Open Access Article
Open Access ArticleElectrochemical operational principles and analytical performance of Pd-based amperometric nanobiosensors†
Y. E.
Silina‡
 *a,
N.
Apushkinskaya
*a,
N.
Apushkinskaya
 a,
N. V.
Talagaeva
b,
M. G.
Levchenko
b and
E. V.
Zolotukhina‡
a,
N. V.
Talagaeva
b,
M. G.
Levchenko
b and
E. V.
Zolotukhina‡
 *bc
*bc
aInstitute for Biochemistry, Zentrum für Human und Molekularbiologie (ZHMB), Campus B 2.2, University of Saarland, 66123, Saarbrücken, Germany. E-mail: yuliya.silina@uni-saarland.de; yuliya.silina@gmx.de
bInstitute of Problems of Chemical Physics RAS, 1 Ac. Semenov avenue, Chernogolovka, 142432, Moscow region, Russia. E-mail: zolek@icp.ac.ru
cMoscow Institute of Physics and Technology, 9 Institutskiy per., Dolgoprudny, 141700, Moscow region, Russia
First published on 7th July 2021
Abstract
Palladium nanoparticles (Pd-NPs) have been approved as an effective catalyst for hydrogen peroxide decomposition which is released during specific enzymatic reactions. However, the general operational principles and electrochemical performance of Pd-NPs-based nanobiosensors have been poorly exploited. Here, the electrochemical behavior of oxidase-associated peroxide oxidation co-catalysis of the modelled microanalytical system based on screen-printed electrodes modified by electroplated Pd-NPs as an electrocatalyst, glucose oxidase (GOx) or alcohol oxidase (AOx) as a bioreceptor and the ionomer Nafion as a polymeric binding agent was studied in detail. The impact of palladium surface oxides and adsorbed oxygen on the activity and product selectivity in an oxidase type of nanobiosensor was ascertained. To avoid PdO and oxygen electroreduction affecting the entire amperometric response of Pd-NPs-based nanobiosensors, a special two-step polarization procedure was proposed. Under the established electrochemical conditions, Pd-NPs-based nanobiosensors with encapsulated oxidases showed a wide dynamic range towards selective bioanalyte detection, excellent basic line stability, accuracy and resistance to the presence of interfering electrochemical species. This work can serve as a guideline for the search and validation of operational principles of novel biosensors based on nanoparticles.
1. Introduction
In the last decade, significant progress has been achieved in the development, application and validation strategies of enzymatic biosensors. Although, the general functioning mechanism of biosensors relies on biorecognition reaction between the enzyme and targeted analyte, the efficiency, completeness and reproducibility of the entire process will depend on the type and stability of the used electromediator (or inorganic electrocatalyst).Oxidase-based amperometric glucose or alcohol sensitive biosensors comprising the enzyme glucose oxidase (GOx) or alcohol oxidase (AOx) as a biorecognition element and recording the enzymatically produced product (H2O2) can be mentioned as an example. The amount of produced hydrogen peroxide will be proportional to the analyte concentration in a sample. Typically, redox mediators (e.g. Prussian Blue and Meldola Blue) can be employed to facilitate electron-transfer between the bioreceptor and transducer, and to decompose the formed hydrogen peroxide via electrocatalytic redox-reaction.1–4 However, some redox mediators (electrocatalysts) used in amperometric glucose or alcohol sensitive biosensors are prone to mechanical instability that leads to their leakage from the electrode surface and degradation of the signal, insufficient sensor sensitivity and selectivity, and low repeatability and accuracy.5
As an alternative solution, nanoparticles (NPs) of noble metals have been explored in amperometric biosensor development.6–8 They are biocompatible, mechanically and chemically stable, they exhibit advanced catalytic properties, and their synthesis is environmentally friendly and often can be conducted in a one-step/one-pot manner.9–11 More importantly, NPs of noble metals in electrochemical biosensors act directly as electrocatalysts in the oxidation/reduction reaction of the co-products (e.g. hydrogen peroxide) formed during biochemical reactions.12,13
A wide spectrum of electrochemical nanobiosensors with NPs of noble metals as electrocatalysts was introduced.14–16 However, the working electrochemical conditions were optimized and validated exclusively for Pt-NPs, Au-NPs and Ag-NPs-based amperometric biosensors.17–21 To the best of our knowledge, almost no attempts were made towards the establishment, validation and systematization of the electrochemical system parameters of Pd-NPs-based nanobiosensors.
In our previous studies, we introduced a novel fabrication approach to produce one-step designed tailored enzymatic nanobiosensors based on Pd-NPs followed by their thorough standardization.5,21 According to this approach, the end-structure of one-step designed nanobiosensors represents a structural model mainly composed of an enzyme incorporated in metal–polymer scaffolds.22 This nanocapsular structure provides on demand controlled release of enzymes (in the presence of an analyte). However, the electrochemical operational principles and guidelines of these novel one-step designed Pd-based amperometric nanobiosensors have been poorly exploited. Meanwhile, specifically, the used working conditions (applied potential, duration of polarization, read-out detection mode, etc.) will affect the performance of the one-step designed Pd-NPs-based nanobiosensors. Thus, in some studies Pd-NPs-based systems were not recommended to be operated in a low potential range (from 0 to −0.4 V) due to oxygen electrodesorption.23 In contrast, electrochemical tuning of the enzymatic activity of Pd-NPs-based biosensors towards alcohol detection in fermentation media under very low applied potentials (from −0.02 to −0.32 V) has recently been reported.24
Hence, to avoid any misinterpretation of the obtained electrochemical results supplied by the one-step produced Pd-NPs-based nanobiosensors, a detailed study on the processes affecting their responses as well as the development of general working principles and operational guidelines are highly necessary.
Here, we conducted a systematic study on the operation of one-step designed Pd-NPs-based nanobiosensors under the complex conditions of a few parallel electrocatalytic reactions. The obtained results were confirmed by a series of advanced analytical techniques, i.e. scanning electron microscopy, laser desorption ionization mass spectrometry, RAMAN spectroscopy, cyclic voltammetry, multi-step amperometry and oxygen minisensor studies. More specifically, the novelty of this study relies on the established mechanism underlying the electrochemical response of one-step designed Pd-NPs-based amperometric nanobiosensors (i); the developed multi-step operational read-out platform (ii); and a novel design of Pd-NPs-based nanobiosensors being operated at anodic potentials which allowed extending the linear response towards glucose detection (iii).
The practical merit of this work is in the development of a time-, cost-effective and simple methodology that can significantly improve the read-out of Pd-NPs-based nanobiosensors and permit their rational design.
2. Experimental part
2.1. Materials and methods
30% H2O2, EtOH, BuOH, glucose, AgCl, K4[Fe(CN)6], K3Fe(CN)6, FeCl3, H2PdCl4, 25% NH4OH, Pd(NH3)4Cl2, K4[Ag2(CN)6], (NH4)2HPO4, and Na2HPO4·12H2O (99% purity) were obtained from Sigma Aldrich. Nafion®117 solution in a mixture of aliphatic alcohols and water, glucose oxidase, GOx, from Aspergillus Niger (EC 1.1.3.4, type VII, 403 U g−1 solid), and alcohol oxidase, AOx, from Pichia pastoris (EC 1.1.3.13, 32 U mg−1), were received from Merck (Darmstadt, Germany). Pd-wire, Ag-wire and Pt-wire (99%, Sigma Aldrich) were used as the anodes for the preparation of palladium nanoparticles (Pd-NPs), silver nanoparticles (Ag-NPs) and Prussian Blue (PB).DRP-110DGPHOX screen printed electrodes (SPEs) were obtained from DropSens (Llanera, Spain). Each sensor consisted of a carbon working electrode (WE) modified with a layer of graphene oxide (referred to as SPE/GO), a carbon counter electrode, and a silver reference electrode. Organic-free, deionized water (DI) was generated with an Elga PureLab (Celle, Germany) water purification system.
2.2. Electrochemical synthesis of Pd-NPs, Ag-NPs and PB
To compare the efficiency of Pd-NPs towards peroxide sensing with alternative catalytic systems, electroplated silver, Ag-NPs, and PB nanoparticles were synthesized on the surface of SPE/GO electrodes. The synthesis of Ag-NPs was performed at −3 mA for 60 s from an AgCl and K4[Fe(CN)6] containing electrolyte according to the following sequence:25| 2AgCl + K4[Fe(CN)6] = K4[Ag2(CN)6] + FeCl2 | (1) |
| FeCl2 + H2O + Na2CO3 = Fe(OH)2 + CO2 + 2NaCl | (2) |
| 2 Fe(OH)2 + 0.5O2 + H2O = 2Fe(OH)3 | (3) |
The formed Fe(OH)3 was removed by filtration. The electroplating of pure Pd-NPs was performed according to an earlier reported protocol.25
For electroplating of PB from an electrolyte containing 0.25 mM K3Fe(CN)6 and 0.25 mM FeCl3, a recently reported procedure was applied.26
2.3. One-step synthesis of enzymatic nanobiosensors
One-step fabrication of nanobiosensors with encapsulated GOx or AOx (depending on the nature of the targeted analyte) and Nafion®117 (further referred to as Nafion or Naf) was carried out directly over the surface of SPE/GO (0.12 cm2).5 Briefly, a multiple electrolyte solution containing a Pd-electrolyte, GOx or AOx dissolved in phosphate buffer and Nafion mixed in the ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v/v was prepared and dropped on SPE/GO. Furthermore, a current of −2.5 mA was passed through the droplet for 30 s in galvanostatic mode.
1 v/v/v was prepared and dropped on SPE/GO. Furthermore, a current of −2.5 mA was passed through the droplet for 30 s in galvanostatic mode.
The electroplating of the Ag-NP-based nanobiosensors was carried out from the multiple electrolyte solution containing a Ag-electrolyte, GOx or AOx dissolved in phosphate buffer and Nafion mixed in the volume ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v/v. The deposition was conducted at the current of −3 mA for 60 s. Remarkably, the electrodeposition performed from the multiple electrolyte solution resulted in the formation of polymer-doped enzymatic Pd-NPs, Ag-NPs or PB hybrids on SPE/GO. The experiments were carried out on a one-channel biologic Potentiostat PalmSens4 (PalmSens, Utrecht, The Netherlands).
1 v/v/v. The deposition was conducted at the current of −3 mA for 60 s. Remarkably, the electrodeposition performed from the multiple electrolyte solution resulted in the formation of polymer-doped enzymatic Pd-NPs, Ag-NPs or PB hybrids on SPE/GO. The experiments were carried out on a one-channel biologic Potentiostat PalmSens4 (PalmSens, Utrecht, The Netherlands).
2.4. Oxygen minisensor studies
To test the electrochemical behavior of pure electroplated Pd-NPs, Ag-NPs, PB-NPs and their polymer-doped enzymatic hybrids towards peroxide, different concentrations of hydrogen peroxide were dropped on the surface of modified electrodes. The obtained responses were recorded in cyclic voltammogram (CV) mode from −0.4 V to 0.4 V (unless stated otherwise). The release of oxygen (μmol L−1) as a product of peroxide decomposition was monitored with an OXR430 retractable needle-type fiber-optic oxygen minisensor (Pyro Science GmbH, Aachen, Germany). The reaction can be summarized as follows (on the electrode):27| H2O2 → O2↑ + 2H+ + 2e− | (4) |
Notably, the detailed mechanism of this reaction is difficult to determine and depends on the nature of the used NPs. Nevertheless, this sequence clearly demonstrates the reason for oxygen release measured with the oxygen minisensor in the designed systems.27
Furthermore, to normalize the rates of oxygen generation to the amount of catalysts, we used a quartz crystal microbalance (QCM-sense) system operated in droplet mode.5 The mass of electroplated co-deposits (mass in ng) was calculated using the Sauerbrey equation.5
2.5. Laser desorption ionization mass spectrometry (LDI-MS)
To ensure enzyme encapsulation together with nanoparticles, LDI-MS screening of the modified SPE/GO surface was used.21 LDI-MS investigations were carried out on a Bruker Esquire 3000 + ESI-ion trap MS (Bruker Daltonics, Bremen, Germany) operated using Bruker Esquire control 5.3 software equipped with an atmospheric pressure (AP-MALDI) ion source and Nd:YAG solid-state laser (355 nm).2.6. Electrochemical characterization of enzymatic Pd-NPs-based nanobiosensors
Hydrogen peroxide (H2O2) is a product of glucose/alcohol oxidation in the presence of oxidases (GOx/AOx) and molecular oxygen.28 For example, for glucose oxidation the reaction can be written as follows:| Glucose + GOx(ox) → GOx(red) + gluconic acid | (5) |
| GOx(red) + O2 → GOx(ox) + H2O2 | (6) |
 | (7) |
H2O2 is an electroactive species which can be reduced or oxidized on the electrode surface depending on the used polarization mode.29 The electrochemical response of the one-step nanobiosensors was recorded in CV mode on the one-channel biologic Potentiostat PalmSens4 (PalmSens, Utrecht, The Netherlands). The CV curves were obtained by placing a 150 μL droplet of pure phosphate buffer or targeted bioanalyte over all three electrodes.
To validate the features of H2O2 decomposition in diluted solutions, we used a glassy carbon electrode, GCE, in disk form (0.07 cm2) as a working electrode. A set of experiments was performed with a conventional three-electrode electrochemical cell (15 mL) equipped with saturated silver chloride and a platinum wire counter electrode. The inert (argon) or air atmosphere was maintained with Schlenk's line. The layer of Pd-NPs was deposited on the GCE surface from a 1 mM solution of Pd(NH3)4Cl2 + 0.1 M KNO3 under the same conditions as those used for the preparation of Pd-NPs modified SPE/GO. All measurements involving the GCE were carried out on the potentiostat Autolab PGSTAT 101 (Metrohm AG, Germany).
2.7. pH measurements
pH measurements were performed in droplet mode with a HORIBA LAQUATWIN PH-22 pH sensor (MMM tech support GmbH&Co KG, Berlin, Germany).2.8. Scanning electron microscopy (SEM) study
The morphology of the synthesized nanobiosensors was investigated by scanning electron microscopy on a 400 FEG SEM (Hillsboro, OR, USA). The images were taken in high-resolution mode.2.9. RAMAN spectroscopy
To verify the formation of Pd-oxides on the surface of the nanobiosensors, RAMAN studies were conducted. The experiments were performed under ambient conditions on a HORIBA Jobin Yvon (Longmujeau, France) Raman microscope equipped with an Nd:YAG laser (Melles Griot, IDEX Optics and Photonics, Albuquerque, NM, USA, wavelength of 532 nm).3. Results and discussion
3.1. Superior electrochemical behavior of electroplated Pd-NPs towards H2O2 sensing
To choose the most sensitive electrocatalyst in the design of oxidase-associated peroxide oxidation co-catalysis, the analytical merit of Pd-, Ag- and PB-based nanoparticles (NPs), viz. the efficiency towards peroxide sensing, was evaluated with the oxygen minisensor. Comparison of various compositions (Fig. S1†) showed a superior behavior of active layers containing Pd-NPs towards H2O2 in a range of 10–100 mM. The overperformance of the Pd-NPs-based electrocatalysts can be explained in terms of kinetics that is expected to be slower on Ag-NPs than on palladium. Prussian Blue nanoparticles (PB-NPs) are usually used as the electrocatalyst for hydrogen peroxide reduction. Depending on the applied potential, hydrogen peroxide can be reduced on the surface of PB-NPs to hydroxide ions that will not affect the dissolved oxygen level in solution. Therefore, the release of oxygen recorded at a high concentration of applied H2O2 on the sensors containing PB-NPs (Fig. S1†) can be explained by the decomposition of hydrogen peroxide on the surface of the SPE/GO electrode.30Notably, the sensing of H2O2 on Pd-NPs did not show any irreversible changes in the sensor response that is an important criterion for the development of long-term nanobiosensors. Thus, no changes in the electrochemical behavior of Pd-NPs were detected during the transition from low to high H2O2 concentration levels and back (see ESI, Fig. S2†).
Hence, the usage of electroplated Pd-NPs as an electrocatalyst in nanobiosensor design can support the efficient sensing of hydrogen peroxide released during specific enzymatic reaction in the presence of a bioanalyte (see section 3.2.2).
3.2. Electrochemical tuning of Pd-NPs-based nanobiosensors at the cathodic potential
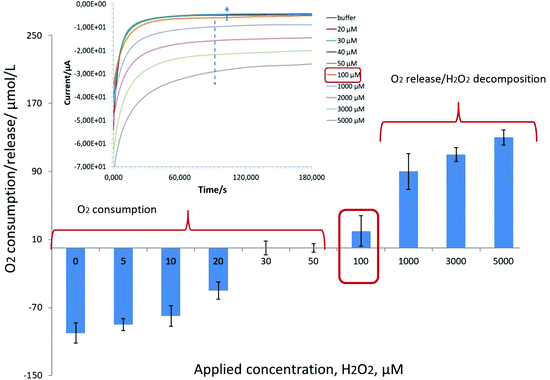
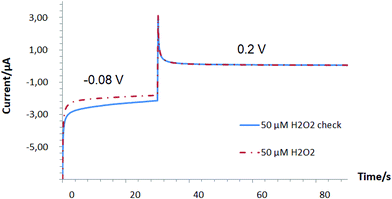
Our results indicate a strong competition in the range of 1 μM–5 mM between the oxygen reduction reaction (ORR) and hydrogen peroxide detection on Pd-NPs, Fig. 1.
Briefly, oxygen consumption by Pd-NPs was recorded in the presence of H2O2 up to a level of 100 μM. Knowing that in hybrid nanobiosensors the amount of Pd-NPs is at least three times less (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v/v mixing, see the Experimental part) vs. pure Pd-NPs (ESI, Fig. S1†) and their efficiency to decompose H2O2 is also several orders of magnitude worse, it is expected that the concentration range of oxygen consumption in the presence of H2O2 can be readily extended. For example, during the detection of alcohols with Pd-based enzymatic biosensors it was shown that at an applied potential of −0.08 V instead of a negative current increase (Fig. 1, see the inset starting from 100 μM) with an increase of loaded analyte concentration (caused by an increase of released hydrogen peroxide as a product of enzymatic reaction), a continuous decrease of the negative current from the hybrid AOx/Nafion/Pd-NPs amperometric biosensors was recorded at least up to the level of 10 mM.24
1 v/v/v mixing, see the Experimental part) vs. pure Pd-NPs (ESI, Fig. S1†) and their efficiency to decompose H2O2 is also several orders of magnitude worse, it is expected that the concentration range of oxygen consumption in the presence of H2O2 can be readily extended. For example, during the detection of alcohols with Pd-based enzymatic biosensors it was shown that at an applied potential of −0.08 V instead of a negative current increase (Fig. 1, see the inset starting from 100 μM) with an increase of loaded analyte concentration (caused by an increase of released hydrogen peroxide as a product of enzymatic reaction), a continuous decrease of the negative current from the hybrid AOx/Nafion/Pd-NPs amperometric biosensors was recorded at least up to the level of 10 mM.24
To sum it up, the observed strong competition between the ORR and peroxide sensing on Pd-NPs will dramatically affect the electrochemical response and analytical merit of the Pd-NPs-based nanobiosensors with encapsulated oxidases (see section 3.2.2).
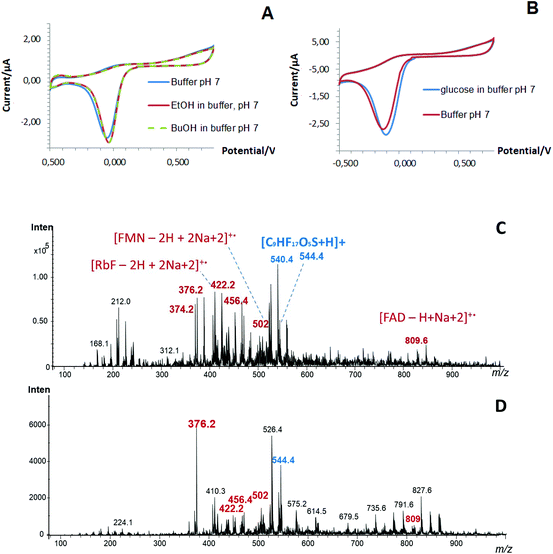
At the same time, the presence of enzymes (evaluated as a co-factor, i.e. flavin adenine dinucleotide, FAD) in the structure of the one-step designed Pd-NPs-based nanobiosensors and the efficiency of the performed co-deposition procedure was confirmed by LDI-MS, Fig. 2C and D. Thus, in addition to Nafion (m/z at 544), FAD (m/z at 809) as a small molecular weight organic compound, its related fragments (FMN and RbF) and their adducts were clearly seen in mass spectra obtained from the one-step nanobiosensors with encapsulated oxidases.22
In other words, despite the obvious presence of enzymes in the end-structure of the Pd-NPs-based nanobiosensors, their operation at the cathodic range of potentials does not seem to be a trivial task. This is in particular true if the hydrogen peroxide concentration is comparable with the concentration of dissolved oxygen (see Fig. 1).
It appears that multiple reactions simultaneously occurring on the surface of Pd-NPs (see section 3.2.1) severely affect the responses supplied by the one-step designed nanobiosensors with encapsulated oxidases. This hypothesis was approved through the standard response studies performed in AM mode at the constant applied potential of −0.05 V with a new droplet of buffer spotted on the surface of the one-step Pd-NPs-based nanobiosensor with encapsulated AOx and Nafion (ESI, Fig. S4†). Briefly, a continuous decrease of the cathodic current in a cycle experiment from droplet to droplet was observed indicating a high instability of the sensor's basic line. This effect can be explained by Pd-oxide formation/reduction (ESI, Fig. S3†) on the one side and by the ORR on the other side (see section 3.3). This type of electrochemical behavior is an illustration of the general problem of oxidase-based biosensors: the dependence on oxygen concentration, which might be quite uncertain in a complex system, viz. hybrid Pd-NPs-based enzymatic nanobiosensors.
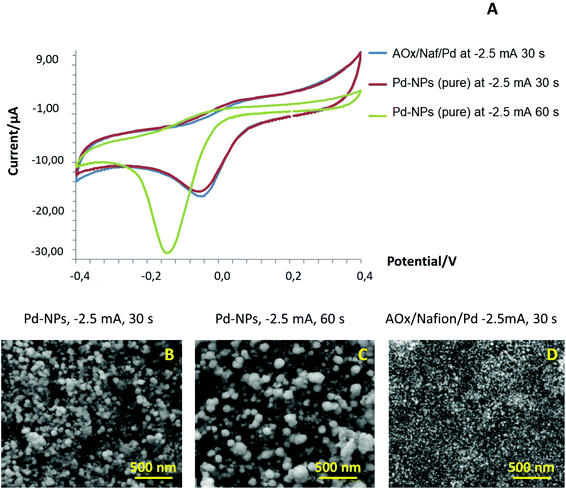
Furthermore, we conducted a blank experiment with pure Pd-NPs deposited on the surface of SPE/GO under different electroplating conditions (deposition time), tested their response in a conventional CV mode, and compared their performance with that of an AOx/Nafion/Pd system. As can be seen from the CV curves (Fig. 3A), even in the absence of an enzyme, pure Pd-NPs produced at −2.5 mA for 30 s showed the same type of electrochemical behavior at the cathodic branch as was observed for one-step designed AOx/Nafion/Pd synthesized under the same conditions. However, the peak potential in the cathodic range of CV for larger particles (−150 mQ, −2.5 mA for 60 s) shifted towards a more negative range vs. smaller Pd-NPs (−75 mQ, −2.5 mA for 30 s). We assume that this effect is caused by the changes in the surface density and size of the Pd-NPs (Fig. 3B and C) that can impact the position of the cathodic peak and amount of the formed PdO. The presence of the polymer and enzyme in the one-step AOx/Naf/Pd nanobiosensor leads to diminution of the end-particle size as compared to pure Pd-NPs synthesized under the same conditions; see Fig. 3D.
In addition, a strong dependency of the cathodic peak position on temperature and solution acidity/basicity was revealed (see ESI, Fig. S5†). This means that the environmental and storage conditions will severely affect the surface chemistry of the Pd-NPs-based nanobiosensors and the amount of the formed palladium oxides. Hence, the one-step designed nanobiosensors with encapsulated enzymes cannot be reliably operated at the cathodic range of potentials. Therefore, the development of a read-out platform and operational guidelines of one-step designed Pd-NPs-based amperometric nanobiosensors is highly desirable (see the next section).
3.3. Development of operational principles of Pd-NPs-based nanobiosensors and their validation with a conventional three-electrode electrochemical cell
To verify the impact of palladium oxides29 on the electrochemical read-out supplied by Pd-NPs, CV studies were conducted for the Pd-NPs-modified GCE under an Ar-atmosphere in the absence of O2 and in the presence of oxygen at different anodic limits, Fig. S6.†The obtained data indicate the increase of both anodic and cathodic currents with the increase of the anodic polarization limit. This means that the presence of molecular oxygen and extension of the anodic polarization limit facilitate palladium oxide formation resulting in the increase of oxygen electroreduction during the backward scan.29 It is important to mention that the cathodic peak is increased in the presence of dissolved oxygen due to the ORR.
Furthermore, to establish the polarization mode where hydrogen peroxide can be selectively and reliable detected by intact Pd-NPs or their enzyme-containing hybrids in the presence of dissolved oxygen, several preliminary tests were performed in a classic three-electrode electrochemical cell with a GCE electrode (see section 2.6).
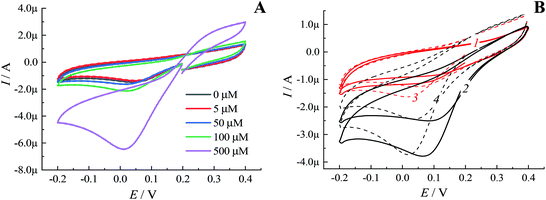
Notably, in solutions with a low concentration of hydrogen peroxide even under an inert atmosphere almost no sensitivity of the cathodic peak current to the presence of H2O2 was observed (Fig. 4A). These results were in line with the dependency revealed by oxygen minisensor studies performed for SPE/GO modified by Pd-NPs (see Fig. 1). Due to the ORR, the presence of molecular oxygen in solution leads to an increase in the cathodic peak current. In addition, the cathodic peak potential is shifted depending on pH (Fig. 4B). Since the concentration of dissolved oxygen strongly depends on the air content and temperature, the cathodic peak current in oxygenated solutions cannot be used to detect hydrogen peroxide with Pd-NPs-based sensors regardless of the used electrode, i.e. SPE/GO or GCE.
In contrast, the current of the anodic branch at a potential of 0.2–0.4 V grows depending on the peroxide concentration due to hydrogen peroxide oxidative decomposition occurring on the Pd surface. Thus, peroxide decomposition facilitates the formation of palladium oxides resulting in the increase of the anodic current in the above-mentioned range of potentials.
Thus, it appears possible to quantitatively detect peroxide in oxygenated aqueous solutions using Pd-NPs-modified electrodes operated at the anodic potentials. However, to refresh the electrode surface, it is highly necessary to reduce Pd-oxides formed during anodic polarization. For this goal, we optimized the following double step procedure: the first step is a polarization at the cathodic peak potential to reduce PdO, and the second step is a polarization at the anodic potential in a range of 0.2–0.4 V to detect/record the current related to hydrogen peroxide decomposition.
Importantly, the potential and duration of the first polarization step depend on the pH of a solution (buffer or fermentation media), the presence of molecular oxygen, intact Pd-NPs size and the nature of organic/bioreceptor compounds in the structure of the functional sensing layer. In this regard, the polarization step performed at the cathodic peak potential should be preliminarily estimated by CV studies for various experimental conditions.
For example, for phosphate buffer with pH 7.5 and pH 5.5 the multi-step amperometric procedure on the Pd-NPs-modified GCE was carried out as follows: the polarization duration at the cathodic peak potential was set to 120 s, and the polarization duration at an anode potential of 0.4 V was 30 s. As is seen from Fig. S7 (ESI),† the current of the second polarization step is strongly proportional to the concentration of the dissolved hydrogen peroxide in solution. At the same time, the change in pH does not significantly affect the current responses at the anodic polarization potentials. More importantly, the presence of molecular oxygen in solutions with low H2O2 concentration has a little impact on the current response; see ESI, Fig. S7.†
Following a similar multi-step polarization procedure as described above, the response of Pd-NPs-modified SPE/GO towards hydrogen peroxide detection at the anodic potential range was validated; see ESI, Fig. S8.† Notably, the electrochemical read-out mode will significantly impact the analytical performance of the Pd-NPs-based amperometric nanobiosensors (see the next section).
3.4. Analytical performance of Pd-NPs-based amperometric nanobiosensors at the anodic potentials
Next, the response of the Pd-NPs synthesized at different electrodeposition parameters (shown for 30 s and 60 s) to hydrogen peroxide detection was evaluated in the optimized multi-step polarization mode. Significantly, the electrocatalytic ability of Pd-NPs-modified SPE/GO changes in the anodic range with Pd-NPs density and size; see ESI, Fig. S9.† Thus, the increase of deposition time from 30 s to 60 s was accompanied by an increase of NPs size from 20 nm to 70–75 nm (see also Fig. 3B and C) followed by the subsequent decrease in H2O2 sensitivity.
Furthermore, the tuning of the analytical performance of Pd-NPs towards H2O2 detection and comparison of their analytical merits at different electrochemical operational modes were performed for nanoparticles produced at −2.5 mA for 30 s. The results of this tuning are summarized in Table 1. Obviously, advanced analytical merits of this sensing system can be achieved by the optimization of the read-out platform delivered by Pd-NPs.
| Applied potential, V | Calibration formula | R 2 | Sensitivity in low range, 0–1000 μM, μA·mM−1 | Linear dynamic range, LDR | LOQ μM | Accuracy, %a | RSDb |
|---|---|---|---|---|---|---|---|
| a Evaluated in a range below 1000 μM. b Reproducibility estimated in a range of 10–1000 μM. | |||||||
| −0.08 | Y = −0.0043x − 4.600 | 0.943 | 4.3 | 100 μM–100 mM | 250 | 30.7 ± 10.2 | Low |
| −0.05 | Y = −0.0051x − 4.330 | 0.994 | 5.1 | 100 μM–100 mM | 150 | 37.7 ± 3.8 | Low |
| 0 | Y = −0.0037x − 2.265 | 0.998 | 3.7 | 100 μM–100 mM | 180 | 45.2 ± 5.1 | Low |
| 0.02 | Y = −0.0011x − 0.167 | 0.999 | 1.1 | 100 μM–100 mM | 180 | 48.3 ± 0.8 | Low |
| 0.08 | Y = −0.0007x − 0.041 | 0.999 | 0.7 | 100 μM–100 mM | 100 | 50.7 ± 6.4 | Low |
| −0.08 V (step 1) | Y = 0.0052x + 0.177 | 0.998 | 5.2 | 5 μM–100 mM | 7 | 101.3 ± 6.1 | High |
| 0.2 V (step 2) | |||||||
| −0.08 V (step 1) | Y = 0.0069x + 0.054 | 0.999 | 6.9 | 1 μM–100 mM | 2 | 101 ± 1.0 | High |
| 0.4 V (step 2) | |||||||
Despite the higher sensitivity of Pd-NPs (−2.5 mA, 30 s) to H2O2 detection revealed at 0.4 V vs. 0.2 V, polarization at 0.2 V should be considered in further experiments to minimize the interferences increasing at 0.4 V from other electroactive species (acetophenone, acetic acid, ascorbic acid, etc.) possibly present in the real samples.
Furthermore, during the validation of the analytical performance of Pd-NPs for H2O2 detection, the accuracy test at a cathodic polarization potential of −0.08 V held for 30 s followed by signal read-out at 0.2 V was performed in multi-step amperometric mode (MAM), Fig. 5. In addition to the excellent accuracy detected at 0.2 V, an advanced basic line stability and tolerance to the presence of interfering electroactive species were observed for Pd-NPs operated at the optimized read-out MAM mode; see ESI, Fig. S10–S12.†
Thus, by gaining closer insights into the designed electrochemical system and by a detailed study of the fundamental processes affecting the generated signal, it was possible to establish and validate the operational conditions of the hybrid Pd-NPs-based nanobiosensors.
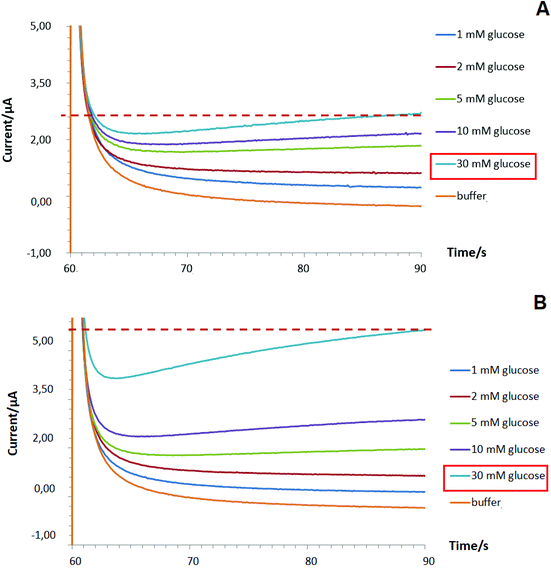
To improve the linear dynamic range of targeted bioanalyte detection (shown for glucose as a case study) by Pd-NPs-based enzymatic nanobiosensors operated at MAM mode, the following modification step to the intact sensor design was implemented. First, a layer of pure Pd-NPs was deposited on the surface of the SPE/GO electrode at −2.5 mA for 30 s in the manner shown in section 3.4.1 (step 1). Next, the encapsulation of GOx with Nafion from the multiple Pd-electrolyte solution (step 2) via one-step deposition with the same electroplating parameters was conducted (will be referred to as two-step designed Pd-NPs-based nanobiosensors). Interestingly, due to the pre-modification step 1 being performed we managed to enhance the linear dynamic range of the proposed glucose biosensing system from 1–10 mM to 1–30 mM, Fig. 6B.
We hypothesize that the first Pd-NPs layer (step 1) allows the increase of the entire electroactive surface area to detect a high concentration of hydrogen peroxide released during enzymatic reaction and, probably, facilitates the electric connection with the Pd-NPs electrocatalyst deposited in the hybrid enzyme contained layer (step 2). Hence, this altogether will help improve the enzymatic activity of the nanobiosensor and significantly improve its overall analytical performance.
Comparison of the analytical merits between the obtained results and data discussed in the literature31–34 on peroxide-based glucose biosensors indicates that the proposed MAM procedure for H2O2 detection permits:
– reduction of the anodic polarization potential value;
– exclusion of the impact of the interfering component (oxygen) on the analytical response of the system;
– enhancement of the linear dynamic range and LOQ;
– improvement of the accuracy and reproducibility of the analysis.
4. Conclusions
Here, a fundamental study on the search and validation of the optimal operating electrochemical parameters of Pd-NPs-based amperometric nanobiosensors was carried out. The role of palladium surface oxides and adsorbed oxygen on the sensitivity and product selectivity in Pd-NPs-based nanobiosensors was confirmed.As a solution, a novel multi-step amperometric read-out mode (the first step is a polarization at the cathodic peak potential to reduce PdO, and the second step is a polarization at the anodic potential to detect the H2O2 oxidation current) that results in a faster reduction of PdO, and reliable, sensitive and selective peroxide sensing was developed. The proposed procedure was fully validated in a conventional three-electrode electrochemical cell under both air and inert atmospheres. Finally, re-design of Pd-NPs allowed us to improve the analytical performance of the Pd-NPs-based nanobiosensors that can be readily considered as a possible alternative to sophisticated, expensive, time- and source-consuming enzyme-related approaches (i.e. amount of enzyme, variation of pH and enzyme strains, immobilization strategies, etc.).
We believe that our results will thus contribute to the future development of noble metal NPs-based nanobiosensors, and also will provide an opportunity to tune their performance based on the requirements of the analytical task.
Author contributions
Y. E. Silina – conceptualized the concept of the research, developed the one-step nanobiosensor approach, was responsible for the design of the manuscript and data analysis, wrote, reviewed, and edited the manuscript, and handled funding acquisition and project administration. N. Apushkinskaya – conducted sensor-related experiments (SPE/GO system), data visualization, validation and formal analysis. N. V. Talagaeva – was responsible for electrochemical experiments utilizing the three-electrode electrochemical cell. M. G. Levchenko – performed sample preparation and took part in electrochemical experiments utilizing the three-electrode electrochemical cell. E. V. Zolotukhina – was responsible for the conceptualization and design of electrochemical experiments, validation strategies, data analysis, writing, reviewing, and editing of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was a part of the research program of Y. E. S. funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation, project 427949628). The authors would like to thank the German-Russian Interdisciplinary Science Center (G-RISC) for the support of collaboration between the University of Saarland (Germany) and Institute of Problems of Chemical Physics RAS (Moscow region, Russia). The authors N. V. Talagaeva, M. G. Levchenko and E. V. Zolotukhina performed their work within the framework of the State assignment (number AAAA-A19-119061890019-5). Y. E. S. also thanks Dr Marcus Koch (INM – Leibniz Institute of New Materials, Saarbrücken, Germany) for the performed SEM studies of Pd-NPs-based nanostructures and Prof. Dr Bruce Morgan (University of Saarland) for continuing support of this research at the department.References
- A. Vasilescu, S. Andreescu, C. Bala, S. C. Litescu, T. Noguer and J. L. Marty, Biosens. Bioelectron., 2003, 18, 781–790 CrossRef CAS PubMed.
- A. M. Titoiu, G. Necula-Petrareanu, D. Visinescu, V. Dinca, A. Bonciu, C. N. Mihailescu, C. Purcarea, R. Boukherroub, S. Szunerits and A. Vasilescu, Microchim. Acta, 2020, 187(10), 550 CrossRef CAS PubMed.
- A. A. Karyakin, Electroanalysis, 2001, 13, 1–7 CrossRef.
- A. A. Karyakin and E. E. Karyakina, Sens. Actuators, B, 1999, 57, 268–273 CrossRef CAS.
- D. Semenova, K. V. Gernaey, B. Morgan and Y. E. Silina, Analyst, 2020, 145, 1014–1024 RSC.
- J. Wang, Microchim. Acta, 2012, 177, 245–270 CrossRef CAS.
- C. Shan, H. Yang, D. Han, Q. Zhang, A. Ivaska and L. Niu, Biosens. Bioelectron., 2010, 25, 1070–1074 CrossRef CAS PubMed.
- S. Azzouzi, L. Rotariu, A. M. Benito, W. K. Maser, M. Ben Ali and C. Bala, Biosens. Bioelectron., 2015, 15(69), 280–286 CrossRef PubMed.
- J. Bai, Y. Li, J. Du, S. Wang, J. Zheng, Q. Yang and X. Chen, Mater. Chem. Phys., 2007, 106, 412–415 CrossRef CAS.
- S. H. Kim, G. H. Jeong, D. Choi, S. Yoon, H. B. Jeon, S. M. Lee and S. W. Kim, J. Colloid Interface Sci., 2013, 389(1), 85–90 CrossRef CAS PubMed.
- H. S. Al-Ghamdi and W. E. Mahmoud, Mater. Lett., 2013, 105, 62–64 CrossRef CAS.
- R. Batool, A. Rhouati, M. H. Nawaz, A. Hayat and J. L. Marty, Biosensors, 2019, 9, 46 CrossRef CAS PubMed.
- M. Holzinger, A. Le Goff and S. Cosnier, Front. Chem., 2014, 2, 1–10 CAS.
- N. Stasyuk, G. Gayda, H. Klepach and M. Gonchar, Sens. Lett., 2016, 14, 1169–1177 CrossRef.
- N. Tujunen, E. Kaivosoja, V. Protopopova, J. J. Valle-Delgado, M. Österberg, J. Koskinen and T. Laurila, Mater. Sci. Eng., C, 2015, 55, 70–78 CrossRef CAS PubMed.
- N. Dimcheva, Curr. Opin. Electrochem., 2020, 19, 35–41 CrossRef CAS.
- V. Mazeiko, A. Kausaite-Minkstimiene, A. Ramanaviciene, Z. Balevicius and A. Ramanavicius, Sens. Actuators, B, 2013, 189, 187–193 CrossRef CAS.
- R. D. O'Neill, S. C. Chang, J. P. Lowry and C. J. McNeil, in Biosensors and Bioelectronics, 2004 Search PubMed.
- M. Topçu Sulak, Ö. Gökdoǧan, A. Gülce and H. Gülce, Biosens. Bioelectron., 2006, 15(21), 1719–1726 CrossRef PubMed.
- Z. Wen, S. Ci and J. Li, J. Phys. Chem. C, 2009, 113, 13482–13487 CrossRef CAS.
- Y. E. Silina and B. Morgan, Talanta, 2020, 223, 121688 CrossRef.
- E. V. Butyrskaya, N. Korkmaz, E. V. Zolotukhina, V. Krasiukova and Y. E. Silina, Analyst, 2021, 146, 2172–2185 RSC.
- R. Rahul, R. K. Singh, B. Bera, R. Devivaraprasad and M. Neergat, Phys. Chem. Chem. Phys., 2015, 17, 15146–15155 RSC.
- D. Semenova, T. Pinto, M. Koch, K. V. Gernaey and H. Junicke, Biosens. Bioelectron., 2020, 170, 112702 CrossRef CAS PubMed.
- Y. E. Silina, F. Meier, V. A. Nebolsin, M. Koch and D. A. Volmer, J. Am. Soc. Mass Spectrom., 2014, 25, 841–851 CrossRef CAS PubMed.
- V. B. Isfahani, H. R. Dizaji, N. Memarian and A. Arab, Mater. Res. Express, 2019, 6, 096449 CrossRef CAS.
- S. B. Hall, E. A. Khudaish and A. L. Hart, Electrochim. Acta, 1998, 43, 579–588 CrossRef CAS.
- K. Pontius, D. Semenova, Y. E. Silina, K. V. Gernaey and H. Junicke, Front. Bioeng. Biotechnol., 2020, 8, 1–15 CrossRef PubMed.
- K. V. Gor'kov, N. V. Talagaeva, S. A. Kleinikova, N. N. Dremova, M. A. Vorotyntsev and E. V. Zolotukhina, Electrochim. Acta, 2020, 345, 136164 CrossRef.
- R. S. Ribeiro, A. M. T. Silva, J. L. Figueiredo, J. L. Faria and H. T. Gomes, Carbon, 2013, 62, 97–108 CrossRef CAS.
- X. B. Sun and Z. F. Ma, Advanced Materials Research, Trans Tech Publications Ltd., 2013, vol. 643, pp. 162–165 Search PubMed.
- Md. M. Rahman, A. J. S. Ahammad, J.-H. Jin, S. J. Ahn and J.-J. Lee, Sensors, 2010, 10, 4855–4886 CrossRef CAS PubMed.
- D. Xiang, L. Yin, J. Ma, E. Guo, Q. Li, Z. Li and K. Liu, Analyst, 2015, 140(2), 644–653 RSC.
- Z. Lu, L. Wu, X. Dai, Y. Wang, M. Sun, C. Zhou, H. Du and H. Rao, J. Hazard. Mater., 2021, 402, 123774 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1an00882j |
| ‡ Authors with equal contribution. |
| This journal is © The Royal Society of Chemistry 2021 |