glucoside (97) and 2-phenylethyl glucoside (98) have been reported.132 Time course studies and isotopic labeling demonstrated that these glycosides form dynamic pools, and so likely represent storage forms of volatiles with glycosylation and glycoside hydrolysis helping to modulate release rates.
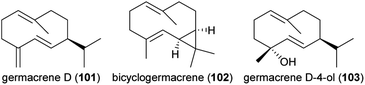
Dudareva and coworkers also discovered an unexpected new dimension in volatile storage and transport between the organs of petunia flowers.133 A blend of sesquiterpene volatiles produced by the petal tube were found to be released inside the bud and to accumulate on the thick waxy cuticle of the stigma prior to floral opening. The compounds, germacrene D (101), bicyclogermacrene (102), and germacrene D-4-ol (103), the major products of a single sesquiterpene synthase expressed in the petal tube, were all mobilized to the stigma where they were shown to promote the development of this organ and enhance subsequent seed yield compared to plants in which this terpene synthase was silenced by RNA interference. These sesquiterpenes may also act to reduce the pathogen load on stigmas. Hence, greater understanding of floral volatile transport and storage can furnish significant new insights on the roles of floral volatiles in the plant. These phenomena can also explain some of the inconsistencies between the results of headspace sampling and solvent extraction, and provide reasons for why expression of biosynthetic genes is sometimes poorly correlated with emission.
5 Effects of abiotic and biotic factors on floral scent emissions
Though floral scents are regarded as being species-specific,4 they also vary among individuals within species.134 Some of this variation has a genetic basis,135 while other variation is due to phenotypic plasticity and is shaped by the environment.136 Both biotic and abiotic factors affect floral scents,137 and here, we, focus on recent studies that demonstrated effects of air pollutants, increased temperature and drought, soil nutrient availability, bacterial, fungi and herbivores on floral scent emissions and on the chemical communication with pollinators.5.1 Air pollution
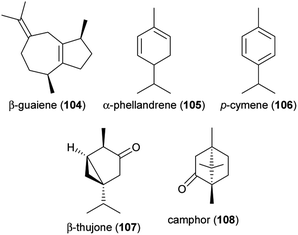
Air pollutants result in lower abundances of pollinators and smaller numbers of flower visits, with negative effects on the seed set of plants.138 One mechanistic explanation for this finding is that air pollutants affect floral scents, the perception of floral scents by pollinators, and thus, the olfactory communication between plants and pollinators. Such effects have been studied over the last decade by fumigating growing plants and pollinators with pollutants (e.g., ozone, diesel exhaust), or by applying the pollutants to scents after their release by the flowers.139Elevated ozone exposure during growth can affect the emission of some floral volatiles, as shown for a few plant species. After 35 days of elevated ozone exposure, roses (Rosa spp.) released higher amounts of sesquiterpenes, while monoterpene emissions were not affected.140 Following 90 days of exposure, the total amount of scent decreased, mainly due to decreased emissions of monoterpenes. There was no effect on total sesquiterpene emissions following this long-time exposure, despite an increase in the sesquiterpene guaiene (104).140 Saunier and Blande141 exposed four Brassicaceae species to elevated ozone levels. None of them showed changes in total scent emissions; however, the emission rates of single compounds changed in three of the four species. The exposure to elevated ozone levels resulted in higher amounts of α-phellandrene (105) in Brassica nigra (black mustard), decreased emission or even a loss of p-cymene (106) in Sinapis alba (white mustard), and a decreased emission of α-pinene (72) and losses of β-thujone (107) and camphor (108) in Brassica napus (oilseed rape). Elevated ozone exposure did not affect the emission of single compounds in S. arvensis (wild mustard).
A recent study showed that increased CO2 levels (800 ppm, predicted as the atmospheric concentration for 2100) also result in altered floral scents.142 CO2 fertilization decreased total scent in Campanula rotundifolia (Scottish bluebell) and increased total scent in Phacelia hastata (silverleaf phacelia). Furthermore, scent composition was altered following CO2 enrichment. In C. rotundifolia 6-methyl-5-hepten-2-one (109), β-pinene (73) and β-myrcene (76), and in P. hastata linalool (60 and/or 61), limonene (75) and (Z)-3-hexenyl acetate (110) were emitted at higher rates from CO2-fertilized plants compared to control plants. In two other study plants (Heterotheca villosa, the hairy goldenaster and Potentilla recta, the sulphur cinquefoil), CO2 enrichment affected neither total scent emission nor scent composition.
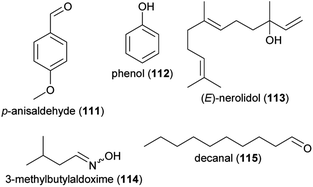
In the examples above, plants were exposed for some time to air pollutants, while floral volatiles were collected at ambient conditions. Other studies, however, exposed the released floral volatiles directly to air pollutants. Volatiles are exposed to many airborne compounds after their release from the flower, among them pollutants in the atmosphere. Several air constituents (e.g., ozone, NO3 radicals) are known to directly interact with floral volatiles and change the composition of scent blends.143–148 Recent advances have been made in quantifying the effects of ozone and diesel exhaust on floral volatiles and how this influences pollinator behavior. Studies on four Brassicaceae species demonstrated that ozone partly degrades some of the floral scent compounds released by the flowers, while others were not affected by ozone.141,149,150 In Brassica nigra, for example, strong negative effects were recorded for p-anisaldehyde (111), phenol (112) and p-cymene (106), as well as for total monoterpenes. Overall, increased levels of ozone affected the relative scent composition in this species, resulting in a decreased attractiveness of floral scent to bumblebee pollinators.149 Negative effects of increased ozone levels on the attraction of pollinators were also shown for the tobacco hawkmoth Manduca sexta and one of its preferred host plants, the jasmine tobacco Nicotiana alata.151 Here, increased ozone levels again affected flower scent composition resulting in drastically reduced amounts of linalool (60 and/or 61), (E)-nerolidol (113), and 3-methylbutylaldoxime (114), but in increased amounts of decanal (115). These changes in scent composition reduced the time hawkmoths spent at the ozone-altered scent sources in wind tunnel bioassays. However, moths were able to learn the ozone-altered blend, which might reduce the negative impact increased ozone levels have on the communication in this pollination system.151 Ozone also affected the flower–florivore interaction between Cucurbita foetidissima (buffalo gourd) and the striped cucumber beetle (Acalymma vittatum). The floral scents, which were not chemically analyzed in detail, were less attractive to the beetle when they were exposed to increased ozone levels.152
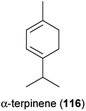
Diesel exhaust (mainly due to nitrogen oxides) affected different well known floral scent compounds differently, based on experiments with two synthetic blends consisting overall of eleven terpenoids and five aromatic compounds.153,154 Exhaust had negative effects on seven terpenoids and two aromatic compounds. The compounds α-terpinene (116), (E,E)-α-farnesene (63), β-ocimene (62) and (E)-β-caryophyllene (64) were most severely affected, as they became undetectable when exposed to diesel exhaust. 64 was completely transformed into its cis-isomer. The amount of only one compound [p-cymene (106)] increased during the experiments, whereas the other six compounds [e.g., α-pinene (72), benzaldehyde (88)] were not affected by diesel exhaust. The alterations of the two diesel exhaust blends reduced the ability of Western honeybees (Apis mellifera; henceforth honeybee) to remember the original blend to which they had been trained.153,154
The chemical degradation of floral volatiles depends on their reactivity with respect to the individual pollutants (e.g., ozone, NO3 radicals).146 Some of the above described results can indeed be explained by the reactivity of the volatiles (e.g. strong ozone effects on linalool), but others not (strong ozone effects on p-cymene), and more research is needed to better understand the observed effects of air pollutants on floral volatiles. Furthermore, it would be interesting to know whether selection is currently occurring on pollinators to rely more on compounds less reactive during the Anthropocene.
Studies on how the perception, learning and memory of flower volatiles is affected when insects are exposed to air pollutants were also performed using ozone. Exposure of honeybees, bumblebees (Bombus terrestris) and fig wasps (Blastophaga psenes) to increased levels of ozone had variable effects on the perception of volatiles in the olfactory circuitry. The strength of the physiological response to a specific compound either decreased [e.g., linalool (60 and/or 61) in fig wasps, benzaldehyde (88) in bumblebees], increased [e.g. benzyl alcohol (99) after short-time ozone exposure of fig wasps], or was not affected [e.g., linalool (60 and/or 61) and 2-phenylethanol (84) in honeybees; 99 after long-time ozone exposure of fig wasps].155–157 In honeybees, ozone exposure not only influenced the perception of floral volatiles, but also resulted in less efficient olfactory learning.155
5.2 Temperature and drought
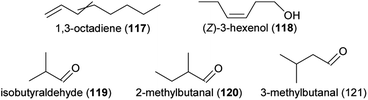
Due to climate change, global mean surface temperatures and the rate as well as the intensity of droughts are increasing across many regions worldwide,158 effecting plant reproduction159 and floral scents (reviewed in Borghi et al.,159 Gérard et al.160). Recent studies confirmed previous results and consistently found that total scent emissions in various plant species are temperature dependent, with maximum emissions typically between 25 °C and 30 °C.161–165 However, in some species flowering during summer in Mediterranean climates, scent maxima were reached at temperatures as high as 40 °C.163 In addition to the variation in total scent, temperature also effected scent composition. Both the number of compounds released163,165 and the absolute as well as relative amounts of single compounds varied among different temperatures.161–163,165–167 The finding that different compounds respond differently to temperature could be traced back to the activity of enzymes involved in the biosynthesis of the compounds in studies on the jasmine Jasminum auriculatum and on Petunia x hybrida.108,161 In J. auriculatum, for example, the enzyme responsible for the production of benzyl acetate (90) had a higher activity at 25 °C than at 30 °C, whereas the enzyme responsible for linalool (60 and/or 61) production had a high activity at both temperatures. These resulted in 90 emissions that peak at 25 °C, whereas high amounts of linalool (60 and/or 61) were emitted both at 25 °C and 30 °C.161In most studied species, drought affected total scent emissions and/or scent composition. Drought increased the total amount of floral scent in Ipomopsis aggregata,168Campanula rotundifolia and Potentilla recta,142,169 but had no effects on total amount of scent in Phacelia hastata,142,169Fagopyrum esculentum (buckwheat)170 and Matthiola livida.171 Regarding scent composition, different compounds were responsible in the various study species for differences between drought and control plants.142,168,169,171,172 For example, in Ipomopsis aggregata, 1,3-octadiene (117) and benzyl alcohol (99) were released in higher amounts in wetter than drier conditions, whereas α-pinene (72), (E)-β-ocimene (62) and (E,E)-α-farnesene (63) were released in highest amounts in drier conditions.168
5.3 Nutrient availability and soil type
Research findings on the effects of nutrient availability and soil type on floral scent emission have become available only in the last few years. In Lithophragma bolanderi, nutrient availability (N, P, K) did not have effects on scent emission,175 whereas in Petunia hybrida, only eugenol (87), out of the 10 volatiles analyzed, increased with increasing nitrogen availability.176 In Arabis alpina, plants with high nutrient availability (N, P, K, Mg, micronutrients) produce more scents, with scent composition not being influenced by nutrient availability.174 A very recent study on Brassica rapa did not find an effect of soil type (limestone versus tuff) on floral scent emissions.1775.4 Bacteria and fungi
In the last decade, there were major advances in our understanding of how microorganisms influence floral scents and the chemical communication between flowers and their visitors. Microorganisms considered were arbuscular mycorrhizal fungi, plant endophytes, such as bacterial pathogens, and especially bacteria and yeasts colonizing flowers.In Polemonium viscosum, resource allocation to scent production vs. to mycorrhizae was shown to be a tradeoff, and when fungicide was used to eliminate mycorrhizae as partners, scent production increased (number of compounds emitted and total scent).178 In strawberry cultivars, mycorrhizal fungi did not have effects on floral scents.179
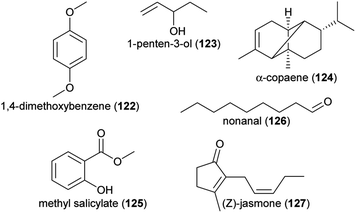
The bacterial plant pathogen Erwinia tracheiphila that causes a fatal wilt disease in wild gourd (Cucurbita pepo ssp. texana), lowered total scent emissions, especially due to an attenuated release of linalool (60 and/or 61), 1,4-dimethoxybenzene (122), and a nonatriene.180Erwinia amylovora, causative agent of the fire blight disease, changed scent emission of apple flowers, which made flowers less attractive to honeybee pollinators.181 Compounds characteristic for apple flowers inoculated with Erwinia amylovora were, among others, 1-penten-3-ol (123), α-copaene (124) and methyl salicylate (125), whereas healthy flowers were characterized by benzaldehyde (88), benzyl alcohol (99), 2-phenylacetonitrile (92), nonanal (126) and (Z)-jasmone (127). Among other potential reasons, the observed changes in scent emission following pathogen attack might be due to the E. amylovora-induced compounds representing defenses of the plant against the pathogen or the use of some but not other volatiles as a carbon source by the pathogen.182
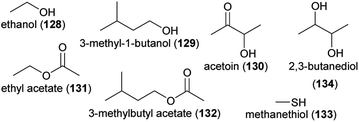
Flowers are colonized by a high number of bacteria and fungi,183,184 and several recent review articles summarized how flower visitors disperse microbes among flowers, which effects flower microbes have on plant fitness, and how flower microbes affect floral phenotypes, such as floral scents, and flower–animal interactions.182,185–189 There is evidence that microbes dwelling in nectar and also microbes colonizing flower or other plant surfaces change the olfactory display of flowers, sometimes with consequences for the behavior of flower visitors. A high number of volatiles is released from nectar dwelling bacteria and fungi, among them various aliphatic alcohols and esters, such as ethanol (128), 3-methyl-1-butanol (129), acetoin (130), ethyl acetate (131), and 3-methylbutyl acetate (132), but also compounds from other chemical classes, such as the aromatic compound 2-phenylethanol (84) and sulfur-bearing methanethiol (133).190–194 Many of these compounds were found to elicit physiological responses in the antennae of pollinators,191,193 and scents released from several nectar dwelling microorganisms were found to be neutral, attractive or repellent to bee pollinators191–193,195,196 or to other flower visitors, such as parasitoids.194,197 Thus, compounds released from nectar-dwelling microorganisms mediate foraging behavior of flower visitors. As shown for Aphidius ervi, an aphid parasitoid that uses nectar as food source, feeding on nectar that contains specific nectar-dwelling microorganisms has negative effects on survival.194
When compared to nectar-dwelling microorganisms, there is much less known about plant-surface inhabiting microorganisms. Helletsgruber et al.198 inoculated Brassica rapa plants with bacterial strains naturally growing on leaves and flowers of this plant species and compared the floral scents of these inoculated plants with the floral scent of plants grown under sterile conditions. They found that the scents strongly differed between the treatments, especially due to higher emission rates of acetoin (130) and 2,3-butanediol (134) in inoculated plants. Peñuelas et al.199 fumigated Sambucus nigra plants with antibiotics and found that fumigation did not affect terpenes in the floral tissue, but negatively affected floral terpene emissions. They concluded that phyllospheric microorganisms contribute to floral terpene emissions and possibly affect the olfactory attraction of thrips pollinators.
5.5 Herbivores
The first studies shedding light on the effects of herbivory/florivory on floral scents and pollinator behavior, were published less than two decades ago. reviewed by 137,200–204 Meanwhile, there are some studies available that link herbivory on leaves and flowers to floral chemistry and pollinator attraction. They have found that herbivory/florivory often, but not always,169,205 has effects on floral scent emissions,177,206 and that different herbivores can have different effects on floral scent emissions.207–209 Further, herbivory-induced changes in scent emissions often translate to differences in flower visitor behaviors169,171,203,207,210–213 and plant reproduction,214but see211 and herbivores exert selective pressures on plants and affect the evolution of floral scents.215,216The results of Kessler et al.213 on herbivore-induced changes in the volatile emission of Nicotiana attenuata, a night-flowering tobacco, are especially remarkable. This species normally releases high amounts of scent, mainly benzylacetone (91), at night to attract nocturnal hawkmoth pollinators, such as Manduca quinquemaculata and M. sexta. Interestingly, these moths not only visit the flowers for nectaring, but females also lay eggs on the leaves, which serve as food for the larvae. When plants are attacked by hawkmoth larvae, they open their flowers in the morning instead of dawn and emit strongly reduced amounts of benzylacetone, likely to avoid further herbivory by hawkmoth larvae. Such plants are then less frequently pollinated by hawkmoths at night, but mainly by hummingbirds during daytime.
In Datura wrightii, feeding of Manduca sexta larvae on leaves did not affect total flower scent emissions, even though the emission rates of methyl benzoate (100) and geraniol (59) were increased.206 Other compounds, such as the ones responsible for attraction of Manduca sexta to flowers [benzaldehyde (88), benzyl alcohol (99), and linalool (60 and/or 61)] were not affected by herbivory.
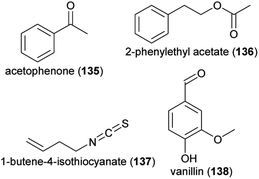
In Brassica rapa, leaf herbivory by both Pieris brassicae and Spodoptera littoralis larvae resulted in reduced scent emission and changed scent floral scent compositions.209 Herbivory by both lepidopteran species reduced amounts of 6-methyl-5-hepten-2-one (109), phenylacetaldehyde (83), acetophenone (135), 2-phenylethanol (84), 2-phenylethyl acetate (136) and decanal (115). Herbivory by P. brassicae additionally reduced p-anisaldehyde (111), whereas this compound increased following herbivory by S. littoralis. Benzaldehyde (88) was (negatively) affected only by P. brassicae. Herbivory by aphids on this plant species also affected floral scent emissions.177 The amounts of 88, 111, methyl benzoate (100) and methyl salicylate (125) were decreased, whereas 1-butene-4-isothiocyanate (137) and (Z)-3-hexenyl acetate (110) increased. For B. rapa, it was also shown that herbivory not only has direct effects on floral scent emissions, but also influences the evolution of floral scents.215,216 In the absence of herbivores, such as Pieris brassicae larvae, but not when herbivores were present, higher amounts of 88, 2-phenylacetonitrile (92), 111, 83, and 100 evolved within a few generations when plants were under selection of bee pollinators. This suggests that there is a trade-off between the attraction of pollinators and the deterrence of herbivores.
In white mustard, Sinapis alba, herbivory by two aphid species (Lipaphis erysimi, Myzus persicae) but not by the moth Plutella xylostella resulted in decreased scent emissions.207 Aphid herbivory resulted in reduced emissions of benzaldehyde (88), benzyl alcohol (99), p-anisaldehyde (111), and methyl salicylate (125). In contrast, 99 and vanillin (138) increased following attack by Plutella xylostella. Attack by aphids did not affect pollinator attraction, whereas an aphid predator, the ladybird Coccinella septempunctata, preferred flower scent of undamaged plants over that of aphid damaged plants, and the aphid parasitoid Diaeretiella rapae preferred flower scent of aphid-damaged plants over that of undamaged plants.207
6 Floral volatiles as attractants for flower visitors
In the last decade, there were major advances in our understanding of the floral volatile constituents involved in the communication between plants and their pollinators. For some pollinator groups (e.g., rodents, Cyclocephalini scarab beetles), the first scent attractants were identified and, for other groups (e.g. flies, bees, moths, and butterflies), new attractants were added to already known ones. In moths, it was demonstrated that olfactory receptors on the tip of the proboscis affect foraging behaviors. We focus here on chemicals that were shown to be biologically active in pollinators and florivores, i.e. elicit behavioral or at least physiological (e.g. stimulate the olfactory circuitry) responses in flower visitors. We especially consider pollinator and flower visitor groups where most advances were made recently, and exclude examples involving orchids, given that orchid volatiles and their importance in attracting pollinators were considered in a recent review.18 First, we will summarize recent studies on beetle attractants, then focus on moths and butterflies, flies, bees and wasps, and finally some groups of flower visitors with only small numbers of species known to interact with flowers.6.1 Beetles
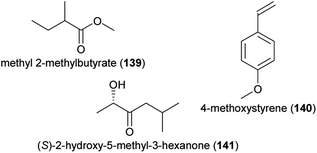
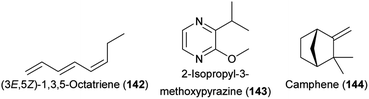
Beetles are the second largest group of pollinator species (following lepidopterans)217 and they are important pollinators of cycads218 as well as of various angiosperms, among them species of early diverging lineages.219 Some beetles are involved in highly specialized pollination systems (e.g. cyclocephalines, weevils), others are generalist pollinators (e.g., oedemeridae), and some beetles are major florivores (e.g., some cyclocephalines). For all of them, attractive floral volatiles were identified in the last decade.Cyclocephaline scarab beetles are a species-rich (c. 500 species) tribe of rhinoceros beetles and some species in this tribe are highly specific pollinators of various neotropical plant species.220,221 Plants pollinated by these beetles have a nocturnal anthesis and thermogenesis, and release extremely strong scents.220 The chemistry of plants pollinated by cyclocephaline beetles is highly variable, as are the compounds involved in the attraction of the beetles, and several new and uncommon floral scents have been identified while studying the chemical communication in these pollination systems. In 2012, the first attractants of cyclocephaline pollinators were identified. Methyl 2-methylbutyrate (139) strongly dominated the flower scent of Magnolia ovata (Magnoliaceae) and was attractive to the single pollinator species of this plant, Cyclocephala literata.222 4-Methyl-5-vinylthiazole (48), a new floral compound that strongly dominated (97–100%) the scents of Annona species (Annonaceae) and was a major compound in Caladium bicolor (Araceae), successfully attracted their pollinators (Cyclocephala atricapilla, C. celata, C. vestita).34 Finally, 4-methoxystyrene (140) from Philodendron form selloum attracted its single pollinator species, Erioscelis emarginata.223 The compound (Z)-jasmone (127) increased the attractiveness of this methoxylated aromatic compound. In 2013, Maia et al.224 identified (S)-2-hydroxy-5-methyl-3-hexanone (141), a rare floral scent, and 7,8-dihydro-β-ionone (96), a more widespread floral scent, as main compounds in the scent of Taccarum ulei (Araceae) and demonstrated that only 141 was attractive to the beetle pollinators (Cyclocephala cearae, Cyclocephala celata).
In 2014, Pereira et al.225 identified 7,8-dihydro-β-ionone (96) as a strongly dominating compound and methyl jasmonate (58) as a minor compound in the scent of Philodendron adamantinum (Araceae) and evidenced that both compounds alone, but especially in a binary mixture, elicit behavioral responses in Erioscelis emarginata, the single beetle pollinator. Just recently, Maia et al.11,226 and Stamm et al.227 identified (S)-dehydrojasmone (5) as a novel natural product and (1S,2S)-isojasmol (3) as a rare floral scent both from the scent of Thaumatophyllum (Philodendron) mello-barretoanum (Araceae), as attractants for C. gravis and C. amblyopsis, respectively.
Key advances were recently made not only in beetle pollination systems that involve Cyclocephalini, but also in other systems with beetles as the only pollinators or as co-pollinators. The African cycad Encephalartos villosus, involved in a highly specialized brood site mutualism mainly with Erotylidae sp. nov. and Porthetes sp. weevils, released the (3E)-isomer of 1,3-octadiene (117) and (3E,5Z)-1,3,5-octatriene (142) as well as 2-isopropyl-3-methoxypyrazine (143) that all elicited physiological responses in the antennae of the beetles.228 Field experiments proved that the two tested compounds 117 and 143 also elicited behavioral responses. In the related cycad E. ghellinckii, Metacucujus goodie (Boganiidae) beetle pollinators were also attracted by the (3E)-isomer of 117, whereas Erotylidae sp. nov. beetles were attracted by camphene (144).229 The weevil Rhopalotria furfuracea, a pollinator of the New World cycad Zamia furfuracea, was shown to be attracted by cone humidity.230
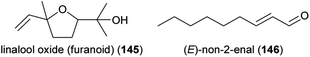
Atrichelaphinis tigrina, which is the most common cetoniine beetle pollinator of sugarbushes (Protea), responded to 15 compounds in electroantennographic analyses, and five thereof were used for behavioral field tests: benzaldehyde (88), methyl benzoate (100), methyl salicylate (125), linalool (60 and/or 61), linalool oxide (furanoid) (145).231 Only 100 and linalool elicited behavioral responses in the Atrichelaphinis tigrina beetles. Antennae of Oedemeridae beetles (Oedemera spp.), generalist pollinators of various plants, responded to several volatiles released from strawberry flowers: nonanal (126), decanal (115), (E)-non-2-enal (146), linalool (60 and/or 61) and 1,4-dimethoxybenzene (122).232
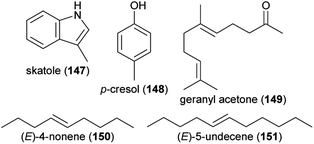
Rove beetle (Staphylinidae) pollinators were attracted by a blend of indole (94), skatole (147), p-cresol (148), 7,8-dihydro-β-ionone (96), geranylacetone (149) and β-ionone (82) to brood-site deceptive Typhonium eliosurum.233 Antennae of the rove beetle Pelecomalium testaceum, main pollinator of the aroid Lysichiton americanus (Western skunk cabbage) responded to (E)-4-nonene (150), (E)-5-undecene (151) and 94, whereas the two hydrocarbons together as well as indole successfully attracted the beetle pollinators in field experiments.234
The compounds α-pinene (72) (S![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) R 16
R 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), sabinene (74), myrcene (76), limonene (75) (S
1), sabinene (74), myrcene (76), limonene (75) (S![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) R 1
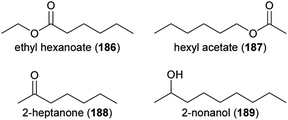
R 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
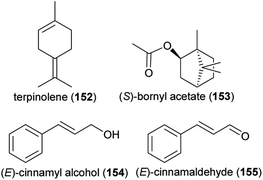
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3), terpinolene (152) and (S)-bornyl acetate (153), which are released from flowers of the wild carrot Daucus carota (Apiaceae), elicited physiological responses in antennae of Acanthoscelides obtectus (Coleoptera: Bruchidae), a global pest of dry beans that visits various plants for their nectar and pollen, among them the wild carrot.235 A synthetic mixture of these compounds was also attractive to this beetle species in lab bioassays, though less than the natural scent of the wild carrot. Another pest of Fabaceae, the bean seed beetle Bruchus rufimanus, was shown to respond to floral volatiles of its principal host plant, the field bean Vicia faba, in physiological and behavioral experiments,236 though the beetle did not necessarily pollinate the flowers. Nine floral scents were active in electroantennographic analyses, whereas a blend of three thereof, (R)-linalool (60), (E)-cinnamyl alcohol (154) and (E)-cinnamaldehyde (155), was as attractive to the beetles in field biotests as a blend of the nine compounds. Similarly, the ladybird beetle Harmonia axyridis visits Sophora japonica flowers to feed on pollen and nectar, with unknown effects on pollination.237 Physiological and behavioral experiments identified nonanal (126) as the floral scent responsible for the attraction of H. axyridis.237
3), terpinolene (152) and (S)-bornyl acetate (153), which are released from flowers of the wild carrot Daucus carota (Apiaceae), elicited physiological responses in antennae of Acanthoscelides obtectus (Coleoptera: Bruchidae), a global pest of dry beans that visits various plants for their nectar and pollen, among them the wild carrot.235 A synthetic mixture of these compounds was also attractive to this beetle species in lab bioassays, though less than the natural scent of the wild carrot. Another pest of Fabaceae, the bean seed beetle Bruchus rufimanus, was shown to respond to floral volatiles of its principal host plant, the field bean Vicia faba, in physiological and behavioral experiments,236 though the beetle did not necessarily pollinate the flowers. Nine floral scents were active in electroantennographic analyses, whereas a blend of three thereof, (R)-linalool (60), (E)-cinnamyl alcohol (154) and (E)-cinnamaldehyde (155), was as attractive to the beetles in field biotests as a blend of the nine compounds. Similarly, the ladybird beetle Harmonia axyridis visits Sophora japonica flowers to feed on pollen and nectar, with unknown effects on pollination.237 Physiological and behavioral experiments identified nonanal (126) as the floral scent responsible for the attraction of H. axyridis.237
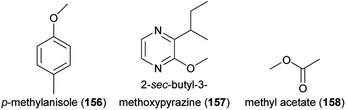
Some plants are not only attractive to beetle pollinators but also to beetle florivores, and in palms (Arecaceae) it was shown that both pollinators and florivores are attracted by the same compounds. The macauba oil palm (Acrocomia aculeata) attracts weevil pollinators of the genus Andranthobius (Curculionidae) by p-methylanisole (156), 2-isopropyl-3-methoxypyrazine (143) and 2-sec-butyl-3-methoxypyrazine (157). Thereof, 156 was also attractive to florivorous Cyclocephala forsteri, and the two pyrazines to another florivorous cyclocephaline beetle, Aspidolea bleuzeni.238 The acuri palm (Attalea phalerata) releases almost exclusively methyl acetate (158) to attract its putative main beetle pollinators (Mystrops spp., Nitidulidae; Andranthobius spp., Curculionidae) and this communication channel is exploited by non-pollinating florivorous beetles, such as palm borers (Paratenthras martinsi, Cerambycidae; Parisoschoenus sp., Belopoeus sp.; Curculionidae).239
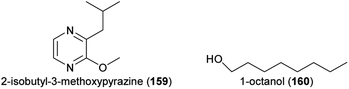
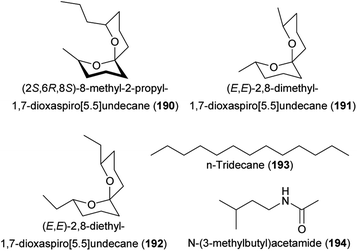
Finally, some recent studies focused only on florivorous beetles and identified the floral attractants. The (3S)-isomer of (E)-nerolidol (113) mediated the attraction of florivorous Cyclocephala paraguayensis beetles to bottle gourd (Lagenaria siceraria, Cucurbitaceae) flowers,240 and 2-isopropyl-3-methoxypyrazine (143) and 2-isobutyl-3-methoxypyrazine (159), major compounds of Acrocomia and Attalea palms, attracted Cyclocephala amazona and C. distincta florivores.241Aulacophora foveicollis (Coleoptera: Chrysomelidae), a beetle pest that feeds on flowers and leaves of two Curcurbitaceae species (Gâc, Momordica cochinchinensis; creeping cucumber, Solena amplexicaulis), was attracted by a blend of linalool oxide (furanoid) (145), 1-octanol (160), and nonanal (126).242 These compounds occurred in flowers of both species and the blend was as attractive as the natural scent. Coccinelidae beetle florivores (Epilachna dodecastigma) of the bitter gourd (Momordica charantia) were attracted to the flowers by sabinene (74), which was as attractive as the total flower scent.243 Studies with transgenic lines of Petunia x hybrida silenced in their ability to produce specific floral compounds revealed that methyl benzoate (100) is attractive to florivorous Diabrotica undecimpunctata.244
6.2 Moths, butterflies
Lepidoptera are the most species-rich group of pollinators,217 and recent advances were made in understanding the molecular basis of olfaction in these insects, i.e. the detection of floral volatiles by olfactory receptors,245,246 and the importance of specific molecules in the attraction of Lepidoptera to flowers. Here, we will summarize the main findings of studies that identified floral volatiles attractive to hawkmoth, settling moth and butterfly pollinators.Some research was also done on Hyles and Agrius hawkmoths. Chemical ecological studies on two closely related Ipomopsis (Polemoniaceae) species, I. aggregata and I. tenuituba, showed that floral scents contribute to the reproductive isolation of the two species. Hyles lineata pollinators were only attracted to I. tenuituba, and key for the selective attraction was indole (94), which was only released from I. tenuituba flowers.251 Quite different compounds seem to be involved in the attraction of H. livornica to flower scents of Dianthus inoxianus.252 Several aliphatic ketones (e.g., 2-undecanone, 2-tridecanone, 2-pentadecanone) and terpenoids [e.g., (E)-β-bergamotene (163), (E,E)-farnesol (164)] elicited physiological responses in antennae of the moths. In elegant studies, von Arx et al.253 and Dahake et al.254 demonstrated that approaching and landing behaviors of foraging H. lineata and M. sexta hawkmoths are influenced by floral humidity. Floral humidity gradients are due to passive (nectar evaporation) and/or active (stomatal conductance) processes and allow the moths to sense nectar availability from a distance and to forage efficiently.253,254 Finally, Agrius convolvuli responded to various compounds of its Gladiolus longicollis host plant, among them aromatic compounds [e.g. benzaldehyde (88), benzyl acetate (90) phenylacetaldehyde (83), eugenol (87)] and nitrogen-bearing compounds [3-methylbutylaldoxime (114), 2-phenylacetonitrile (92), 2-phenylnitroethane (93)].255
Epicephala moths are obligate nursery pollinators of several Phyllanthaceae and the first attractants were identified in 2010. Flowers of both sexes of monoecious Breynia vitis-idaea released similar scent compounds, though in sex-specific ratios, and several main compounds in at least one of the sexes [geranylacetone (149), 2-phenylethanol (84), 2-phenylacetonitrile (92)] but also minor compounds [2-phenylethyl acetate (136), indole (94)] elicited signals in antennae of the Epicephala moths.26284 and 92 were used in bioassays, which revealed that 84 but not 92 was attractive to the moths when tested alone. Still, only the mixture of both compounds was as attractive as the natural scent.262 Further studies in Phyllanthaceae species pollinated by Epicephala moths revealed that the scents are highly dimorphic between female and male flowers263–266 and that moths are able to discriminate between them.264,265 This allows the moths to carry out different behaviors on different sex flowers: pollen collection on male flowers, active pollination and oviposition on female flowers. Regarding the interaction between Yucca and yucca moths, it has been known for a while that plants release strong floral scents that are attractive to the moth pollinators.267 Just recently, however, the first Yucca compounds were identified that are biologically active in yucca moths. Among them are several compounds new for science.29 Antennae of Tegeticula mexicana pollinating moths and the parasitic, non-pollinating bogus yucca moth, Prodoxus tamaulipellus, responded in electrophysiological experiments to (Z)-filamentolide (24) of Yucca treculeana, whereas antennae of pollinating T. yucasella and T. cassandra, and parasitic P. decipiens responded to 24, (Z)-filamentol (22), (Z)-filamental (23), (E)-4,8-dimethyl-1,3,7-nonatriene (80), and (Z)-9-nonadecene (168) of Y. filamentosa. 24 was used for field bioassays and was capable of attracting parasitic Prodoxus species, but the attraction of pollinating yucca moths to their host plants still needs to be resolved in the future.29
6.3 Flies
Many plants rely on flies as pollinators,217,273,274 and, in the last decade, various flower volatile attractants were identified in mutualistic as well as in deceptive pollination systems, and new olfactory based strategies of deceptive plants that exploit flies as pollinators were introduced.Mosquitoes, among them species that are vectors of diseases (e.g. malaria, dengue), also frequently visit flowers for nectaring. Several recent reviews focused on (floral) volatiles attractive to mosquitoes and how such volatiles could be implemented in strategies for management of disease vectors.276–282 Here a few examples for studies that successfully identified compounds that elicit behavioral responses in mosquitoes are given. Otienoburu et al.283 analysed floral scents in the common milkweed Asclepias syriaca, which is visited by the northern house mosquito, Culex pipiens, and demonstrated that a blend of benzaldehyde (88), phenylacetaldehyde (83), and (E)-2-nonenal (177) is key in the attraction of the mosquitoes to the flowers. Similarly, acetophenone (135) from sweet alyssum (Lobularia maritima) flowers successfully attracted the yellow fever mosquito, Aedes aegypti.284 In a remarkable study, Peach et al.285 demonstrated that the northern house mosquito and the yellow fever mosquito are not only attracted by the floral scents (and visual plant cues) of the common tansy (Tanacetum vulgare), but also by plant-derived CO2 (178). After sunset, plants became net producers of 178, which increased the attractiveness of a synthetic floral blend that consisted of 20 compounds, among them benzaldehyde (88), acetophenone (135), artemisia ketone (179), and 2- and 3-methylbutanoic acid (180 and 181).
Various miterwort species (Mitella spp.) are mainly pollinated by nectaring fungus gnats, either by Gnoriste mikado, a long-tongued species, or by short-tongued species. The flowers of these plant species release mainly terpenoids, with the scent of species pollinated by G. mikado being dominated by lilac aldehyde (165).286 This compound was found to repel short-tongued gnats, but to frequently elicit nectaring behavior in G. mikado.
The globeflower Trollius europaeus is involved in a highly specific brood site mutualism with Chiastocheta (Anthomyiidae) flies. Its flowers release mainly terpenoids, but also a few fatty acid derivatives and miscellaneous cyclic and aromatic compounds, of which the terpenes (E)-β-caryophyllene (64), germacrene D (101), (E,E)-α-farnesene (63) and linalool (60 and/or 61), the aromatic compound methyl salicylate (125), and the fatty acid derivative (Z)-jasmone (127) elicited antennal responses in Chiastocheta fly pollinators.287 In another plant involved in a brood site mutualism with flies, i.e. Rheum nobile, the Sikkim rhubarb, the floral scents are dominated by methyl 2-methylbutyrate (139) and α-pinene (72). Bradysia fungus gnat pollinators were attracted by a blend of these two compounds and by 139 alone, but not by 72 alone.288
A very different set of chemicals is involved in plants that chemically mimic fermenting organic plant material such as fruits and vegetables, and target drosophilid flies as pollinators.301 In an extraordinary work, Stökl et al.37 decoded the mimicry strategy of the Solomon's lily, Arum palaestinum (Araceae). This brood-site deceptive trap plant attracted various drosophilid species, mostly Drosophila simulans, by using fermentation-associated volatiles. The floral scent was, to the human nose, reminiscent of a fruity wine and contained 14 compounds that elicited antennal responses in drosophilids. The most consistent responses were elicited by 2,3-butanediol acetate (55), acetoin acetate (56), ethyl hexanoate (186), hexyl acetate (187), 2-phenylethanol (84) and 2-phenylethyl acetate (136), compounds that also occurred in fermenting fruits.37 Studies in the brain of Drosophila revealed the olfactory receptors involved.37 As a synthetic mixture, these compounds were as attractive to the flies as the odor released from a fermenting banana in behavioral assays.
Studies on the chemical ecology of pollination in Aristolochia (Aristolochiaceae) and Ceropegia (Apocynaceae) species also revealed a new intriguing deceptive strategy in these plants, called kleptomyiophily. Species in these genera build trap flowers that mimic volatiles released from preyed-upon insects to attract and exploit kleptoparasitic flies as pollinators. Kleptoparasitic Desmometopa flies (Milichiidae) are main pollinators of the giant Ceropegia, C. sandersonii. These flies are well known to feed on honeybees that are eaten by spiders. Heiduk et al.22 found that the flies use compounds of the alarm pheromone of honeybees, that they release following an attack, to find appropriate feeding sites. These same compounds are released from Ceropegia sandersonii flowers to attract these flies as pollinators. Compounds that were released from both honeybees under simulated attack and C. sandersonii flowers, that elicited antennal responses in Desmometopa and that attracted the flies as a blend were geraniol (59), 2-heptanone (188), 2-nonanol (189) and (E)-2-octen-1-yl acetate (2).22
A congeneric species, C. dolichophylla, deceives kleptoparasitic Desmometopa flies by using different chemicals. Its flower scent is dominated by spiroacetals, mainly (2S,6R,8S)-8-methyl-2-propyl-1,7-dioxaspiro[5.5]undecane (190), (E,E)-2,8-dimethyl-1,7-dioxaspiro[5.5]undecane (191) and (E,E)-2,8-diethyl-1,7-dioxaspiro[5.5]undecane (192), as well as by n-tridecane (193) and N-(3-methylbutyl)acetamide (194). Thereof, the spiroacetals and 194 elicited antennal responses and the compounds most attractive to the flies in bioassays were 190 and mixtures that contained this compound, with 194 being less attractive.302 Isomers of 190 and 194 are known from venom glands of paper wasps, suggesting that C. dolichophylla mimics pheromones of paper wasps, a potential food source of its pollinators.302
Aristolochia rotunda also has a kleptomyiophilous pollination strategy, but mimics volatiles of true bugs (Miridae) to deceive kleptoparasitic chloropid flies. Specifically, Oelschlägel et al.303 demonstrated that A. rotunda mimics pheromones/defensive compounds of the bugs. Compounds that overlapped between the flowers and the bugs and elicited physiological and behavioral responses (as a blend) in the chloropid pollinators (mainly Trachysiphonella ruficeps) were aliphatic esters, among them hexyl butyrate (195), (E)-2-hexenyl butyrate (196), and (E)-2-hexenyl hexanoate (197).303
Bioactive compounds, among them novel natural products, were also described from flowers of another deceptive Ceropegia species, C. stenantha. This species is pollinated by scatopsid flies and releases various aromatic compounds.31 Several thereof elicited antennal responses in fly pollinators, among them widespread floral scents [e.g. benzyl alcohol (99), 2-phenylethanol (84), 1-phenyl-1,2-propandione (198)], but also compounds so far only known from C. stenantha (37 and 38).
6.4 Bees
Bees are the most important pollinators when it comes to the number of plants pollinated,217 and it has been known for a long time that they use floral scent for locating host plants, often together with visual floral cues.2,6,304 While some bees are generalist flower visitors that visit (mainly during the day, but sometimes at night) various species to collect pollen (and nectar) provisions for their offspring, others collect pollen only from plants of a single family or genus (oligoleges).305 Bees that collect fatty oils from oil-secreting flowers (oil-collecting bees) or those that collect floral volatiles (male euglossine bees) are highly specialized.305 For all these types of bees, major advances were recently made in our understanding of the specific chemicals involved in the communication between plants and their bee pollinators.In Brassica rapa, Knauer and Schiestl308 identified an honest volatile signal for the availability of rewards. The flowers of this species released four compounds that elicited consistent antennal responses in Bombus terrestris pollinators. The aromatics phenylacetaldehyde (83), acetophenone (135) and p-anisaldehyde (111), and the terpene (E,E)-α-farnesene (63). The amount of 83 positively correlated with pollen and nectar sugar availability, and while foraging on the flowers, bumblebees developed a strong preference for this compound over other EAD-active compounds. In Salix caprea, the temporal emission pattern of floral scent was found to correlate with the olfactory preferences of diurnal bee and nocturnal moth pollinators. Honeybee pollinators were strongly attracted to 1,4-dimethoxybenzene (122), a compound emitted in much higher amounts during the day than at night. They preferred this compound over lilac aldehyde (165) (strong attractant for moths, see above), which attracted only a few individuals in a choice setting and is released in higher amounts during night than day.257 Compounds bioactive to bees were, in the last decade, also described from some crop plants. In three highbush blueberry (Vaccinium corymbosum) cultivars, floral scents are dominated by (E)-cinnamyl alcohol (154), but various aliphatic compounds and terpenes also are released.309 A synthetic blend consisting of 16 compounds, among them 154, α-pinene (72), β-pinene (73), linalool (60 and/or 61), and (Z)-3-hexenyl acetate (110), was attractive to honeybee pollinators, whereas single compounds were inactive. El-Sayed et al.272 identified pollinator attractants of stone fruits. 4-Oxoisophorone (169) from apricot (Prunus armeniaca), cherry (P. avium), and plum (P. domestica), and 3,5-dimethoxytoluene (176) from peach (P. persica) and cherry successfully attracted bee pollinators in field trapping experiments.272 Floral scents that elicit electroantennographic responses in honeybee pollinators were recently identified in the vegetable crops, apple and European pear. Compounds eliciting antennal responses were 2-aminobenzaldehyde (166), benzaldehyde (88), decanal (115) and nonanal (126) in Chinese cabbage (Brassica rapa breeds), nonanal (126), methyl salicylate (125), and phenylacetaldehyde (83) in carrot (Daucus carota sativus), and nonanal (126), 169, 83 and 2-phenylethanol (84) in radish (Raphanus raphanistrum sativus).310 In apple (Malus domestica), antennal responses were elicited by 88, benzyl alcohol (99) (dominated the scent bouquet), (E)-cinnamyl alcohol (154), eugenol (87), methyleugenol (175), and two unknown compounds (Rachersberger et al. 2019).311 About 20 compounds in the flower scent of pear (Pyrus communis) elicited antennal responses in honeybees, among them common [e.g., (E)-β-ocimene (62), 4-oxoisophorone (169)], but also rare [N-isobutylformamide (199), (E)-N-(3-methylbutyl)-1-phenylmethanimine (200)] floral scents and even compounds newly identified from nature [(E)-N-(2-methylbutyl)- and (E)-N-(3-methylbutyl)-1-(pyridin-3-yl)methanimine (46 and 47)].312
Finally, one study tested for correlations between floral volatiles of greenhouse tomato (Lycopersicon esculentum, Solanaceae) and pollination by bumblebees.313 Tomato flowers released β-phellandrene (201), 2-carene (202), α-pinene (72) and p-cymene (106), and bumblebees preferably pollinated flowers that produced less 201 and 202 than flowers producing more of these compounds. The repellent properties of these compounds may protect flowers from damage (bees grasp the flowers with their mandibles for extracting pollen by buzzing) caused by over-pollination.313
Recently, it was shown that many of these compounds also occur in Malva moschata and Geranium sanguineum, plants also occasionally visited by Chelostoma rapunculi.319Campanula species are also frequently visited by the second generation of Andrena bicolor, a bivoltine bee. While the first generation of this species visits various different plant species in spring, among them Taraxacum officinale, the second generation in summer prefers Campanula spp.320 Despite this difference, bees of the two generations did not differ in their floral preference when being offered T. officinale and C. trachelium in a choice setting, and also responded to the same compounds in electroantennographic measurements [T. officinale: benzaldehyde (88), acetophenone (135), benzoic acid (210), β-copaene (68); C. trachelium: 2-phenylethyl acetate (136), terpinolene (152), β-elemene (211), (E)-β-caryophyllene (64); both species: linalool (60 and/or 61), (E)-β-ocimene (62)], with Campanula-spiroacetals eliciting no responses in either generation of this bee species.320 In a comparative study on the olfactory systems of Chelostoma rapunculi and other bee species specialized on Campanula (Chelostoma campanularum, Hoplitis mitis) as well as of generalist bees (Andrena bicolor, Bombus terrestris, Apis mellifera), Brandt et al.321 found that Campanula specialists are more sensitive to spiroacetals suggesting that they are well adapted in locating their specific host plants effectively. Campanula species release spiroacetals only in small amounts and the sensitivity of these bees might border on a pheromone-like attraction.318
A similar conclusion was drawn on Andrena vaga, a bee specialized on willows (Salix). Measurements of the neural activity in the brain (antennal lobe) demonstrated that A. vaga is more sensitive to 1,4-dimethoxybenzene (122), a behavior-mediating odorant of Salix host flowers,322 than honey bees.323 Schäffler et al.19 identified with diacetin [1,2- + 1,3-diacetin (53 and 54)] the first olfactory mediator of the interaction between oil-secreting plants and oil-collecting bees. This compound was key in attracting Macropis fulvipes to Lysimachia punctata host flowers, with other compounds [heptanoic acid (212), (E)-2-dodecenal (213)] increasing the attractiveness of diacetin. Further, diacetin occurs not only in L. punctata but also in other Lysimachia species pollinated by M. fulvipes or other Macropis species,324 as well as in other oil-secreting plants pollinated by other oil-collecting bees.19,325 As this compound elicited antennal responses only in oil bees (M. fulvipes, Redivia neliana), but not in non-oil bees (Apis mellifera, Melitta haemorrhoidalis), it is a private communication channel between oil-secreting plants and oil-collecting bees.19
The aromatic 1,4-benzoquinone (214), a compound otherwise known to deter arthropods, was shown to be used by oligolectic Hoplitis adunca to recognize its Echium host plants.326,327 The flowers of E. vulgare released various compounds that were electroantennographically active, among them aromatics [e.g.214, 2-phenylethanol (84)] and terpenoids [(Z)- and (E)-β-ocimene (62), linalool (60 and/or 61)]. Synthetic mixtures of these EAD-active compounds only elicited attractive responses in the bees when 214 was present. Further, 214 alone was as attractive to flower-inexperienced bees as a complete synthetic mixture and a natural headspace sample demonstrating the key importance of this compound in host recognition of Hoplitis adunca.327
The narrowly oligolectic bee Protodiscelis palpalis is specialized on the aquatic plant Hydrocleys martii (Alismataceae), which itself depends on this bee species for sexual reproduction. The flower scent was strongly dominated by p-methylanisole (156), which successfully attracted the bees in bioassays. Compounds that occurred in smaller amounts [3,4-dimethoxytoluene (215), 3,4,5-trimethoxytoluene (216)] or not at all [2-methoxy-4-methylphenol (217)] in H. martii, but in higher amounts in the closely-related species H. nymphoides, a plant species not used by P. palpalis as host plant, were not attractive to P. palpalis.328
6.5 Wasps
Around 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 species of wasps are pollinators,217 and in the last decade bioactive compounds were identified for both social and solitary species. Brodmann et al.332 studied the olfactory attractiveness of Scrophularia umbrosa (Scrophulariaceae) host plants to social Vespula vulgaris wasps and found that only a blend of antennal-active (Z)-3-hexenyl acetate (110), 1-hexanol (203), and (Z)-3-hexenol (118) was capable of attracting the wasps, whereas a blend of other antennal-active compounds [benzaldehyde (88), decanal (115), geranyl acetate (218), 6-methyl-5-hepten-2-one (109), nonanal (126), octanal (219), 1-octen-3-ol (220)] was inactive. In another study, social Vespula wasps responded to a high number of floral volatiles of their Heracleum sphondylium and Hedera helix host plants in electroantennographic measurements, among them aliphatic [e.g., 2-heptanone (188)], aromatic (e.g., 88) and nitrogen-bearing [e.g., 2-phenylacetonitrile (92)] compounds as well as terpenoids [e.g., β-myrcene (76), 4-oxoisophorone (169)].312 For H. helix experiments with synthetic compounds were also performed and it was found that a synthetic mixture resembling the floral scent of this species was attractive to the wasps, but less than the natural olfactory cues. Besides social Vespula wasps, advances were made in the host finding of solitary fig wasps, which are involved in the highly specialized brood-site mutualisms with figs, and in generalist parasitoids. The Mediterranean fig, Ficus carica, attracts its highly specialized pollinator species, the fig wasp Blastophaga psenes, with volatiles, and, among the five major volatiles released by receptive figs, linalool (60 and/or 61) and oxides thereof (145, 174), the (−)-isomer of (E)-β-caryophyllene (64), and germacrene D (101) elicited antennal responses in B. psenes, whereas α-copaene (124) was inactive.333 In another fig species, Ficus curtipes, the scent is dominated by 6-methyl-5-hepten-2-ol (221) and 6-methyl-5-hepten-2-one (109). Both compounds elicited behavioral responses in the fig wasp pollinator Eupristina sp., but elicited different responses: 221 attracted the wasp from a distance, whereas 109 induced fig-entry behavior in the wasp.334 The egg parasitoid Trissolcus basalis uses buckwheat for nectaring and was successfully attracted by a blend of four floral [butanoic acid (222), 2-methylbutanoic acid (180), 3-methylbutanoic acid (181), pentanoic acid (223)] and two vegetative [(Z)-3-hexenyl acetate (110), (E,E)-α-farnesene (63)] volatiles.335
000 species of wasps are pollinators,217 and in the last decade bioactive compounds were identified for both social and solitary species. Brodmann et al.332 studied the olfactory attractiveness of Scrophularia umbrosa (Scrophulariaceae) host plants to social Vespula vulgaris wasps and found that only a blend of antennal-active (Z)-3-hexenyl acetate (110), 1-hexanol (203), and (Z)-3-hexenol (118) was capable of attracting the wasps, whereas a blend of other antennal-active compounds [benzaldehyde (88), decanal (115), geranyl acetate (218), 6-methyl-5-hepten-2-one (109), nonanal (126), octanal (219), 1-octen-3-ol (220)] was inactive. In another study, social Vespula wasps responded to a high number of floral volatiles of their Heracleum sphondylium and Hedera helix host plants in electroantennographic measurements, among them aliphatic [e.g., 2-heptanone (188)], aromatic (e.g., 88) and nitrogen-bearing [e.g., 2-phenylacetonitrile (92)] compounds as well as terpenoids [e.g., β-myrcene (76), 4-oxoisophorone (169)].312 For H. helix experiments with synthetic compounds were also performed and it was found that a synthetic mixture resembling the floral scent of this species was attractive to the wasps, but less than the natural olfactory cues. Besides social Vespula wasps, advances were made in the host finding of solitary fig wasps, which are involved in the highly specialized brood-site mutualisms with figs, and in generalist parasitoids. The Mediterranean fig, Ficus carica, attracts its highly specialized pollinator species, the fig wasp Blastophaga psenes, with volatiles, and, among the five major volatiles released by receptive figs, linalool (60 and/or 61) and oxides thereof (145, 174), the (−)-isomer of (E)-β-caryophyllene (64), and germacrene D (101) elicited antennal responses in B. psenes, whereas α-copaene (124) was inactive.333 In another fig species, Ficus curtipes, the scent is dominated by 6-methyl-5-hepten-2-ol (221) and 6-methyl-5-hepten-2-one (109). Both compounds elicited behavioral responses in the fig wasp pollinator Eupristina sp., but elicited different responses: 221 attracted the wasp from a distance, whereas 109 induced fig-entry behavior in the wasp.334 The egg parasitoid Trissolcus basalis uses buckwheat for nectaring and was successfully attracted by a blend of four floral [butanoic acid (222), 2-methylbutanoic acid (180), 3-methylbutanoic acid (181), pentanoic acid (223)] and two vegetative [(Z)-3-hexenyl acetate (110), (E,E)-α-farnesene (63)] volatiles.3356.6 Other pollinators
Next to the common pollinators and flower visitors mentioned above, attractive compounds also were recently identified for uncommon pollinators/flower visitors. For example, ant pollinators (e.g., Aphaenogaster senilis) of Cytinus hypocistis (Cytinaceae) were shown to be attracted by (E)-cinnamaldehyde (155) and (E)-cinnamyl alcohol (154).336 Thrips (Frankliniella occidentalis) flower visitors of Arabidopsis thaliana were attracted by linalool (60 and/or 61) and repelled by lilac aldehyde (165) and lilac alcohol (224),59,337 and thrips were also shown to be attracted by (S)-verbenone (225), a compound released from pollen host plants (Pinus spp.).338 In a very recent intriguing study, Etl et al.26 described a new plant bug pollination system, likely evolved from a cyclocephaline beetle pollination system, and decoded the chemical communication involved. They found that the aroid Syngonium hastiferum specializes on a single plant bug pollinator species, Neella sp. nov. (Miridae), that is attracted by the scent of the inflorescences. The scent was strongly dominated by the previously unknown natural product (Z)-3-isopropylpent-3-en-1-ol (12) (gambanol). In field bioassays, this compound specifically attracted the pollinator species.26 Congeners of Neella sp. nov., such as N. floridula, visit cyclocephaline beetle pollinated aroids as non-pollinating florivores.339 One of those plants is Dieffenbachia aurantiaca. The floral scent of this species attracted as many plant bug florivores as natural inflorescences. It was dominated by (Z)-jasmone (127), and this compound was highly effective in attracting the plant bugs in field bioassays.339The first floral attractant for ground-dwelling mammal pollinators was identified from Cytinus visseri (Cytinaceae). This species is pollinated by the striped fieldmouse, R. pumilio, the pygmy mouse, Mus minutoides, and the short-snouted elephant shrew, E. brachyrhynchus.340 Whole flowers of this species as well as separated nectar release a plastic-like scent, due to the emission of 1-hexen-3-one (226). Only whole flowers released another major compound, i.e. 3-hexanone (227). Behavioral experiments with the striped fieldmouse and the short-snouted elephant shrew showed that the natural floral scent is highly attractive to these mammal pollinators. Y-Maze experiments with the striped fieldmouse further demonstrated that this mouse species is attracted by 227, whereas 226 has repellent properties, but only when tested alone and not when tested in combination with 227.340 The African pineapple lily Eucomis regia, similar to C. visseri, is also pollinated by mice and elephant-shrews that are attracted by the scent of the plant species. Flowers of this lily released common [e.g. 2-heptanone (188), 2-heptanol (228)] and uncommon [e.g. exo-brevicomin (229), methional (230)] scents, of which 230 was responsible for the boiled-potato-like smell of the flowers and of separated nectar. This compound successfully attracted Namaqua rock mouse (Micaelamys namaquensis) pollinators in y-maze experiments.341 Just recently, Johnson and Govender342 tested the effects of various volatiles found in rodent pollinated plants and in insect pollinated congeners (aromatic compounds) on their attractiveness to mouse pollinators. Several compounds were attractive to the mice, among them the aliphatic ketones 227, 188 and acetoin (130), the aliphatic ester ethyl butyrate (231) and the aliphatic acid butanoic acid (222). Dimethyl disulfide (182) and the aromatic compounds [benzaldehyde (88), methyl benzoate (100), 2-phenylethanol (84), phenylacetaldehyde (83)] were neutral or repellent to the mice. Adding unattractive compounds to attractive ones resulted in a non-attractive signal.342
Finally, bioactive compounds were identified for crab spiders that visit flowers to hunt for prey (pollinators, florivores). Knauer et al.343 asked whether the crab spider Thomisus onustus, which frequently visits flowers of the buckler mustard (Biscutella laevigata), is attracted by floral volatiles. Indeed, the spider was attracted by β-ocimene (62), whereas the other two main floral scent compounds [p-anisaldehyde (111), 2-aminobenzaldehyde (166)] did not elicit a behavioral response in the spiders. Florivores induced the emission of (62). Accordingly, spiders had an olfactory preference for plants infested with florivores.343
7 Functions of floral volatiles other than pollinator attraction
Flower volatiles did not only evolve to attract pollinators, but have various other functions,344–346 such as to repel florivores, to fight against pathogenic microorganisms, or to attract enemies of plant pests. Several of these functions have only recently been understood, and here we summarize the main advances and present the compounds involved.7.1 Repelling florivores
Ants are frequent florivores of flowers, and in a study on Phlox paniculata and Lasius niger ants it was demonstrated that floral scent is repellent to the ants.347 The ant repellent properties were lost after inhibition of terpene biosynthesis pointing to the importance of terpenes in repelling the ants. Indeed, linalool (60 and/or 61), which was strongly inhibited347 and known to be repellent to ants before,348 repelled the ants in olfactometer assays. However, linalool (60 and/or 61) was only repellent when offering the compound in concentrations higher than released from the flowers, suggesting that other compounds are also involved in repelling ants from the flowers.347 Other flower volatiles known to be repellent to ants are p-anisaldehyde (111) from Petasites fragrans flowers349 and 2-phenylethanol (84) from flowers of the alpine skypilot Polemonium viscosum.350 In the latter species, the flowers are polymorphic in the amount of 84 emitted. Ants avoided high emitters in the field and died when exposed to 84 in the laboratory.350 Other florivores than ants shown to be repelled by floral scents were beetles and crickets. By using genetically modified Nicotiana attenuata, Kessler et al.351 demonstrated that benzylacetone (91) is repellent to the florivorous cucumber beetles Diabrotica undecimpunctata, and similar experiments with Petunia x hybrida revealed that isoeugenol (232) and benzyl benzoate (233) are repellent to D. undecimpunctata and the snowy tree cricket, Oecanthus fultoni.244 The bush cricket Metrioptera bicolor, which mainly feeds on vegetative plant parts, but occasionally also on flowers, was repelled by the common floral scents linalool (60 and/or 61) and β-caryophyllene (64).3528 Chirality in flower scents and its importance for interspecific interactions
Many floral scent components are optically active and it is known that chirality is of relevance in flower–animal interactions.5,357–359 However, despite this knowledge there are very few recent studies that determined the absolute configuration of compounds from floral scents and tested single enantiomers or reconstructed enantiomeric ratios on flower visitors in physiological and/or behavioral experiments (see also Perkins et al.18).In the pollinator-plant and herbivore-plant association between Manduca sexta and Datura wrightii it was shown that floral (S)-linalool (61) elicited oviposition behavior, while floral (R)-linalool (60) had negative effects on oviposition behaviour.360 Neither the single enantiomers nor mixtures of the two enantiomers had effects on the feeding behavior of the moths, which is triggered by other floral volatiles (mixture of benzaldehyde (88), benzyl alcohol (99), geraniol (59) and nerol). Field bean (Vicia faba) flowers only released 60, and this enantiomer attracted the bean seed beetle Bruchus rufimanus, a pest of the field bean.236 In the pollination mutualism between Phyllanthaceae and Epicephala moths, the different enantiomers of linalool (60 and 61) might elicit different behaviors in the moths. In both Glochidion obovatum and Glochidion rubrum female flowers released (S)-linalool (61) and male flowers (R)-linalool (60). Okamoto et al.265 speculated that this sexual difference in the emission of linalool enantiomers allows moth pollinators to discriminate between the flower sexes and collect pollen on male flowers and actively pollinate and oviposit on female flowers (see above). In another nursery pollination mutualism, between Lithophragma bolanderi and Greya politella moths, it was just recently demonstrated that the flowers produce only the (6R,10R)-enantiomer of 6,10,14-trimethylpentadecan-2-one (167), and that this enantiomer is elicits antennal responses in the moth pollinators.261 This same compound was also detected in tibial fragrances of male euglossine bees (Euglossa spp.), which likely obtained this compound from aroid or orchid flowers, and in behavioral assays, it attracted more individuals and species of euglossines than a racemate of 167.361
Another optically active attractant for male euglossines is ipsdienol. This compound is released from various male euglossine bee pollinated plants,331,362 and it was demonstrated for Euglossa cyanura males that only (−)-ipsdienol (237) is attractive, whereas the (+)-isomer (238) is ignored by the bees.363 Further, antennae of these bees showed larger physiological responses to the (−)-isomer than to the (+)-isomer. Another bee species, E. mixta, which is not attracted by ipsdienol, did not discriminate between the enantiomers in electroantennographic measurements.363 New data were also obtained for carvone epoxide. This compound is released from plants of several families (e.g., Annonaceae, Araceae, Gesneriaceae, Orchidaceae) and is a main attractant for male euglossine bees of the genus Eulaema.331 The plants mainly release (E)-carvone epoxides, though some also release smaller amounts of the (Z)-isomers (summarized by Brandt et al.364). For each diastereomer two enantiomers are possible, summing up to a total of four stereoisomers of carvone epoxide (239–242). All five orchid species recently studied for the presence of the stereoisomers of carvone epoxide released only the (−)-(E)-isomer (239). In Eulaema bees, this enantiomer elicited the highest antennal responses and was the most attractive.364
9 Conclusions
Our review provides clear evidence that the number of described floral volatiles is still increasing. While this increase is due partly to advances in analytical procedures, it results mainly from the increasing number of plant species being studied motivated by both basic research on pollination biology and the search for new volatiles by the flavor and fragrance industry. Some of the newly discovered volatiles are derived from common floral volatiles, such as (1S,2S)-isojasmol (3), (1S,2S)-isojasmyl acetate (4), and (S)-dehydrojasmone (5) detected in various Araceae species, which are derived from (Z)-jasmone (Fig. 25).11Another example is a group of new volatiles of Yucca species, including (Z)- and (E)-filamentol (22 and 25), (Z)- and (E)-filamental (23 and 26), (Z)- and (E)-filamentolide (24 and 27) and filamentone (28), and of (E)-4,8-dimethyl-1,3,7-nonatrien-5-yl acetate (29) and the (Z)-isomer thereof from Philodendron squamiferum, all derived from the widespread 4,8-dimethyl-1,3,7-nonatriene (see Fig. 26).11,29
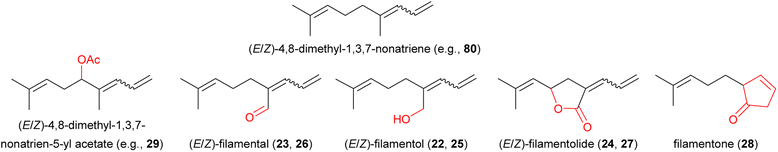 | ||
| Fig. 26 The widespread floral scent (E/Z)-4,8-dimethyl-1,3,7-nonatriene (e.g., 80) and uncommon putative derivatives thereof. | ||
The continuous increase in the number of known floral volatiles also shows that our understanding of the floral scent chemistry is far from being complete, calling for studies that analyse floral volatiles (including their absolute configuration) from plant lineages and species where no data are available so far.
Major advances during the period of this review were also made in understanding the biosynthesis of floral volatiles and the regulation of volatile emissions. Several new genes and biosynthetic routes were discovered, among them alternative pathways, which sometimes work in parallel in specific flowers, e.g. for production of 2-phenylethanol (84), of one of the most widespread floral volatiles. Similarly, several new transcription factors and other factors that regulate floral scent production and emission, such as epigenetic influences and long non-coding RNAs, were discovered. A breakthrough was the identification of a transporter protein that exports floral volatiles from cells.
Major advances were also achieved in our understanding of how floral scents are shaped by abiotic factors, such as air pollution and increased temperatures, but how, e.g. temperature-induced changes in scent translate into pollinator behavior is less well understood. Several studies demonstrated in the last years that not only herbivores but also microorganisms (e.g., mycorrhizal fungi, plant endophytes, flower-dwelling bacteria and yeasts) have effects on flower scent emissions and sometimes also on the communication with flower visitors. Most of these studies were performed in the laboratory or using experimental settings, and it remains to be tested how microorganisms affect floral scents and flower–animal interactions in natural settings.
Since 2010, a large number of studies identified compounds attractive to flower visitors. The behaviorally active compounds were main compounds of the released scent in some pollination systems,e.g.,365 while they were only minor or even trace compounds in others. We now have a much better understanding how plants communicate with beetle, lepidopteran, bee and fly pollinators, and the first attractants were also identified for less common pollinators, such as rodents, shrews and true bugs. Despite these advances, our knowledge about the communication between flowers and their visitors is still very limited, especially when considering the high number of plants pollinated by animals and thef high number of animals involved. Also, most advances were made in specialized pollination systems, and more studies are required to elucidate olfactory communication in generalist systems. Considering the chemical properties of the compounds involved in pollinator attraction, such as volatility and the functional groups present (e.g., alcohol, olefin, ester, ketone), there seem to be no trends for different groups of pollinators (bees, beetles, flies, Lepidoptera, etc.). However, there are some trends regarding overall biosynthetic class. For instance, among reported beetle attractants fatty acid derivatives are well represented with a smaller number of compounds from other classes.
Lepidoptera seem to respond mainly to phenylpropanoids/benzenoids, terpenes and nitrogen-bearing compounds, and flies additionally to fatty acid derivatives and sulphur-bearing compounds. For bees, reported attractants were mainly phenylpropanoids/benzenoids, terpenes, and fatty acid derivatives, but only a few nitrogen- and no sulphur-bearing compounds have been described. For wasps, there are a lot of reports of fatty acid derivatives, but also several terpenes and some compounds from other classes (see also ref. 18), and for ground-dwelling mammals mainly fatty acid derivatives are listed as attractants with the conspicuous absence of terpenes and phenylpropanoids/benzenoids. In fact, phenylpropanoids/benzenoids, which are otherwise common among floral scents and attractive to other pollinators [e.g. phenylacetaldehyde (83), 2-phenylethanol (84)] were even shown to be repellent to rodents. Finally, a few floral volatiles were identified during the period of this review that serve plants in functions other than pollinator attraction, such as by repelling florivores, inhibiting pathogenic microorganisms, and attracting enemies of herbivores. Considering the abundance and chemical diversity of floral volatiles, there are relatively few studies in this “young” field of research and many new insights can be expected in the near future.
10 Author contributions
Both authors worked together on the manuscript.11 Conflicts of interest
There are no conflicts to declare.12 Acknowledgements
SD thanks Roman Fuchs for help with the chemical structures and Karin Gross, Danae Laina, Monica Barman and other members of the Plant Ecology group as well as Robert R. Junker for comments on an earlier version of the manuscript. JG thanks Kirsten Heinrichs for help in literature searching, and Eran Pichersky for comments on an earlier draft. SD and JG thank two anonymous reviewers for helpful comments on an earlier version of the manuscript.13 References
- H. Bouwmeester, R. C. Schuurink, P. M. Bleeker and F. Schiestl, Plant J., 2019, 100, 892–907 CrossRef CAS PubMed.
- H. E. M. Dobson, in Biology of Floral Scent, ed. N. Dudareva and E. Pichersky, CRC Press, Boca Raton, 2006, pp. 147–198 Search PubMed.
- R. R. Junker and A. L. Parachnowitsch, J. Indian Inst. Sci., 2015, 95, 43–67 Search PubMed.
- J. T. Knudsen, R. Eriksson, J. Gershenzon and B. Ståhl, Bot. Rev., 2006, 72, 1–120 CrossRef.
- R. A. Raguso, Annu. Rev. Ecol. Evol. Syst., 2008, 39, 549–569 CrossRef.
- R. A. Raguso, in Biology of Plant Volatiles, eds. E. Pichersky and N. Dudareva, CRC Press, Boca Raton, 2020, pp. 297–325 Search PubMed.
- J. Ollerton, R. Winfree and S. Tarrant, Oikos, 2011, 120, 321–326 CrossRef.
- A. M. Klein, B. E. Vaissière, J. H. Cane, I. Steffan-Dewenter, S. A. Cunningham, C. Kremen and T. Tscharntke, Proc. R. Soc. B, 2007, 274, 303–313 CrossRef PubMed.
- B. Bohman, M. M. Y. Tan, R. D. Phillips, A. Scaffidi, A. N. Sobolev, S. A. Moggach, G. R. Flematti and R. Peakall, Angew. Chem., Int. Ed., 2020, 59, 1124–1128 CrossRef CAS PubMed.
- K. Lukas, T. Harig, S. Schulz, J. Hadersdorfer and S. Dötterl, Chemoecology, 2019, 29, 211–223 CrossRef.
- A. C. D. Maia, C. Grimm, M. Schubert, F. Etl, E. G. Gonçalves, D. M. Do Amaral Ferraz Navarro, S. Schulz and S. Dötterl, J. Chem. Ecol., 2019, 45, 204–213 CrossRef CAS PubMed.
- H. Teichert, S. Dötterl and G. Gottsberger, Plant Syst. Evol., 2018, 304, 831–839 CrossRef CAS.
- L. A. Burkle and J. B. Runyon, Funct. Ecol., 2019, 33, 2116–2129 CrossRef.
- R. R. Junker, N. Höcherl and N. Blüthgen, J. Anim. Ecol., 2010, 79, 818–823 Search PubMed.
- A. Kantsa, R. A. Raguso, A. G. Dyer, J. M. Olesen, T. Tscheulin and T. Petanidou, Nat. Commun., 2018, 9, 1041 CrossRef PubMed.
- B. Boachon, Y. Burdloff, J.-X. Ruan, R. Rojo, R. R. Junker, B. Vincent, F. Nicolè, F. Bringel, A. Lesot, L. Henry, J.-E. Bassard, S. Mathieu, L. Allouche, I. Kaplan, N. Dudareva, S. Vuilleumier, L. Miesch, F. André, N. Navrot, X.-Y. Chen and D. Werck-Reichhart, Plant Cell, 2019, 31, 2947–2972 CrossRef CAS PubMed.
- H. M. Li, W. B. Liu, L. L. Yang, H. Q. Cao, P. Pelosi, G. R. Wang and B. Wang, J. Agric. Food Chem., 2020, 68, 12212–12220 CrossRef CAS PubMed.
- J. Perkins, T. Hayashi, R. Peakall, G. R. Flematti and B. Bohman, Nat. Prod. Rep., 2023, 40, 819–839 RSC.
- I. Schäffler, K. E. Steiner, M. Haid, S. S. van Berkel, G. Gerlach, S. D. Johnson, L. Wessjohann and S. Dötterl, Sci. Rep., 2015, 5, 12779 CrossRef PubMed.
- P. Milet-Pinheiro, A. Domingos-Melo, J. B. Olivera, N. S. L. Albuquerque, A. C. G. Costa, S. Albuquerque-Lima, M. F. R. Silva, D. M. A. F. Navarro, A. C. D. Maia, L.-L. Gundersen, M. Schubert, S. Dötterl and I. C. Machado, Curr. Biol., 2021, 31, 860–868 CrossRef CAS PubMed.
- T. Hayashi, B. Bohman, A. Scaffidi, R. Peakall and G. R. Flematti, Curr. Biol., 2021, 31, 1954–1961 CrossRef CAS PubMed.
- A. Heiduk, I. Brake, M. von Tschirnhaus, M. Göhl, A. Jürgens, S. D. Johnson, U. Meve and S. Dötterl, Curr. Biol., 2016, 26, 2787–2793 CrossRef CAS PubMed.
- P. Stamm, F. Etl, A. C. D. Maia, S. Dötterl and S. Schulz, J. Org. Chem., 2021, 86, 5245–5254 CrossRef CAS PubMed.
- R. Kaiser, Scent of the vanishing flora, Wiley VCH, 2011 Search PubMed.
- C. Cohen, W. R. Liltved, J. F. Colville, A. Shuttleworth, J. Weissflog, A. Svatoš, B. Bytebier and S. D. Johnson, Curr. Biol., 2021, 31, 1962–1969 CrossRef CAS PubMed.
- F. Etl, C. Kaiser, O. Reiser, M. Schubert, S. Dötterl and J. Schönenberger, Curr. Biol., 2022, 32, 4688–4698 CrossRef CAS PubMed.
- B. Bohman, A. M. Weinstein, R. D. Phillips, R. Peakall and G. R. Flematti, J. Nat. Prod., 2019, 82, 1107–1113 CrossRef CAS PubMed.
- R. Peakall, D. Ebert, J. Poldy, R. A. Barrow, W. Francke, C. C. Bower and F. P. Schiestl, New Phytol., 2010, 188, 437–450 CrossRef CAS PubMed.
- A. Tröger, G. P. Svensson, H.-M. Galbrecht, R. Twele, J. M. Patt, S. Bartram, P. H. G. Zarbin, K. A. Segraves, D. M. Althoff, S. von Reuss, R. A. Raguso and W. Francke, J. Chem. Ecol., 2021, 47, 1025–1041 CrossRef PubMed.
- C. E. P. Nunes, M. F. G. V. Peñaflor, J. M. S. Bento, M. J. Salvador and M. Sazima, Oecologia, 2016, 182, 933–946 CrossRef PubMed.
- A. Heiduk, J.-P. Haenni, U. Meve, S. Schulz and S. Dötterl, J. Comp. Physiol., A, 2019, 205, 301–310 CrossRef PubMed.
- B. Bohman, L. Jeffares, G. Flematti, L. T. Byrne, B. W. Skelton, R. D. Phillips, K. W. Dixon, R. Peakall and R. A. Barrow, J. Nat. Prod., 2012, 75, 1589–1594 CrossRef CAS PubMed.
- B. Bohman, L. Jeffares, G. Flematti, R. D. Phillips, K. W. Dixon, R. Peakall and R. A. Barrow, Org. Lett., 2012, 14, 2576–2578 CrossRef CAS PubMed.
- A. C. D. Maia, S. Dötterl, R. Kaiser, I. Silberbauer-Gottsberger, H. Teichert, M. Gibernau, D. M. do Amaral Ferraz Navarro, C. Schlindwein and G. Gottsberger, J. Chem. Ecol., 2012, 38, 1072–1080 CAS.
- B. Bohman, R. D. Phillips, G. R. Flematti, R. A. Barrow and R. Peakall, Angew. Chem., Int. Ed., 2017, 56, 8455–8458 CrossRef CAS PubMed.
- S. Wakamura, N. Arakaki, D. Moriyama, S. Kanayama, M. Oike, A. Kimura, S. Wajima, H. Ono and H. Yasui, Chemoecology, 2020, 30, 49–57 CrossRef CAS.
- J. Stökl, A. Strutz, A. Dafni, A. Svatos, J. Doubsky, M. Knaden, S. Sachse, B. S. Hansson and M. C. Stensmyr, Curr. Biol., 2010, 20, 1846–1852 CrossRef PubMed.
- F. Chen, J. C. D'Auria, D. Tholl, J. R. Ross, J. Gershenzon, J. P. Noel and E. Pichersky, Plant J., 2003, 36, 577–588 CrossRef CAS PubMed.
- Y. Jiang, G. Liu, W. Zhang, C. Zhang, X. Chen, Y. Chen, C. Yu, D. Yu, J. Fu and F. Chen, Phytochemistry, 2021, 191, 112899 CrossRef CAS PubMed.
- Q. Xu, S. Wang, H. Hong and Y. Zhou, BMC Genomics, 2019, 20, 125 CrossRef PubMed.
- F. Chen, D. Tholl, J. Bohlmann and E. Pichersky, Plant J., 2011, 66, 212–229 CrossRef CAS PubMed.
- J. Degenhardt, T. G. Köllner and J. Gershenzon, Phytochemistry, 2009, 70, 1621–1637 CrossRef CAS PubMed.
- X. Zeng, C. Liu, R. Zheng, X. Cai, J. Luo, J. Zou and C. Wang, Front. Plant Sci., 2016, 6, 1232 Search PubMed.
- M. Lv, X. Sun, D. Li, G. Wei, L. Liu, F. Chen, Y. Cai and H. Fan, Ind. Crops Prod., 2022, 182, 114875 CrossRef CAS.
- J. Zhao, Z. Wang, Z. Li, J. Shi, L. Meng, G. Wang, J. Cheng and Y. Du, J. Asia-Pac. Entomol., 2020, 23, 1023–1029 CrossRef.
- H. Huang, Y.-W. Kuo, Y.-C. Chuang, Y.-P. Yang, L.-M. Huang, M.-F. Jeng, W.-H. Chen and H.-H. Chen, Front. Plant Sci., 2021, 12, 700958 CrossRef PubMed.
- C. Zhao, Z. Yu, J. A. T. d. Silva, C. He, H. Wang, C. Si, M. Zhang, D. Zeng and J. Duan, Int. J. Mol. Sci., 2020, 21, 7005 CrossRef CAS PubMed.
- Y. Yue, R. Yu and Y. Fan, Planta, 2014, 240, 745–762 CrossRef CAS PubMed.
- G. Liu, M. Yang, X. Yang, X. Ma and J. Fu, J. Plant Physiol., 2021, 258–259, 153358 CrossRef CAS PubMed.
- T. Bao, K. Shadrack, S. Yang, X. Xue, S. Li, N. Wang, Q. Wang, L. Wang, X. Gao and Q. Cronk, Plant Cell Physiol., 2020, 61, 1733–1749 CrossRef CAS PubMed.
- J. Jin, M. J. Kim, S. Dhandapani, J. G. Tjhang, J.-L. Yin, L. Wong, R. Sarojam, N.-H. Chua and I.-C. Jang, J. Exp. Bot., 2015, 66, 3959–3975 CrossRef CAS.
- E. Pichersky and E. Lewinsohn, Annu. Rev. Plant Biol., 2011, 62, 549–566 CrossRef CAS PubMed.
- J.-i. Hattan, K. Shindo, T. Sasaki, F. Ohno, H. Tokuda, K. Ishikawa and N. Misawa, Sci. Rep., 2018, 8, 12474 CrossRef PubMed.
- A. Fähnrich, A. Brosemann, L. Teske, M. Neumann and B. Piechulla, Plant Mol. Biol., 2012, 79, 537–553 CrossRef PubMed.
- K. J. R. P. Byers, J. P. Vela, F. Peng, J. A. Riffell and H. D. Bradshaw Jr, Plant J., 2014, 80, 1031–1042 CrossRef CAS PubMed.
- J.-L. Magnard, A. Roccia, J.-C. Caissard, P. Vergne, P. Sun, R. Hecquet, A. Dubois, L. Hibrand-Saint Oyant, F. Jullien, F. Nicolè, O. Raymond, S. Huguet, R. Baltenweck, S. Meyer, P. Claudel, J. Jeauffre, M. Rohmer, F. Foucher, P. Hugueney, M. Bendahmane and S. Baudino, Science, 2015, 349, 81–83 CrossRef CAS PubMed.
- L. Sheng, S. Zang, J. Wang, T. Wei, Y. Xu and L. Feng, PeerJ, 2021, 9, e11098 CrossRef PubMed.
- J.-F. Ginglinger, B. Boachon, R. Höfer, C. Paetz, T. G. Köllner, L. Miesch, R. Lugan, R. Baltenweck, J. Mutterer, P. Ullmann, F. Beran, P. Claudel, F. Verstappen, M. J. C. Fischer, F. Karst, H. Bouwmeester, M. Miesch, B. Schneider, J. Gershenzon, J. Ehlting and D. Werck-Reichhart, Plant Cell, 2013, 25, 4640–4657 CrossRef CAS PubMed.
- B. Boachon, R. R. Junker, L. Miesch, J.-E. Bassard, R. Höfer, R. Caillieaudeaux, D. E. Seidel, A. Lesot, C. Heinrich, J.-F. Ginglinger, L. Allouche, B. Vincent, D. S. C. Wahyuni, C. Paetz, F. Beran, M. Miesch, B. Schneider, K. Leiss and D. Werck-Reichhart, Plant Cell, 2015, 27, 2972–2990 CAS.
- S. Lee, S. Badieyan, D. R. Bevan, M. Herde, C. Gatz and D. Tholl, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 21205–21210 CrossRef CAS PubMed.
- A. Richter, C. Schaff, Z. Zhang, A. E. Lipka, F. Tian, T. G. Köllner, C. Schnee, S. Preiß, S. Irmisch, G. Jander, W. Boland, J. Gershenzon, E. S. Buckler and J. Degenhardt, Plant Cell, 2016, 28, 2651–2665 CrossRef CAS PubMed.
- R. Sohrabi, J.-H. Huh, S. Badieyan, L. H. Rakotondraibe, D. J. Kliebenstein, P. Sobrado and D. Tholl, Plant Cell, 2015, 27, 874–890 CrossRef CAS PubMed.
- S. Baldermann, M. Kato, M. Kurosawa, Y. Kurobayashi, A. Fujita, P. Fleischmann and N. Watanabe, J. Exp. Bot., 2010, 61, 2967–2977 CrossRef CAS PubMed.
- A. Rubio-Moraga, J. L. Rambla, A. Fernández-de-Carmen, A. Trapero-Mozos, O. Ahrazem, D. Orzáez, A. Granell and L. Gómez-Gómez, Plant Mol. Biol., 2014, 86, 555–569 CrossRef CAS PubMed.
- Y. Kaminaga, J. Schnepp, G. Peel, C. M. Kish, G. Ben-Nissan, D. Weiss, I. Orlova, O. Lavie, D. Rhodes, K. Wood, D. M. Porterfield, A. J. L. Cooper, J. V. Schloss, E. Pichersky, A. Vainstein and N. Dudareva, J. Biol. Chem., 2006, 281, 23357–23366 CrossRef CAS PubMed.
- M. Farhi, O. Lavie, T. Masci, K. Hendel-Rahmanim, D. Weiss, H. Abeliovich and A. Vainstein, Plant Mol. Biol., 2010, 72, 235–245 CrossRef CAS PubMed.
- M. Gutensohn, A. Klempien, Y. Kaminaga, D. A. Nagegowda, F. Negre-Zakharov, J.-H. Huh, H. Luo, R. Weizbauer, T. Mengiste, D. Tholl and N. Dudareva, Plant J., 2011, 66, 591–602 CrossRef CAS PubMed.
- M. Sakai, H. Hirata, H. Sayama, K. Sekiguchi, H. Itano, T. Asai, H. Dohra, M. Hara and N. Watanabe, Biosci., Biotechnol., Biochem., 2007, 71, 2408–2419 CrossRef CAS PubMed.
- X.-M. Chen, H. Kobayashi, M. Sakai, H. Hirata, T. Asai, T. Ohnishi, S. Baldermann and N. Watanabe, J. Plant Physiol., 2011, 168, 88–95 CrossRef CAS PubMed.
- A. Roccia, L. Hibrand-Saint Oyant, E. Cavel, J.-C. Caissard, J. Machenaud, T. Thouroude, J. Jeauffre, A. Bony, A. Dubois, P. Vergne, J. Szécsi, F. Foucher, M. Bendahmane and S. Baudino, Plant Physiol., 2019, 179, 1064–1079 CrossRef CAS PubMed.
- T. Koeduka, Y. Ueyama, S. Kitajima, T. Ohnishi and K. Matsui, J. Plant Physiol., 2020, 252, 153245 CrossRef CAS PubMed.
- H. Hirata, T. Ohnishi, K. Tomida, H. Ishida, M. Kanda, M. Sakai, J. Yoshimura, H. Suzuki, T. Ishikawa, H. Dohra and N. Watanabe, Sci. Rep., 2016, 6, 20234 CrossRef CAS PubMed.
- L. Sheng, Y. Zeng, T. Wei, M. Zhu, X. Fang, X. Yuan, Y. Luo and L. Feng, Genes, 2018, 9, 576 CrossRef PubMed.
- S. Dhandapani, J. Jin, V. Sridhar, N.-H. Chua and I.-C. Jang, Plant Physiol., 2019, 180, 171–184 CrossRef CAS PubMed.
- J. Günther, N. D. Lackus, A. Schmidt, M. Huber, H.-J. Stödtler, M. Reichelt, J. Gershenzon and T. G. Köllner, Plant Physiol., 2019, 180, 767–782 CrossRef PubMed.
- F. Dong, Z. Yang, S. Baldermann, Y. Kajitani, S. Ota, H. Kasuga, Y. Imazeki, T. Ohnishi and N. Watanabe, J. Plant Physiol., 2012, 169, 217–225 CrossRef CAS PubMed.
- F. Dong, Y. Zhou, L. Zeng, Q. Peng, Y. Chen, L. Zhang, X. Su, N. Watanabe and Z. Yang, Molecules, 2016, 21, 1106 CrossRef PubMed.
- Y. Zhou, Q. Peng, L. Zeng, J. Tang, J. Li, F. Dong and Z. Yang, Food Chem., 2018, 258, 352–358 CrossRef CAS PubMed.
- Y. Zhou, Q. Peng, L. Zhang, S. Cheng, L. Zeng, F. Dong and Z. Yang, Food Chem., 2019, 280, 27–33 CrossRef CAS PubMed.
- T. Koeduka, E. Fridman, D. R. Gang, D. G. Vassão, B. L. Jackson, C. M. Kish, I. Orlova, S. M. Spassova, N. G. Lewis, J. P. Noel, T. J. Baiga, N. Dudareva and E. Pichersky, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 10128–10133 CrossRef CAS PubMed.
- N. F. M. Shaipulah, J. K. Muhlemann, B. D. Woodworth, A. Van Moerkercke, J. C. Verdonk, A. A. Ramirez, M. A. Haring, N. Dudareva and R. C. Schuurink, Plant Physiol., 2015, 170, 717–731 CrossRef PubMed.
- T. Zhang, T. Huo, A. Ding, R. Hao, J. Wang, T. Cheng, F. Bao and Q. Zhang, PLoS One, 2019, 14, e0223974 CrossRef CAS PubMed.
- H. Yan, S. Baudino, J.-C. Caissard, N. Florence, H. Zhang, K. Tang, S. Li and S. Lu, Plant Physiol. Biochem., 2018, 129, 21–26 CrossRef CAS PubMed.
- J. R. Widhalm and N. Dudareva, Mol. Plant, 2015, 8, 83–97 CrossRef CAS PubMed.
- A. V. Qualley, J. R. Widhalm, F. Adebesin, C. M. Kish and N. Dudareva, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 16383–16388 CrossRef CAS PubMed.
- T. A. Akhtar and E. Pichersky, Plant Physiol., 2013, 162, 52–62 CrossRef CAS PubMed.
- K. Van Gelder, T. Forrester and T. A. Akhtar, Planta, 2020, 252, 3 CrossRef CAS PubMed.
- A. K. Gupta, T. A. Akhtar, A. Widmer, E. Pichersky and F. P. Schiestl, BMC Plant Biol., 2012, 12, 158 CrossRef CAS PubMed.
- T. Koeduka, M. Kajiyama, T. Furuta, H. Suzuki, T. Tsuge and K. Matsui, J. Biosci. Bioeng., 2016, 122, 679–684 CrossRef CAS PubMed.
- T. Koeduka, M. Kajiyama, H. Suzuki, T. Furuta, T. Tsuge and K. Matsui, Planta, 2016, 244, 725–736 CrossRef CAS PubMed.
- Y. Wang, H. Zhang, C. Wan, X. He, J. Huang, M. Lyu, Y. Yuan and B. Wu, Plants, 2022, 11, 13 CrossRef CAS PubMed.
- M. Kang, Y. Choi, H. Kim and S.-G. Kim, New Phytol., 2022, 234, 527–544 CrossRef CAS PubMed.
- H. Guo, N. D. Lackus, T. G. Köllner, R. Li, J. Bing, Y. Wang, I. T. Baldwin and S. Xu, Mol. Biol. Evol., 2019, 37, 1090–1099 CrossRef PubMed.
- I. Abe, Y. Takahashi, H. Morita and H. Noguchi, Eur. J. Biochem., 2001, 268, 3354–3359 CrossRef CAS PubMed.
- R. A. Raguso, A. Kelber, M. Pfaff, R. A. Levin and L. A. McDade, Ann. Mo. Bot. Gard., 2007, 94, 236–257 CrossRef.
- L. J. Nielsen and B. L. Møller, Phytochemistry, 2015, 120, 3–18 CrossRef CAS PubMed.
- S. Irmisch, A. Clavijo McCormick, G. A. Boeckler, A. Schmidt, M. Reichelt, B. Schneider, K. Block, J.-P. Schnitzler, J. Gershenzon, S. B. Unsicker and T. G. Köllner, Plant Cell, 2013, 25, 4737–4754 CrossRef CAS PubMed.
- M. Sørensen, E. H. J. Neilson and B. L. Møller, Mol. Plant, 2018, 11, 95–117 CrossRef PubMed.
- S. Irmisch, A. Clavijo McCormick, J. Günther, A. Schmidt, G. A. Boeckler, J. Gershenzon, S. B. Unsicker and T. G. Köllner, Plant J., 2014, 80, 1095–1107 CrossRef CAS PubMed.
- J. Martínez-Harms, A.-C. Warskulat, B. Dudek, G. Kunert, S. Lorenz, B. S. Hansson and B. Schneider, ChemBioChem, 2018, 19, 1553–1562 CrossRef PubMed.
- E. C. Tatsis, H. Böhm and B. Schneider, Phytochemistry, 2013, 92, 105–112 CrossRef CAS PubMed.
- J. C. Verdonk, M. A. Haring, A. J. van Tunen and R. C. Schuurink, Plant Cell, 2005, 17, 1612–1624 CrossRef CAS PubMed.
- U. Klahre, A. Gurba, K. Hermann, M. Saxenhofer, E. Bossolini, P. M. Guerin and C. Kuhlemeier, Curr. Biol., 2011, 21, 730–739 CrossRef CAS PubMed.
- B. Spitzer-Rimon, E. Marhevka, O. Barkai, I. Marton, O. Edelbaum, T. Masci, N.-K. Prathapani, E. Shklarman, M. Ovadis and A. Vainstein, Plant Cell, 2010, 22, 1961–1976 CrossRef CAS PubMed.
- A. Van Moerkercke, M. A. Haring and R. C. Schuurink, Plant J., 2011, 67, 917–928 CrossRef CAS PubMed.
- B. Spitzer-Rimon, M. Farhi, B. Albo, A. Cna’ani, M. M. Ben Zvi, T. Masci, O. Edelbaum, Y. Yu, E. Shklarman, M. Ovadis and A. Vainstein, Plant Cell, 2012, 24, 5089–5105 CrossRef CAS PubMed.
- M. R. Boersma, R. M. Patrick, S. L. Jillings, N. F. M. Shaipulah, P. Sun, M. A. Haring, N. Dudareva, Y. Li and R. C. Schuurink, Plant J., 2022, 109, 1134–1151 CrossRef CAS PubMed.
- A. Cna'ani, B. Spitzer-Rimon, J. Ravid, M. Farhi, T. Masci, J. Aravena-Calvo, M. Ovadis and A. Vainstein, New Phytol., 2015, 208, 708–714 CrossRef PubMed.
- F. Liu, Z. Xiao, L. Yang, Q. Chen, L. Shao, J. Liu and Y. Yu, New Phytol., 2017, 215, 1490–1502 CrossRef CAS PubMed.
- J. Ravid, B. Spitzer-Rimon, Y. Takebayashi, M. Seo, A. Cna'ani, J. Aravena-Calvo, T. Masci, M. Farhi and A. Vainstein, New Phytol., 2017, 215, 411–422 CrossRef CAS PubMed.
- J. H. Lynch, Y. C. Qian, L. Y. Guo, I. Maoz, X. Q. Huang, A. S. Garcia, G. Louie, M. E. Bowman, J. P. Noel, J. A. Morgan and N. Dudareva, Nat. Chem. Biol., 2020, 16, 850–856 CrossRef CAS PubMed.
- M. M. Ben Zvi, E. Shklarman, T. Masci, H. Kalev, T. Debener, S. Shafir, M. Ovadis and A. Vainstein, New Phytol., 2012, 195, 335–345 CrossRef CAS PubMed.
- Z. Yang, Y. Li, F. Gao, W. Jin, S. Li, S. Kimani, S. Yang, T. Bao, X. Gao and L. Wang, J. Exp. Bot., 2020, 71, 4140–4158 CrossRef CAS PubMed.
- G.-J. Hong, X.-Y. Xue, Y.-B. Mao, L.-J. Wang and X.-Y. Chen, Plant Cell, 2012, 24, 2635–2648 CrossRef CAS PubMed.
- Z. Yu, G. Zhang, J. A. Teixeira da Silva, C. Zhao and J. Duan, Plant Sci., 2021, 309, 110952 CrossRef CAS PubMed.
- F. Abbas, Y. Ke, Y. Zhou, Y. Yu, M. Waseem, U. Ashraf, C. Wang, X. Wang, X. Li, Y. Yue, R. Yu and Y. Fan, Front. Plant Sci., 2021, 12, 623742 CrossRef PubMed.
- Y. Ke, F. Abbas, Y. Zhou, R. Yu and Y. Fan, Front. Plant Sci., 2021, 12, 710826 CrossRef PubMed.
- F. Abbas, Y. Ke, Y. Zhou, Y. Yu, M. Waseem, U. Ashraf, X. Li, R. Yu and Y. Fan, Plant Cell Rep., 2021, 40, 1269–1284 CrossRef CAS PubMed.
- W. Ding, Q. Ouyang, Y. Li, T. Shi, L. Li, X. Yang, K. Ji, L. Wang and Y. Yue, Tree Physiol., 2019, 40, 557–572 CrossRef PubMed.
- H.-Y. Li, Y.-Z. Yue, W.-J. Ding, G.-W. Chen, L. Li, Y.-L. Li, T.-T. Shi, X.-L. Yang and L.-G. Wang, Genes, 2020, 11, 353 CrossRef CAS PubMed.
- Y.-Y. Yang, B. Ma, Y.-Y. Li, M.-Z. Han, J. Wu, X.-F. Zhou, J. Tian, W.-H. Wang, P.-S. Leng and Z.-H. Hu, Front. Plant Sci., 2022, 13, 1021576 CrossRef PubMed.
- K. Yoshida, N. Oyama-Okubo and M. Yamagishi, Mol. Breed., 2018, 38, 144 CrossRef.
- J. Cai, P. Zu and F. P. Schiestl, Sci. Rep., 2016, 6, 36966 CrossRef CAS PubMed.
- M. Oliva, R. Ovadia, A. Perl, E. Bar, E. Lewinsohn, G. Galili and M. Oren-Shamir, Plant Biotechnol. J., 2015, 13, 125–136 CrossRef CAS PubMed.
- V. Kumar, Y. Elazari, R. Ovadia, E. Bar, A. Nissim-Levi, N. Carmi, E. Lewinsohn and M. Oren-Shamir, Postharvest Biol. Technol., 2021, 181, 111657 CrossRef CAS.
- F. Abbas, X. Nian, Y. Zhou, Y. Ke, L. Liu, R. Yu and Y. Fan, Plant Physiol. Biochem., 2021, 167, 619–629 CrossRef CAS PubMed.
- R. M. Patrick, X.-Q. Huang, N. Dudareva and Y. Li, J. Exp. Bot., 2021, 72, 3704–3722 CrossRef CAS PubMed.
- S. Shi, S. Zhang, J. Wu, X. Liu and Z. Zhang, Front. Plant Sci., 2022, 13, 996474 CrossRef PubMed.
- J. R. Widhalm, R. Jaini, J. A. Morgan and N. Dudareva, Trends Plant Sci., 2015, 20, 545–550 CrossRef CAS PubMed.
- F. Adebesin, J. R. Widhalm, B. Boachon, F. Lefèvre, B. Pierman, J. H. Lynch, I. Alam, B. Junqueira, R. Benke, S. Ray, J. A. Porter, M. Yanagisawa, H. Y. Wetzstein, J. A. Morgan, M. Boutry, R. C. Schuurink and N. Dudareva, Science, 2017, 356, 1386–1388 CrossRef CAS PubMed.
- P. Liao, S. Ray, B. Boachon, J. H. Lynch, A. Deshpande, S. McAdam, J. A. Morgan and N. Dudareva, Nat. Chem. Biol., 2021, 17, 138–145 CrossRef CAS PubMed.
- A. Cna’ani, R. Shavit, J. Ravid, J. Aravena-Calvo, O. Skaliter, T. Masci and A. Vainstein, Front. Plant Sci., 2017, 8, 1898 CrossRef PubMed.
- B. Boachon, J. H. Lynch, S. Ray, J. Yuan, K. M. P. Caldo, R. R. Junker, S. A. Kessler, J. A. Morgan and N. Dudareva, Nat. Chem. Biol., 2019, 15, 583–588 CrossRef CAS PubMed.
- R. Delle-Vedove, B. Schatz and M. Dufay, Ann. Bot., 2017, 120, 1–20 CrossRef PubMed.
- P. J. Zu, W. U. Blanckenhorn and F. P. Schiestl, New Phytol., 2016, 209, 1208–1219 CrossRef CAS PubMed.
- F. P. Schiestl, New Phytol., 2015, 206, 571–577 CrossRef PubMed.
- I. Paul, M. P. Sarkar and P. B. S. Bhadoria, Chemoecology, 2022, 32, 49–68 CrossRef CAS.
- J. M. W. Ryalls, B. Langford, N. J. Mullinger, L. M. Bromfield, E. Nemitz, C. Pfrang and R. D. Girling, Environ. Pollut., 2022, 297, 118847 CrossRef CAS PubMed.
- E. Agathokleous, Z. Feng and J. Peñuelas, Trends Ecol. Evol., 2022, 37, 939–941 CrossRef CAS PubMed.
- X. Y. Yuan, Z. Z. Feng, C. F. Hu, K. Zhang, L. Y. Qu and E. Paoletti, Environ. Pollut., 2021, 291, 118141 CrossRef CAS PubMed.
- A. Saunier and J. D. Blande, Environ. Pollut., 2019, 255, 113257 CrossRef CAS PubMed.
- W. R. Glenny, J. B. Runyon and L. A. Burkle, New Phytol., 2018, 220, 785–798 CrossRef CAS PubMed.
- R. Atkinson, J. Arey, S. M. Aschmann, S. B. Corchnoy and Y. H. Shu, Int. J. Chem. Kinet., 1995, 27, 941–955 CrossRef CAS.
- J. D. Blande, Curr. Opin. Environ. Sci. Health, 2021, 19, 100228 CrossRef.
- J. D. Blande, J. K. Holopainen and U. Niinemets, Plant, Cell Environ., 2014, 37, 1892–1904 CrossRef CAS PubMed.
- J. D. Fuentes, M. Chamecki, T. Roulston, B. C. Chen and K. R. Pratt, Atmos. Environ., 2016, 141, 361–374 CrossRef CAS.
- A. Jürgens and M. Bischoff, Funct. Ecol., 2017, 31, 56–64 CrossRef.
- Q. S. McFrederick, J. D. Fuentes, T. Roulston, J. C. Kathilankal and M. Lerdau, Oecologia, 2009, 160, 411–420 CrossRef PubMed.
- G. Farré-Armengol, J. Peñuelas, T. Li, P. Yli-Pirilä, I. Filella, J. Llusià and J. D. Blande, New Phytol., 2016, 209, 152–160 CrossRef PubMed.
- A. Saunier, P. Grof-Tisza and J. D. Blande, Environ. Pollut., 2023, 316, 120573 CrossRef CAS PubMed.
- B. Cook, A. Haverkamp, B. S. Hansson, T. Roulston, M. Lerdau and M. Knaden, J. Chem. Ecol., 2020, 46, 987–996 CrossRef CAS PubMed.
- J. D. Fuentes, T. H. Roulston and J. Zenker, Environ. Res. Lett., 2013, 8, 014048 CrossRef CAS.
- R. D. Girling, I. Lusebrink, E. Farthing, T. A. Newman and G. M. Poppy, Sci. Rep., 2013, 3, 2779 CrossRef PubMed.
- I. Lusebrink, R. D. Girling, E. Farthing, T. A. Newman, C. W. Jackson and G. M. Poppy, J. Chem. Ecol., 2015, 41, 904–912 CrossRef CAS PubMed.
- F. Démares, L. Gibert, P. Creusot, B. Lapeyre and M. Proffit, Sci. Total Environ., 2022, 827, 154342 CrossRef PubMed.
- S. Dötterl, M. Vater, T. Rupp and A. Held, J. Chem. Ecol., 2016, 42, 486–489 CrossRef PubMed.
- M. Vanderplanck, B. Lapeyre, M. Brondani, M. Opsommer, M. Dufay, M. Hossaert-McKey and M. Proffit, Antioxidants, 2021, 10, 636 CrossRef CAS PubMed.
- IPCC, Climate Change 2021: The Physical Science Basis. Contribution of Working Group I to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change, ed. V. Masson-Delmotte, P. Zhai, A. Pirani, S. L. Connors, C. Péan, S. Berger, N. Caud, Y. Chen, L. Goldfarb, M. I. Gomis, M. Huang, K. Leitzell, E. Lonnoy, J. B. R. Matthews, T. K. Maycock, T. Waterfield, O. Yelekçi, R. Yu and B. Zhou, Cambridge University Press, Cambridge, United Kingdom, New York, NY, USA, 2021 Search PubMed.
- M. Borghi, L. P. de Souza, T. Yoshida and A. R. Fernie, New Phytol., 2019, 224, 1425–1441 CrossRef PubMed.
- M. Gérard, M. Vanderplanck, T. Wood and D. Michez, Emerging Top. Life Sci., 2020, 4, 77–86 CrossRef PubMed.
- M. Barman and A. Mitra, Environ. Exp. Bot., 2021, 181, 104296 CrossRef CAS.
- A. Cna'ani, J. K. Mühlemann, J. Ravid, T. Masci, A. Klempien, T. T. H. Nguyen, N. Dudareva, E. Pichersky and A. Vainstein, Plant, Cell Environ., 2015, 38, 1333–1346 CrossRef PubMed.
- G. Farré-Armengol, I. Filella, J. Llusià, U. Niinemets and J. Peñuelas, Global Change Biol., 2014, 20, 3660–3669 CrossRef PubMed.
- G. Farre-Armengol, I. Filella, J. Llusià, U. Niinemets and J. Peñuelas, Funct. Plant Biol., 2015, 42, 851–857 CrossRef CAS PubMed.
- Z. H. Hu, H. X. Zhang, P. S. Leng, J. Zhao, W. H. Wang and S. D. Wang, Acta Physiol. Plant., 2013, 35, 1691–1700 CrossRef CAS.
- S. H. Cheng, X. M. Fu, X. Mei, Y. Zhou, B. Du, N. Watanabe and Z. Y. Yang, Plant Physiol. Biochem., 2016, 107, 1–8 CrossRef CAS PubMed.
- J. X. Fu, D. Hou, C. Zhang, Z. Y. Bao, H. B. Zhao and S. Q. Hu, Molecules, 2017, 22, 430 CrossRef PubMed.
- D. R. Campbell, P. Sosenski and R. A. Raguso, Ann. Bot., 2019, 123, 601–610 CrossRef CAS PubMed.
- L. A. Burkle and J. B. Runyon, Global Change Biol., 2016, 22, 1644–1654 CrossRef PubMed.
- C. C. Rering, J. G. Franco, K. M. Yeater and R. E. Mallinger, Ecosphere, 2020, 11, e03254 CrossRef.
- I. N. A. Salman, A. Cna'ani, V. Tzin and M. Seifan, Entomol. Exp. Appl., 2022, 170, 666–680 CrossRef CAS.
- C. C. Jaworski, B. Geslin, M. Zakardjian, C. Lecareux, P. Caillault, G. Nève, J. Y. Meunier, S. Dupouyet, A. C. T. Sweeney, O. T. Lewis, L. V. Dicks and C. Fernandez, J. Ecol., 2022, 110, 2628–2648 CrossRef CAS.
- R. J. Höfer, M. Ayasse and J. Kuppler, Front. Plant Sci., 2021, 11, 564802 CrossRef PubMed.
- V. J. Luizzi, M. Friberg and H. Petrén, Funct. Ecol., 2021, 35, 1655–1665 CrossRef CAS.
- M. Friberg, M. T. Waters and J. N. Thompson, Ann. Bot., 2017, 120, 471–478 CrossRef CAS PubMed.
- C. J. Majetic, A. M. Fetters, O. M. Beck, E. F. Stachnik and K. M. Beam, Flora, 2017, 232, 183–193 CrossRef.
- T. Dorey and F. P. Schiestl, Evolution, 2022, 76, 2930–2944 Search PubMed.
- K. M. Becklin, G. Gamez, B. Uelk, R. A. Raguso and C. Galen, Am. J. Bot., 2011, 98, 1299–1308 CrossRef PubMed.
- N. Wei, R. L. Whyle, T.-L. Ashman and M. A. Jamieson, Hortic. Res., 2022, 9, uhab005 CrossRef PubMed.
- L. Shapiro, C. M. De Moraes, A. G. Stephenson and M. C. Mescher, Ecol. Lett., 2012, 15, 1430–1438 CrossRef PubMed.
- A. Cellini, V. Giacomuzzi, I. Donati, B. Farneti, M. T. Rodriguez-Estrada, S. Savioli, S. Angeli and F. Spinelli, ISME J., 2019, 13, 847–859 CrossRef CAS PubMed.
- R. R. Junker and D. Tholl, J. Chem. Ecol., 2013, 39, 810–825 CrossRef CAS PubMed.
- P. Gaube, R. R. Junker and A. Keller, Basic Appl. Ecol., 2021, 50, 1–15 CrossRef.
- R. R. Junker and A. Keller, FEMS Microbiol. Ecol., 2015, 91, fiv097 CrossRef PubMed.
- A. Crowley-Gall, C. C. Rering, A. B. Rudolph, R. L. Vannette and J. J. Beck, Curr. Opin. Insect. Sci., 2021, 44, 23–34 CrossRef PubMed.
- E. D. Fenner, T. Scapini, M. da Costa Diniz, A. Giehl, H. Treichel, S. Álvarez-Pérez and S. L. Alves Jr., J. Fungi, 2022, 8, 984 CrossRef CAS PubMed.
- J. S. Francis, A. R. Tatarko, S. K. Richman, A. D. Vaudo and A. S. Leonard, Curr. Opin. Insect. Sci., 2021, 44, 16–22 CrossRef PubMed.
- J. Klaps, B. Lievens and S. Álvarez-Pérez, Fungal Biol. Biotechnol., 2020, 7, 1 CrossRef PubMed.
- R. L. Vannette, Annu. Rev. Ecol. Evol. Syst., 2020, 51, 363–386 CrossRef.
- A. M. Golonka, B. Obi Johnson, J. Freeman and D. W. Hinson, East. biol., 2014, 3, 1–26 Search PubMed.
- C. C. Rering, J. J. Beck, G. W. Hall, M. M. McCartney and R. L. Vannette, New Phytol., 2018, 220, 750–759 CrossRef CAS PubMed.
- C. C. Rering, A. B. Rudolph and J. J. Beck, Environ. Microbiol., 2021, 23, 4141–4150 CrossRef CAS PubMed.
- R. N. Schaeffer, C. C. Rering, I. Maalouf, J. J. Beck and R. L. Vannette, Biol. Lett., 2019, 15, 20190132 CrossRef CAS PubMed.
- I. S. Sobhy, D. Baets, T. Goelen, B. Herrera-Malaver, L. Bosmans, W. Van den Ende, K. J. Verstrepen, F. Wäckers, H. Jacquemyn and B. Lievens, Front. Plant Sci., 2018, 9, 1009 CrossRef PubMed.
- A. Colda, S. Bossaert, C. Verreth, B. Vanhoutte, O. Honnay, W. Keulemans and B. Lievens, PLoS One, 2021, 16, e0250203 CrossRef CAS PubMed.
- R. L. Vannette, M. P. L. Gauthier and T. Fukami, Proc. R. Soc. B: Biol. Sci., 2013, 280, 20122601 CrossRef PubMed.
- A. Cusumano, P. Bella, E. Peri, M. Rostás, S. Guarino, B. Lievens and S. Colazza, Microb. Ecol., 2023, 86, 364–376 CrossRef CAS PubMed.
- C. Helletsgruber, S. Dötterl, U. Ruprecht and R. R. Junker, J. Chem. Ecol., 2017, 43, 1073–1077 CrossRef CAS PubMed.
- J. Peñuelas, G. Farré-Armengol, J. Llusia, A. Gargallo-Garriga, L. Rico, J. Sardans, J. Terradas and I. Filella, Sci. Rep., 2014, 4, 6727 CrossRef PubMed.
- L. Huang, Y. Liu, L. Dou, S. Pan, Z. Li, J. Zhang and J. Li, PeerJ, 2022, 10, e14107 CrossRef PubMed.
- A. Kessler and A. Chautá, Emerging Top. Life Sci., 2020, 4, 33–43 CrossRef PubMed.
- D. Lucas-Barbosa, Trends Plant Sci., 2016, 21, 125–133 CrossRef CAS PubMed.
- D. Lucas-Barbosa, J. J. A. van Loon and M. Dicke, Phytochemistry, 2011, 72, 1647–1654 CrossRef CAS PubMed.
- Q. Rusman, D. Lucas-Barbosa, E. H. Poelman and M. Dicke, Trends Plant Sci., 2019, 24, 725–740 CrossRef CAS PubMed.
- P. Tunes, S. Dötterl and E. Guimarães, Front. Plant Sci., 2022, 13, 813418 CrossRef PubMed.
- C. E. Reisenman, J. A. Riffell, K. Duffy, A. Pesque, D. Mikles and B. Goodwin, J. Chem. Ecol., 2013, 39, 76–89 CrossRef CAS PubMed.
- M. Pareja, E. Qvarfordt, B. Webster, P. Mayon, J. Pickett, M. Birkett and R. Glinwood, PLoS One, 2012, 7, e31971 CrossRef CAS PubMed.
- Q. Rusman, E. H. Poelman, F. Nowrin, G. Polder and D. Lucas-Barbosa, Plant, Cell Environ., 2019, 42, 1882–1896 CrossRef CAS PubMed.
- F. P. Schiestl, H. Kirk, L. Bigler, S. Cozzolino and G. A. Desurmont, New Phytol., 2014, 203, 257–266 CrossRef CAS PubMed.
- M. Bruinsma, D. Lucas-Barbosa, C. J. M. ten Broeke, N. M. van Dam, T. A. van Beek, M. Dicke and J. J. A. van Loon, J. Chem. Ecol., 2014, 40, 39–49 CrossRef CAS PubMed.
- M. Hoffmeister, N. Wittkopper and R. R. Junker, Oikos, 2016, 125, 1241–1249 CrossRef.
- A. Kessler, R. Halitschke and K. Poveda, Ecology, 2011, 92, 1769–1780 CrossRef PubMed.
- D. Kessler, C. Diezel and I. T. Baldwin, Curr. Biol., 2010, 20, 237–242 CrossRef CAS PubMed.
- S. Cozzolino, S. Fineschi, M. Litto, G. Scopece, J. Trunschke and F. P. Schiestl, J. Chem. Ecol., 2015, 41, 622–630 CrossRef CAS PubMed.
- S. E. Ramos and F. P. Schiestl, Science, 2019, 364, 193–196 CrossRef CAS PubMed.
- S. E. Ramos and F. P. Schiestl, Front. Ecol. Evol., 2020, 8, 30 CrossRef.
- J. Ollerton, Annu. Rev. Ecol. Evol. Syst., 2017, 48, 353–376 CrossRef.
- D. Schneider, M. Wink, F. Sporer and P. Lounibos, Naturwissenschaften, 2002, 89, 281–294 CrossRef CAS PubMed.
- G. Gottsberger, Plant Divers. Evol., 2016, 131, 263–362 CrossRef.
- M. R. Moore, R. D. Cave and M. A. Branham, Zookeys, 2018, 745, 1–99 CrossRef PubMed.
- M. R. Moore and M. L. Jameson, J. Insect Sci., 2013, 13, 100 CrossRef PubMed.
- G. Gottsberger, I. Silberbauer-Gottsberger, R. S. Seymour and S. Dötterl, Flora, 2012, 207, 107–118 CrossRef.
- S. Dötterl, A. David, W. Boland, I. Silberbauer-Gottsberger and G. Gottsberger, J. Chem. Ecol., 2012, 38, 1539–1543 CrossRef PubMed.
- A. C. D. Maia, M. Gibernau, S. Dötterl, D. M. D. F. Navarro, K. Seifert, T. Müller and C. Schlindwein, Phytochemistry, 2013, 93, 71–78 CrossRef CAS PubMed.
- J. Pereira, C. Schlindwein, Y. Antonini, A. C. D. Maia, S. Dötterl, C. Martins, D. M. D. F. Navarro and R. Oliveira, Biol. J. Linn. Soc., 2014, 111, 679–691 CrossRef.
- A. C. D. Maia, C. Grimm, M. Schubert, F. Etl, E. G. Gonçalves, D. M. D. F. Navarro, S. Schulz and S. Dötterl, J. Chem. Ecol., 2019, 45, 214–215 CrossRef CAS PubMed.
- P. Stamm, F. Etl, A. C. D. Maia, S. Dötterl and S. Schulz, J. Org. Chem., 2021, 86, 5245–5254 CrossRef CAS PubMed.
- T. N. Suinyuy, J. S. Donaldson and S. D. Johnson, Proc. R. Soc. B: Biol. Sci., 2015, 282, 20152053 CrossRef PubMed.
- T. N. Suinyuy and S. D. Johnson, Bot. J. Linn. Soc., 2021, 195, 233–248 CrossRef.
- S. Salzman, A. Dahake, W. Kandalaft, W. A. Valencia-Montoya, M. Calonje, C. D. Specht and R. A. Raguso, Curr. Biol., 2023, 33, 1654–1664 CrossRef CAS PubMed.
- S. L. Steenhuisen, A. Jürgens and S. D. Johnson, J. Chem. Ecol., 2013, 39, 438–446 CrossRef CAS PubMed.
- L. Blažytė-Čereškienė, V. Apšegaitė and V. Būda, Arthropod-Plant Interact., 2019, 13, 735–743 CrossRef.
- T. D. J. Sayers, M. J. Steinbauer, K. Farnier and R. E. Miller, Bot. J. Linn. Soc., 2020, 193, 375–401 CrossRef.
- B. S. Brodie, A. Renyard, R. Gries, H. M. Zhai, S. Ogilvie, J. Avery and G. Gries, Arthropod-Plant Interact., 2018, 12, 591–599 CrossRef.
- J. Vuts, C. M. Woodcock, J. C. Caulfield, S. J. Powers, J. A. Pickett and M. A. Birkett, Pest Manage. Sci., 2018, 74, 2069–2075 CrossRef CAS PubMed.
- T. J. A. Bruce, J. L. Martin, L. E. Smart and J. A. Pickett, Pest Manage. Sci., 2011, 67, 1303–1308 CrossRef CAS PubMed.
- C. L. Xiu, B. Xu, H. S. Pan, W. Zhang, Y. Z. Yang and Y. H. Lu, J. Integr. Agric., 2019, 18, 873–883 CrossRef CAS.
- A. C. D. Maia, L. K. Reis, D. M. D. F. Navarro, F. Aristone, C. A. Colombo, J. Carreño-Barrera, L. A. Núñez-Avellaneda and G. K. N. Santos, J. Appl. Entomol., 2020, 144, 33–40 CrossRef CAS.
- A. C. D. Maia, D. M. D. F. Navarro, L. A. Núñez-Avellaneda, J. Carreño-Barrera, L. Iannuzzi, J. Cardona-Duque and W. A. G. Nantes, Sci. Nat., 2021, 108, 21 CrossRef CAS PubMed.
- A. P. Favaris, A. C. Tuler, W. D. Silva, S. R. Rodrigues, W. S. Leal and J. M. S. Bento, PLoS One, 2020, 15, e0235028 CrossRef CAS PubMed.
- A. C. D. Maia, G. K. N. Santos, E. G. Gonçalves, D. M. D. F. Navarro and L. A. Nuñez-Avellaneda, Pest Manage. Sci., 2018, 74, 2053–2058 CrossRef CAS PubMed.
- A. Karmakar, A. Mukherjee and A. Barik, Entomol. Exp. Appl., 2016, 158, 133–141 CrossRef CAS.
- N. Sarkar, S. Mitra and A. Barik, Int. J. Pest Manage., 2017, 63, 138–145 CrossRef.
- D. Kessler, C. Diezel, D. G. Clark, T. A. Colquhoun and I. T. Baldwin, Ecol. Lett., 2013, 16, 299–306 CrossRef PubMed.
- A. de Fouchier, W. B. Walker, N. Montagné, C. Steiner, M. Binyameen, F. Schlyter, T. Chertemps, A. Maria, M. C. François, C. Monsempes, P. Anderson, B. S. Hansson, M. C. Larsson and E. Jacquin-Joly, Nat. Commun., 2017, 8, 5709 Search PubMed.
- M. B. Guo, L. X. Du, Q. Y. Chen, Y. L. Feng, J. Zhang, X. X. Zhang, K. Tian, S. Cao, T. Y. Huang, E. Jacquin-Joly, G. R. Wang and Y. Liu, Mol. Biol. Evol., 2021, 38, 1413–1427 CrossRef CAS PubMed.
- J. A. Riffell, H. Lei, L. Abrell and J. G. Hildebrand, Science, 2013, 339, 200–204 CrossRef CAS PubMed.
- A. Haverkamp, F. Yon, I. W. Keesey, C. Missbach, C. Koenig, B. S. Hansson, I. T. Baldwin, M. Knaden and D. Kessler, eLife, 2016, 5, e15039 CrossRef PubMed.
- W. W. Zhou, A. Kuegler, E. McGale, A. Haverkamp, M. Knaden, H. Guo, F. Beran, F. Yon, R. Li, N. Lackus, T. G. Köllner, J. Bing, M. C. Schuman, B. S. Hansson, D. Kessler, I. T. Baldwin and S. Q. Xu, Curr. Biol., 2017, 27, 1336–1341 CrossRef CAS PubMed.
- E. Adam, B. S. Hansson and M. Knaden, J. Exp. Biol., 2021, 224, jeb242780 CrossRef PubMed.
- M. Bischoff, R. A. Raguso, A. Jürgens and D. R. Campbell, Evolution, 2015, 69, 1–13 CrossRef PubMed.
- F. Balao, J. Herrera, S. Talavera and S. Dötterl, Phytochemistry, 2011, 72, 601–609 CrossRef CAS PubMed.
- M. von Arx, J. Goyret, G. Davidowitz and R. A. Raguso, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 9471–9476 CrossRef CAS PubMed.
- A. Dahake, P. Jain, C. C. Vogt, W. Kandalaft, A. D. Stroock and R. A. Raguso, Nat. Commun., 2022, 13, 7773 CrossRef CAS PubMed.
- A. Shuttleworth and S. D. Johnson, Entomol. Exp. Appl., 2020, 168, 191–197 CrossRef.
- C. Eby, M. G. T. Gardiner, R. Gries, G. J. R. Judd, G. Khaskin and G. Gries, Entomol. Exp. Appl., 2013, 147, 82–92 CrossRef CAS.
- A. Jürgens, U. Glück, G. Aas and S. Dötterl, Bot. J. Linn. Soc., 2014, 175, 624–640 CrossRef.
- B. P. Molnár, Z. Kárpáti, A. Nagy, I. Szarukán, J. Csabai, S. Koczor and M. Tóth, J. Chem. Ecol., 2019, 45, 657–666 CrossRef PubMed.
- G. Thöming and G. K. Knudsen, Phytochemistry, 2014, 100, 66–75 CrossRef PubMed.
- M. Friberg, C. Schwind, L. C. Roark, R. A. Raguso and J. N. Thompson, J. Chem. Ecol., 2014, 40, 955–965 CrossRef CAS PubMed.
- F. P. Schiestl, E. A. Wallin, J. J. Beck, M. Friberg and J. N. Thompson, Arthropod-Plant Interact., 2021, 15, 209–221 CrossRef CAS.
- G. P. Svensson, T. Okamoto, A. Kawakita, R. Goto and M. Kato, New Phytol., 2010, 186, 995–1004 CrossRef CAS PubMed.
- D. H. Huang, J. Mamut, X. F. Yang, F. C. Shi and H. H. Li, Diversity, 2022, 14, 266 CrossRef.
- T. Okamoto, in Obligate Pollination Mutualism. Ecological Research Monographs, ed. M. Kato and A. Kawakita, Springer, Tokyo, 2017 Search PubMed.
- T. Okamoto, A. Kawakita, R. Goto, G. P. Svensson and M. Kato, Proc. R. Soc. B, 2013, 280, 20132280 CrossRef PubMed.
- T. Okamoto, G. P. Svensson, R. Goto, A. Kawakita and M. Kato, Plant Species Biol., 2022, 37, 197–208 CrossRef.
- G. P. Svensson, O. Pellmyr and R. A. Raguso, Oikos, 2011, 120, 1577–1583 CrossRef CAS.
- M. Kinoshita, F. J. Stewart and H. Omura, BioEssays, 2017, 39, 1600086 CrossRef PubMed.
- S. Lehner, S. Schulz and S. Dötterl, Front. Plant Sci., 2022, 13, 994851 CrossRef PubMed.
- S. Andersson, Chemoecology, 2003, 13, 13–20 CrossRef CAS.
- S. Andersson, L. A. Nilsson, I. Groth and G. Bergström, Bot. J. Linn. Soc., 2002, 140, 129–153 CrossRef.
- A. M. El-Sayed, A. Sporle, K. Colhoun, J. Furlong, R. White and D. M. Suckling, Chemoecology, 2018, 28, 39–49 CrossRef CAS.
- R. A. Raguso, Appl. Entomol. Zool., 2020, 55, 1–7 CrossRef.
- T. S. Woodcock, B. M. H. Larson, P. G. Kevan, D. W. Inouye and K. Lunau, J. Pollinat. Ecol., 2014, 12, 63–94 CrossRef.
- C. Primante and S. Dötterl, J. Chem. Ecol., 2010, 36, 1207–1210 CrossRef CAS PubMed.
- L. Dormont, M. Mulatier, D. Carrasco and A. Cohuet, J. Chem. Ecol., 2021, 47, 351–393 CrossRef CAS PubMed.
- R. Ignell and S. R. Hill, Curr. Opin. Insect. Sci., 2020, 40, 6–10 CrossRef PubMed.
- E. K. Lutz, C. Lahondère, C. Vinauger and J. A. Riffell, Curr. Opin. Insect. Sci., 2017, 20, 75–83 CrossRef PubMed.
- A. Mafra-Neto and T. Dekker, Curr. Opin. Insect. Sci., 2019, 34, 105–111 CrossRef PubMed.
- J. M. Muema, J. L. Bargul, S. N. Njeru, J. O. Onyango and S. S. Imbahale, Parasites Vectors, 2017, 10, 184 CrossRef PubMed.
- V. O. Nyasembe and B. Torto, Phytochem. Lett., 2014, 8, 196–201 CrossRef CAS PubMed.
- M. Wooding, Y. Naudé, E. Rohwer and M. Bouwer, Parasites Vectors, 2020, 13, 80 CrossRef PubMed.
- P. E. Otienoburu, B. Ebrahimi, P. L. Phelan and W. A. Foster, J. Chem. Ecol., 2012, 38, 873–881 CrossRef CAS PubMed.
- S. von Oppen, H. Masuh, S. Licastro, E. Zerba and P. Gonzalez-Audino, Entomol. Exp. Appl., 2015, 155, 184–192 CAS.
- D. A. H. Peach, R. Gries, H. M. Zhai, N. Young and G. Gries, Sci. Rep., 2019, 9, 3908 CrossRef PubMed.
- T. Okamoto, Y. Okuyama, R. Goto, M. Tokoro and M. Kato, J. Evol. Biol., 2015, 28, 590–600 CrossRef CAS PubMed.
- S. Ibanez, S. Dötterl, M. C. Anstett, S. Baudino, J. C. Caissard, C. Gallet and L. Despres, New Phytol., 2010, 188, 451–463 CrossRef CAS PubMed.
- B. Song, G. Chen, J. Stöcklin, D. L. Peng, Y. Niu, Z. M. Li and H. Sun, New Phytol., 2014, 203, 1109–1118 CrossRef CAS PubMed.
- A. Jürgens, S. L. Wee, A. Shuttleworth and S. D. Johnson, Ecol. Lett., 2013, 16, 1157–1167 CrossRef PubMed.
- G. Chen, X. K. Ma, A. Jürgens, J. Lu, E. X. Liu, W. B. Sun and X. H. Cai, J. Chem. Ecol., 2015, 41, 808–815 CrossRef CAS PubMed.
- M. Moré, A. A. Cocucci and R. A. Raguso, Int. J. Plant Sci., 2013, 174, 863–876 CrossRef.
- M. Moré, P. Mulieri, M. Battán-Horenstein, A. A. Cocucci and R. A. Raguso, Arthropod-Plant Interact., 2019, 13, 375–386 CrossRef.
- A. Shuttleworth and S. D. Johnson, Proc. R. Soc. B: Biol. Sci., 2010, 277, 2811–2819 CrossRef CAS PubMed.
- S. L. Wee, S. B. Tan and A. Jürgens, Phytochemistry, 2018, 153, 120–128 CrossRef CAS PubMed.
- P. Zito, M. Sajeva, A. Raspi and S. Dötterl, Chemoecology, 2014, 24, 261–267 CrossRef CAS.
- A. Shuttleworth, S. D. Johnson and A. Jürgens, Flora, 2017, 232, 92–103 CrossRef.
- P. Zito, S. Dötterl and M. Sajeva, J. Chem. Ecol., 2015, 41, 340–349 CrossRef CAS PubMed.
- S. D. Johnson, J. Sivechurran, S. Doarsamy and A. Shuttleworth, New Phytol., 2020, 228, 1662–1673 CrossRef CAS PubMed.
- G. Chen, A. Jürgens, L. D. Shao, Y. Liu, W. B. Sun and C. F. Xia, J. Chem. Ecol., 2015, 41, 244–252 CrossRef CAS PubMed.
- P. Zito, S. Guarino, E. Peri, M. Sajeva and S. Colazza, Arthropod-Plant Interact., 2013, 7, 485–489 CrossRef.
- K. R. Goodrich and A. Jürgens, New Phytol., 2018, 217, 74–81 CrossRef PubMed.
- A. Heiduk, H. Kong, I. Brake, M. von Tschirnhaus, T. Tolasch, A. G. Tröger, E. Wittenberg, W. Francke, U. Meve and S. Dötterl, Front. Ecol. Evol., 2015, 3, 66 Search PubMed.
- B. Oelschlägel, M. Nuss, M. von Tschirnhaus, C. Pätzold, C. Neinhuis, S. Dötterl and S. Wanke, New Phytol., 2015, 206, 342–351 CrossRef PubMed.
- S. Dötterl and N. Vereecken, Can. J. Zool., 2010, 88, 668–697 CrossRef.
- C. D. Michener, The bees of the world, The John Hopkins University Press, Baltimore, 2nd edn, 2007 Search PubMed.
- R. Kubo and M. Ono, Entomol. Sci., 2014, 17, 432–434 CrossRef.
- G. Rabeschini, P. J. Bergamo and C. E. P. Nunes, J. Chem. Ecol., 2021, 47, 444–454 CrossRef CAS PubMed.
- A. C. Knauer and F. P. Schiestl, Ecol. Lett., 2015, 18, 135–143 CrossRef CAS PubMed.
- C. Rodriguez-Saona, L. Parra, A. Quiroz and R. Isaacs, Ann. Bot., 2011, 107, 1377–1390 CrossRef PubMed.
- F. Mas, A. Harper, R. Horner, T. Welsh, P. Jaksons and D. M. Suckling, J. Sci. Food Agric., 2018, 98, 4445–4453 CrossRef CAS PubMed.
- M. Rachersberger, G. D. Cordeiro, I. Schäffler and S. Dötterl, J. Agric. Food Chem., 2019, 67, 13221–13227 CrossRef CAS PubMed.
- K. Lukas, S. Dötterl, M. Ayasse and H. Burger, Front. Ecol. Evol., 2020, 8, 1–14 CrossRef.
- A. Morse, P. Kevan, L. Shipp, S. Khosla and B. McGarvey, Environ. Entomol., 2012, 41, 855–864 CrossRef.
- G. D. Cordeiro, M. Pinheiro, S. Dötterl and I. Alves-dos-Santos, Plant Biol., 2017, 19, 132–139 CrossRef CAS PubMed.
- C. A. Martínez-Martínez, G. D. Cordeiro, H. O. J. Martins, R. O. A. C. Kobal, P. Milet-Pinheiro, M. A. Stanton, E. L. Franco, C. Krug, S. Mateus, C. Schlindwein, S. Dötterl and I. Alves-dos-Santos, Front. Ecol. Evol., 2021, 9, 676743 CrossRef.
- C. Krug, G. D. Cordeiro, I. Schäffler, C. I. Silva, R. Oliveira, C. Schlindwein, S. Dötterl and I. Alves-dos-Santos, Front. Plant Sci., 2018, 9, 1072 CrossRef PubMed.
- C. Krug, M. V. B. Garcia and F. B. Gomes, Apidologie, 2015, 46, 164–166 CrossRef.
- P. Milet-Pinheiro, M. Ayasse, H. E. M. Dobson, C. Schlindwein, W. Francke and S. Dötterl, J. Chem. Ecol., 2013, 39, 1347–1360 CrossRef CAS PubMed.
- H. Burger, N. Joos and M. Ayasse, Front. Ecol. Evol., 2021, 9, 682960 CrossRef.
- P. Milet-Pinheiro, K. Herz, S. Dötterl and M. Ayasse, BMC Ecol., 2016, 16, 20 CrossRef PubMed.
- K. Brandt, S. Dötterl, W. Francke, M. Ayasse and P. Milet-Pinheiro, J. Chem. Ecol., 2017, 43, 4–12 CrossRef CAS PubMed.
- S. Dötterl, U. Füssel, A. Jürgens and G. Aas, J. Chem. Ecol., 2005, 31, 2993–2998 CrossRef PubMed.
- H. Burger, M. Ayasse, S. Dötterl, S. Kreissl and C. G. Galizia, J. Comp. Physiol., A, 2013, 199, 751–761 CrossRef CAS PubMed.
- I. Schäffler, F. Balao and S. Dötterl, Ann. Bot., 2012, 110, 125–138 CrossRef PubMed.
- M. Castañeda-Zárate, S. D. Johnson and T. van der Niet, Curr. Biol., 2021, 31, 238–246 CrossRef PubMed.
- H. Burger, M. Ayasse, C. M. Häberlein, S. Schulz and S. Dötterl, S. Afr. J. Bot., 2010, 76, 788–795 CrossRef CAS.
- H. Burger, S. Dötterl, C. Häberlein, S. Schulz and M. Ayasse, Oecologia, 2012, 168, 727–736 CrossRef PubMed.
- A. T. Carvalho, S. Dötterl and C. Schlindwein, J. Chem. Ecol., 2014, 40, 1126–1134 CrossRef CAS PubMed.
- T. Eltz, D. W. Roubik and M. W. Whitten, Physiol. Entomol., 2003, 28, 251–260 CrossRef CAS.
- S. Vogel, Österr. Bot. Z., 1966, 113, 302–361 CrossRef.
- S. Ramírez, R. L. Dressler and M. Ospina, Biota Colomb., 2002, 3, 7–118 Search PubMed.
- J. Brodmann, D. Emer and M. Ayasse, Plant Biol., 2012, 14, 500–505 CrossRef CAS PubMed.
- C. C. L. Soler, M. Proffit, J.-M. Bessière, M. Hossaert-McKey and B. Schatz, Ecol. Lett., 2012, 15, 978–985 CrossRef PubMed.
- D. Gu, D.-R. Yang, P. Yang, Y.-Q. Peng and Z.-J. Wang, Chem. Ecol., 2016, 32, 733–741 CrossRef CAS.
- M. C. Foti, M. Rostás, E. Peri, K. C. Park, T. Slimani, S. D. Wratten and S. Colazza, J. Pest Sci., 2017, 90, 299–310 CrossRef.
- C. de Vega, C. M. Herrera and S. Dötterl, Perspect. Plant Ecol. Evol. Syst., 2014, 16, 32–42 CrossRef.
- T. Yang, G. Stoopen, M. Thoen, G. Wiegers and M. A. Jongsma, Plant Biotechnol. J., 2013, 11, 875–882 CrossRef CAS PubMed.
- Z. S. Abdullah, K. J. Ficken, B. P. J. Greenfield and T. M. Butt, J. Chem. Ecol., 2014, 40, 534–540 CrossRef CAS PubMed.
- F. Etl, A. Berger, A. Weber, J. Schönenberger and S. Dötterl, J. Chem. Ecol., 2016, 42, 300–304 CrossRef CAS PubMed.
- S. D. Johnson, P. M. Burgoyne, L. D. Harder and S. Dötterl, Proc. R. Soc. B, 2011, 278, 2303–2310 CrossRef CAS PubMed.
- P. Wester, S. D. Johnson and A. Pauw, New Phytol., 2019, 222, 1624–1637 CrossRef CAS PubMed.
- S. D. Johnson and K. Govender, Philos. Trans. R. Soc., B, 2022, 377, 20210167 CrossRef CAS PubMed.
- A. C. Knauer, M. Bakhtiari and F. P. Schiestl, Nat. Commun., 2018, 9, 1367 CrossRef PubMed.
- A. Hammerbacher, T. A. Coutinho and J. Gershenzon, Plant, Cell Environ., 2019, 42, 2827–2843 CrossRef CAS PubMed.
- R. R. Junker and N. Blüthgen, Ann. Bot., 2010, 105, 777–782 CrossRef PubMed.
- J. Li, H. Hu, J. Mao, L. Yu, G. Stoopen, M. Wang, R. Mumm, N. C. A. de Ruijter, M. Dicke, M. A. Jongsma and C. Wang, New Phytol., 2019, 223, 1607–1620 CrossRef CAS PubMed.
- R. R. Junker, J. Gershenzon and S. B. Unsicker, J. Chem. Ecol., 2011, 37, 1323–1331 CrossRef CAS PubMed.
- R. R. Junker and N. Blüthgen, Evol. Ecol. Res., 2008, 10, 295–308 Search PubMed.
- J. G. Pattrick, T. Shepherd, W. Hoppitt, N. S. Plowman and W. Pat, Arthropod-Plant Interact., 2017, 11, 623–627 CrossRef.
- C. Galen, R. Kaczorowski, S. L. Todd, J. Geib and R. A. Raguso, Am. Nat., 2011, 177, 258–272 CrossRef PubMed.
- D. Kessler, J. Bing, A. Haverkamp and I. T. Baldwin, Funct. Ecol., 2019, 33, 1223–1232 CrossRef.
- R. R. Junker, I. M. M. Heidinger and N. Blüthgen, J. Orthoptera Res., 2010, 19, 69–74 CrossRef.
- M. Huang, A. M. Sanchez-Moreiras, C. Abel, R. Sohrabi, S. Lee, J. Gershenzon and D. Tholl, New Phytol., 2012, 193, 997–1008 CrossRef CAS PubMed.
- R. C. F. Burdon, R. R. Junker, D. G. Scofield and A. L. Parachnowitsch, Chemoecology, 2018, 28, 11–19 CrossRef CAS PubMed.
- R. R. Junker, C. Loewel, R. Gross, S. Dötterl, A. Keller and N. Blüthgen, Plant Biol., 2011, 13, 918–924 CrossRef CAS PubMed.
- E. A. Dannon, M. Tamò, A. Van Huis and M. Dicke, J. Chem. Ecol., 2010, 36, 1083–1091 CrossRef CAS PubMed.
- M. Ayasse, F. P. Schiestl, H. F. Paulus, F. Ibarra and W. Francke, Proc. R. Soc. B, 2003, 270, 517–522 CrossRef CAS PubMed.
- S. Dötterl, D. Burkhardt, B. Weißbecker, A. Jürgens, S. Schütz and A. Mosandl, J. Chromatogr. A, 2006, 1113, 231–238 CrossRef PubMed.
- R. A. Raguso, Curr. Opin. Plant Biol., 2016, 32, 31–36 CrossRef CAS PubMed.
- C. E. Reisenman, J. A. Riffell, E. A. Bernays and J. G. Hildebrand, Proc. R. Soc. B, 2010, 277, 2371–2379 CrossRef CAS PubMed.
- T. Eltz, E. Hedenström, J. Bång, E. A. Wallin and J. Andersson, J. Chem. Ecol., 2010, 36, 1322–1326 CrossRef CAS PubMed.
- M. W. Whitten, H. G. Hills and N. H. Williams, Phytochemistry, 1988, 27, 2759–2760 CrossRef.
- D. L. P. Schorkopf, L. Mitko and T. Eltz, J. Chem. Ecol., 2011, 37, 953–960 CrossRef CAS PubMed.
- K. Brandt, S. Dötterl, R. Fuchs, D. M. A. F. Navarro, I. C. S. Machado, D. Dobler, O. Reiser, M. Ayasse and P. Milet-Pinheiro, J. Chem. Ecol., 2019, 45, 464–473 CrossRef CAS PubMed.
- F. Etl, W. Francke, J. Schönenberger and S. Dötterl, J. Chem. Ecol., 2022, 48, 263–269 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2023 |