Open Access Article
Open Access ArticleTunable fluorescent probes for detecting aldehydes in living systems†
Rachel
Wills‡
a,
Rajendra
Shirke‡
a,
Hannah
Hrncir
b,
John M.
Talbott
 a,
Kirti
Sad
c,
Jennifer M.
Spangle
c,
Adam D.
Gracz
b and
Monika
Raj
a,
Kirti
Sad
c,
Jennifer M.
Spangle
c,
Adam D.
Gracz
b and
Monika
Raj
 *a
*a
aDepartment of Chemistry, Emory University, Atlanta, GA 30322, USA. E-mail: Monika.Raj@emory.edu
bDepartment of Digestive Diseases, Department of Medicine, Emory University, Atlanta, GA 30322, USA
cDepartment of Radiation Oncology, Winship Cancer Institute of Emory University School of Medicine, Atlanta, GA 30322, USA
First published on 19th March 2024
Abstract
Aldehydes, pervasive in various environments, pose health risks at elevated levels due to their collective toxic effects via shared mechanisms. Monitoring total aldehyde content in living systems is crucial due to their cumulative impact. Current methods for detecting cellular aldehydes are limited to UV and visible ranges, restricting their analysis in living systems. This study introduces an innovative reaction-based trigger that leverages the exceptional selectivity of 2-aminothiophenol for aldehydes, leading to the production of dihydrobenzothiazole and activating a fluorescence response. Using this trigger, we developed a series of fluorescent probes for aldehydes by altering the fluorophore allowing for excitation and emission wavelengths across the visible to near-infrared spectral regions without compromising the reactivity of the bioorthogonal moiety. These probes exhibit remarkable aldehyde chemoselectivity, rapid kinetics, and high quantum yields, enabling the detection of diverse aldehyde types, both exogenous and endogenous, within complex biological contexts. Notably, we employed the most red-shifted near-infrared probe from this series to detect aldehydes in living systems, including biliary organoids and mouse organs. These probes provide valuable tools for exploring the multifaceted roles of aldehydes in biological functions and diseases within living systems, laying the groundwork for further investigations.
Introduction
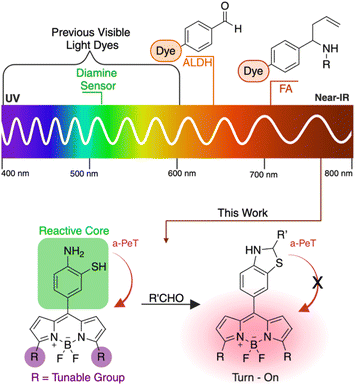
Aldehydes, in normal physiological concentrations, function as antioxidants and signal molecules to maintain system homeostasis.1 However, excessive aldehydes in the cellular environment can create direct crosslinks between biological nucleophiles, including proteins, lipids, and DNA, which reduces DNA repair capabilities and causes chromosomal abnormalities.2 Furthermore, aldehydes work together in a synergistic manner,3 compounding their impact. Accurate monitoring and quantification of total aldehyde content plays a crucial role in understanding their impact on disease states. However, due to the volatile nature4 and synergistic behavior of these compounds, assessing their pathogenesis in living systems with single-cell or blood specimens may not be sufficient. Therefore, it is imperative to develop a precise method for detecting and quantifying total aldehyde content in complex living systems.In recent years, diverse detection methodologies for aldehydes have emerged, encompassing both analytical techniques5,6 and bioorthogonal fluorescence probes.7–12 However, a comprehensive tool specifically designed for in vitro/in vivo detection and monitoring of total aldehyde content within physiological relevant systems like organoids and tissues are lacking. Notably, near-infrared (NIR) probes, spanning from 650 nm to 900 nm,13 have exhibited superior efficacy in living systems as compared to traditional fluorescent dyes.14 Their extended wavelengths minimize harm to living tissues and cells,15 enabling deeper tissue penetration for enhanced imaging while reducing light scattering.16 Furthermore, these NIR dyes' red-shifted properties mitigate autofluorescence occurring in the 200–600 nm range, commonly originating from biological metabolites like hemoglobin.17
Currently, two primary techniques utilizing NIR probes for aldehyde detection exist. The first involves monitoring Aldehyde Dehydrogenase enzymes (ALDH) (Fig. 1).18,19 However, this method lacks a direct correlation between ALDH enzyme activity and aldehyde abundance, leading to inaccuracies in assessing the role of aldehydes in disease conditions. The second method is the direct detection of formaldehyde (FA) in living systems through hydrazine or aza-cope chemistry (Fig. 1).20–23 While effective for singular aldehyde detection, it falls short in quantifying overall aldehyde content, leaving the broader impact of total aldehydes in disease conditions largely unexplored. Herein, we have developed a series of cutting-edge fluorophores featuring a reactive bioorthogonal core, 2-aminothiophenol, recognized for its high selectivity towards aldehydes, facilitating the generation of dihydrobenzothiazole and initiating fluorescence (Fig. 1). The distinct feature of these fluorogenic probes is their tunable emission of fluorescence spanning from the visible to NIR spectrum, while maintaining their reactivity and selectivity towards aldehydes. We have synthesized a series of BODIPY fluorophore probes generating a highly reactive 2-aminothiophenol bioorthogonal moiety (Scheme 1). Furthermore, we have strategically modified the 3,7-dimethyl positions of the BODIPY core to tailor the probes, emitting fluorescence within the visible to near-IR range (Fig. 1). Our study successfully detected both endogenous and exogenous total cellular aldehydes within live cells, organoids, and tissues using these innovative fluorogenic probes.
Results and discussion
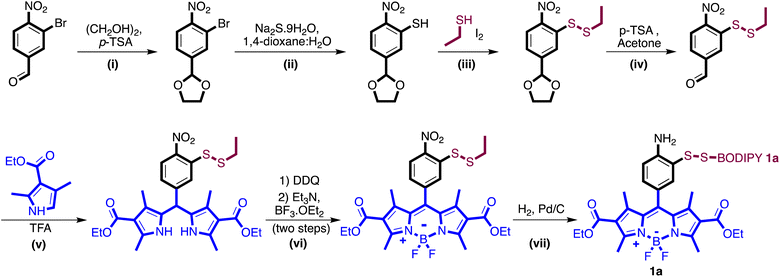
Development of novel dihydrobenzothiazole probe
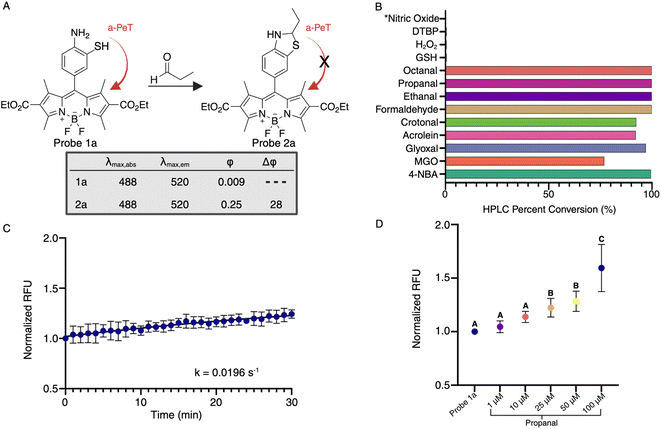
The varied types of aldehydes display a noteworthy collective impact, necessitating the simultaneous tracking of these compounds within live cells to fully understand their biological implications. Unfortunately, the lack of a chemical framework enabling real-time detection and quantification of multiple aldehyde types in live cells hinders this pursuit. Previously, our research group employed a phenyl diamine BODIPY turn-on probe, designed to selectively monitor total aliphatic aldehyde content in live cells.24 However, limitations arose due to its reactivity towards another biological metabolite, nitric oxide, and its selectivity restricted solely to aliphatic aldehydes, without any response towards aromatic aldehydes. In an attempt to develop a more selective moiety for aldehyde detection, we capitalized on the enhanced nucleophilicity provided by sulfur in combination with an amine, employing 2-aminothiophenol to exclusively capture aldehydes by forming dihydrobenzothiazole. To our satisfaction, the inclusion of the thiol group not only expanded the range of reactivity to various aldehyde types, including aromatic and α,β-unsaturated aldehydes, but also significantly accelerated the reaction kinetics.Our primary challenge revolved around successfully integrating thiol and amine moieties into the BODIPY structure. We initiated synthesis with 3-bromo-4-nitrobenzaldehyde or 4-bromo-3-nitrobenzaldehyde and carried out reactions with ethyl 2-methyl-1H-pyrrole-3-carboxylate producing two fluorophores, 4-amino-3-thiophenol-BODIPY 1a and 3-amino-4-thiophenol-BODIPY 1b (Scheme 1, Fig. S1†). Following our synthesis, we evaluated the photophysical characteristics of probes, 4-amino-3-thiophenol-BODIPY 1a and 3-amino-4-thiophenol-BODIPY 1b. Surprisingly, in DMSO only 4-amino-3-thiophenol-BODIPY 1a demonstrated attenuated fluorescence, exhibiting a quantum yield of 0.009 owing to photo-induced electron transfer (PeT). After the reaction with propanal, the conjugate 2a displayed activated fluorescence, emitting at 520 nm wavelength due to dihydrobenzothiazole formation, with a quantum yield of 0.25 (Fig. 2A). In aqueous conditions, the quantum yields of 1a and 2a are 0.0027 and 0.22, respectively (Fig. S2†). This observation showcased a remarkable 80-fold increase in fluorescence post-aldehyde reaction.
Selectivity towards aldehydes
Our subsequent study involved examining the cross-reactivity of probe 1a with various biological metabolites including glutathione (GSH), pyruvate, hydrogen peroxide (H2O2), di-tert-butyl peroxide (DTBP), and nitric oxide at high concentrations (50 mM). This investigation underscored the exclusive selectivity of our 4-amino-3-thiol moiety solely for aldehydes, as no product formation occurred with any other biological metabolite, including nitric oxide, under physiological conditions (Fig. 2B). To gauge the capacity of probe 1a in detecting total cellular aldehyde concentrations, we conducted experiments involving a spectrum of aldehydes, encompassing aliphatic, aromatic, α,β-unsaturated, and keto-aldehydes. Encouragingly, our 4-amino-3-thiophenol moiety exhibited a robust reactivity towards all forms of aldehydes, generating stable dihydrobenzothiazole BODIPY products that significantly augmented fluorescence compared to the baseline. This successful detection of diverse aldehydes in living systems aligned with our primary objective. Further exploration delved into determining the sensitivity and responsiveness of probe 1a to aldehydes, by incubating 10 μM of the probe with 1 μM propanal and measuring changes in fluorescence over time. Analysis of this data yielded a calculated pseudo-first-order rate constant of 0.0196 s−1 for 1a (Fig. 2C), marking a two-fold enhancement compared to the previously employed diamine probe, which demonstrated a pseudo-first-order rate constant of 0.0103 s−1.24 This enhancement was attributed to the potent nucleophilicity of the thiol group in probe 1a.Live cell compatibility and imaging of aldehydes
To evaluate the working capacity of probe 1a inside live cells, we incubated probe 1a with diverse cell types, including both cancerous and non-cancerous breast epithelial cell lines (T-47D and MCF10A, respectively) at 10 μM for 24 hours. Flow cytometry analysis revealed no increase in cell death compared to the naive control within the tested concentration range (Fig. S3†). Utilizing exogenous aldehydes, we determined the detection limit of probe 1a in live cells. T-47D cells treated with probe 1a (10 μM) were exposed to varying levels of propanal (0 μM, 1 μM, 10 μM, 25 μM, 50 μM, and 100 μM) for 1 hour, and fluorescence was measured using a microwell plate reader. Results indicate that probe 1a has a high dynamic range for detecting aldehydes in living cells with statistical differences in higher quantities of aldehyde (25–100 μM) (Fig. 2D), which is a notable improvement over previously reported probes24 and underscores its potential in detecting aldehydes within live cellular environments.Live cell imaging and monitoring of endogenous aldehyde levels in preclinically relevant cancer models
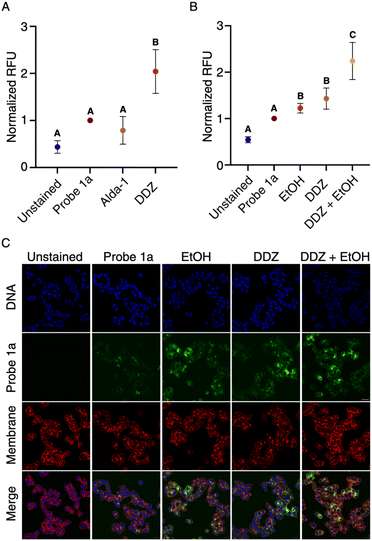
To evaluate the capability of our probe 1a in monitoring endogenous aldehyde levels, we utilized an activator (Alda-1) and an inhibitor (DDZ) of ALDH2, known to mimic distinct disease states by modulating aldehyde concentrations. DDZ, an inhibitor, induces a buildup of aldehyde concentrations, mirroring the effects of an ALDH2*2 mutation.25 We treated T-47D cells with DDZ or Alda-1 at varying concentrations for an hour, followed by co-treatment with 10 μM probe 1a. The cells were then imaged on a confocal microscope, and pixel intensity was quantified (Fig. S4†). As anticipated, Alda-1 treatment (50 μM) resulted in decreased fluorescence signals, indicative of increased endogenous aldehyde metabolism. Conversely, DDZ treatment (20 μM) led to higher fluorescence signals, reflecting aldehyde accumulation due to inhibited ALDH2 activity (Fig. 3A). Similar responses to DDZ and Alda-1 treatments were observed in other cell lines, including MCF10A. These outcomes highlight the sensitivity of our probe 1a in monitoring disease states resulting from alterations in aldehyde content, indicating its utility in diverse pathogenic conditions. Furthermore, to assess the probe's capacity to detect natural aldehyde production during metabolic processes within live cells, we incubated T-47D cells with DDZ, ethanol, or their combination, followed by co-treatment with 10 μM of probe 1a. Subsequent confocal microscope analysis after a 1 hour incubation period revealed an increase in fluorescence in both the DDZ and ethanol-only trials, with a notably larger increase observed in the DDZ/ethanol combination (Fig. 3B and C). These results affirm the high sensitivity and rapid kinetics of probe 1a, qualifying it for the identification of natural aldehyde production in both diseased and non-diseased cellular states.Design and synthesis of NIR probes for detecting aldehydes
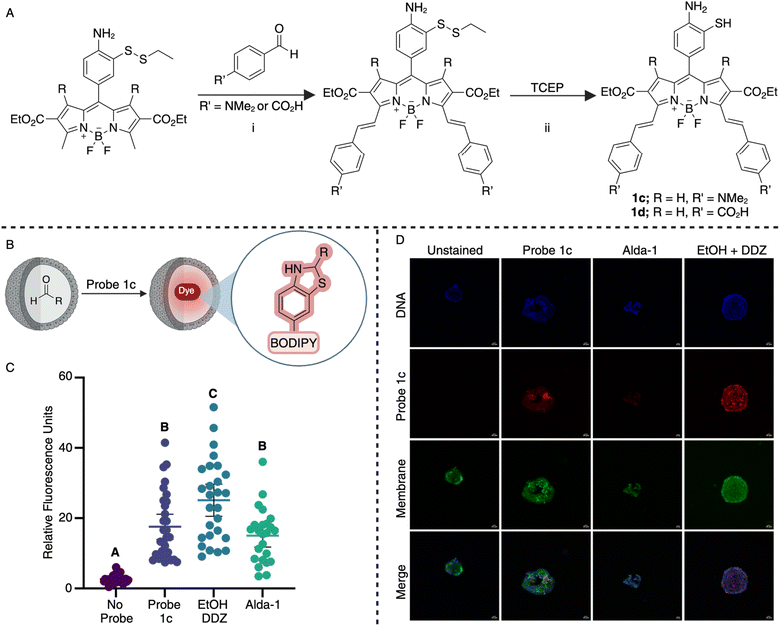
The successful demonstration of biocompatibility, chemoselectivity, and fluorescence enhancement exhibited by probe 1a spurred the development of our near-infrared (NIR) probes. To craft these probes, we employed substituted benzaldehydes to condense with the 2-methylpyrrole position of the BODIPY core (Fig. 4A), elongating conjugation and enhancing system rigidity to augment emission wavelength. This Knoevenagel-type condensation26 allows probe tunability without compromising the reactivity of aminothiol moiety toward aldehydes, thereby maintaining chemoselectivity, substrate scope, and reaction kinetics. Following this synthesis, we generated two near-infrared (NIR) probes 1c (Fig. S5†) and 1d (Fig. S6†) using 4-dimethylamino benzaldehyde or 4-formylbenzoic acid, respectively starting from intermediate from Scheme 1 (Fig. S5 and S6†). Initial assessment revealed that probe 1c manifested a more pronounced red-shifted wavelength with a larger Stokes shift, exciting at 725 nm and emitting at 780 nm upon dihydrobenzothiazole formation, compared to 1d with absorption and emission wavelengths at 650 nm and 710 nm, respectively (Fig. S7†). Consequently, we selected probe 1c for further investigations in cells, organoids, and tissues.To ascertain the viability of probe 1c within living systems, we conducted flow cytometry-based cell viability assays utilizing the T-47D cell line. Results revealed 80.8% cell viability at a concentration of 15 μM (Fig. S8†). Notably, this cell viability of 1c falls within the non-cytotoxic classification according to ISO 10993-5 standards.27
In cellular settings, we assessed the responsiveness of probe 1c to exogenously introduced levels of aldehydes within T-47D cells. These cells were exposed to varying concentrations of propanal (20 μM and 100 μM) for one hour to facilitate cellular uptake before subsequent treatment with 15 μM probe 1c for an additional hour. Confocal microscopy was employed to visualize the cells and determine fluorescence intensity (Fig. S9†). Encouragingly, a substantial increase in fluorescence was observed compared to trials utilizing the probe only. This successful outcome motivated us to progress toward more intricate experiments.
Detection of aldehydes in living systems
Organoids are three-dimensional cell cultures28 that model complex tissues and can develop into various cell types, thus acting as valuable tools for studying organ properties while maintaining disease or patient-specific phenotypes.29 Our investigation focused on utilizing murine biliary organoids derived from primary bile duct epithelium to assess the efficacy of our NIR probe 1c in detecting aldehydes in complex living systems with cellular heterogeneity. DDZ and ethanol were employed to replicate ALDH2*2 mutation, while Alda-1 was utilized to mirror a decrease in antioxidant capabilities or a disruption to cellular signaling.1Live biliary organoids were pre-treated with DDZ and ethanol or Alda-1 for 1 hour to achieve cellular penetration followed by the co-treatment with these compounds along with 15 μM of probe 1c (Fig. 4B). Following a four-hour incubation for probe 1c to permeate organoids, we imaged the live organoids using a confocal microscope. At least twenty organoids were analyzed in each trial to gauge fluctuations in fluorescence intensity. Remarkably, probe 1c effectively highlighted the increased aldehyde accumulation induced by DDZ and ethanol. Moreover, it demonstrated high sensitivity by revealing the decreased aldehyde levels in Alda-1-treated organoids compared to those treated solely with probe (Fig. 4C and D). The bimodal distribution in the two treatment groups points to the biological heterogeneity of aldehyde handling in primary biliary epithelium, which may warrant follow up studies. The dynamic response of probe 1c to both heightened and reduced aldehyde content enables the monitoring of disease states arising from antioxidant imbalances, ALDH enzyme alterations, and exposure to external aldehydes. Our results indicate the capability of probe 1c to function in intricate systems, mirroring disease pathogenesis with high spatial and temporal resolution.
Encouraged by these outcomes in organoids, we progressed to assess the functionality of probe 1c in whole tissue samples, serving as a model for complex biological specimens. Due to the lung's heightened susceptibility to aldehyde toxicity,30 murine lung tissue was selected for ex-vivo investigation. Surgically resected mouse lungs were submerged in 15 μM probe 1c in buffer for 6 or 24 hours. Tissues were then cryopreserved, sectioned, and subsequently analyzed via confocal imaging (Fig. 5A).
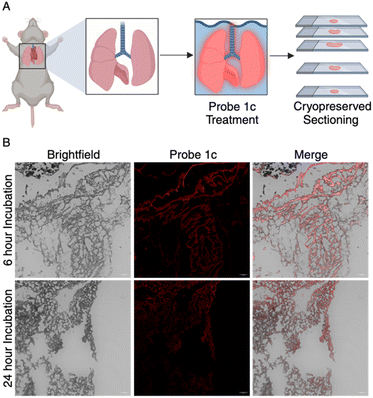 | ||
| Fig. 5 (A) Representation of whole tissue experimentation. (B) Surgically resected lung tissues ex vivo treated with probe 1c for 6 h and 24 h were imaged to reveal increased fluorescence intensity. | ||
Gratifyingly, fluorescence signals emanated from the tissue's peripheral areas. Interestingly, tissues treated for 6 hours showed increased fluorescence as compared to the 24 hour trial (Fig. 5B). We believe that this is due to the efflux of the dihydrobenzothiazole product out of the tissues over 24 hours. These results suggest the probe's ability to penetrate cells and detect aldehydes within whole tissue samples, achieving our objective of detecting total aldehyde content in complex biological specimens. We believe the perfusion of the probes into tissues or direct treatment in living mice would greatly increase the efficacy of the system. These findings effectively showcase the ability of probe 1c as a valuable diagnostic tool for monitoring aldehydes in intricate biological systems.
Conclusions
In this research endeavor, we have successfully synthesized an innovative 4-amino-3-thiophenol core in conjunction with a versatile BODIPY dye that exhibits fluorescence in both visible and IR regions, facilitating the detection of aldehydes within living systems. The 4-amino-3-thiophenol component of these adaptable probes demonstrates exceptional selectivity towards diverse aldehydes, boasting rapid kinetics (0.0196 s−1) ideal for real-time monitoring of aldehyde dynamics in complex biological systems. The reaction between the 4-amino-3-thiophenol moiety and various aldehyde types generates dihydrobenzothiazoles, resulting in a remarkable 80-fold increase in fluorescence. Notably, these probes exhibit a wide dynamic range (1–100 μM), enabling the detection of fluctuations in aldehyde concentrations across a broad spectrum. Additionally, we demonstrated that probe 1a has high efficiency in detecting both exogenous and endogenous aldehydes in live cells. This was further compounded by the probe's ability to detect aldehyde fluctuation in disease models. This sensitivity and real-time measurement capability make it a promising candidate for exploring the role of aldehydes in various cellular processes.The synthesis of near-infrared (NIR) probes was achieved through the modification of the BODIPY core, preserving the reactivity of the 2-aminothiophenol core via a Knoevenagel-type condensation, resulting in two novel NIR probes. These probes exhibited a far red-shifted wavelength with the best emitting at 780 nm, prompting further testing in cells, organoids, and ex vivo tissues. Dynamic responses were observed across all systems, including organoids, showcasing the versatility of these probes in detecting both aldehyde-deficient and aldehyde-abundant disease models. Furthermore, the application of these probes in fresh mouse lungs demonstrates their utility in complex biological systems, marking them as valuable tools for biomedical research.
Collectively, the 2-aminothiophenol moiety of our probes offers a robust tool for investigating aldehyde dynamics in biological systems, free from interference by other biological metabolites. This capability allows for a deeper exploration of cellular aldehyde processes, fostering research into aldehyde-related pathogenesis. This work adds to the expanding array of fluorescent probes and lays a foundation for future investigations to explore potential applications and enhancements of such probes across diverse biological contexts.
Data availability
The data supporting this article have been uploaded as part of the ESI.†Author contributions
All authors have given approval to the final version of the manuscript.Conflicts of interest
There are no other conflicts to declare.Acknowledgements
This research was supported by NIH (No. 1R35GM133719-01) and NSF (Grant No. CHE-1752654 and CHE-2108774) to M. R. NIH/NIDDK (Grant No. R01-DK132653) to A. D. G. and NIH/NIDDK (Grant No. F31-DK134199) to H. R. H. Monika Raj, PhD was supported by a Research Scholar Grant, RSG-22-025-01-CDP, from the American Cancer Society. This work was supported by the Emory University Emory Integrated Cellular Imaging Core Facility (RRID:SCR_023534). All the images are created with http://biorender.com. All mouse experiments were conducted in accordance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) of Emory University School of Medicine (EUCM).Notes and references
- K. S. Fritz and D. R. Petersen, Free Radical Biol. Med., 2013, 59, 85 CrossRef CAS PubMed.
- S. Vijayraghavan and N. Saini, Chem. Res. Toxicol., 2023, 36, 983 Search PubMed.
- R. M. LoPachin and T. Gavin, Chem. Res. Toxicol., 2014, 27, 1081 Search PubMed.
- R. P. Dator, M. J. Solivio, P. W. Villalta and S. Balbo, Toxics, 2019, 7, 32 Search PubMed.
- N. Kishikawa, M. H. El-Maghrabey and N. Kuroda, J. Pharm. Biomed. Anal., 2019, 175, 112782 CrossRef CAS PubMed.
- C. E. Baños and M. Silva, J. Chromatogr. B, 2010, 878, 653 CrossRef PubMed.
- A. Abu-Rayyan, I. Ahmad, N. H. Bahtiti, T. Muhmood, S. Bondock, M. Abohashrh, H. Faheem, N. Tehreem, A. Yasmeen, S. Waseem, T. Arif, A. H. Al-Bagawi and M. M. Abdou, ACS Omega, 2023, 8, 14859 CrossRef CAS PubMed.
- T. F. Brewer and C. J. Chang, J. Am. Chem. Soc., 2015, 137, 10886 CrossRef CAS PubMed.
- K. J. Bruemmer, R. R. Walvoord, T. F. Brewer, G. Burgos-Barragan, N. Wit, L. B. Pontel, K. J. Patel and C. J. Chang, J. Am. Chem. Soc., 2017, 139, 5338 CrossRef CAS PubMed.
- L. H. Yuen, N. S. Saxena, H. S. Park, K. Weinberg and E. T. Kool, ACS Chem. Biol., 2016, 11, 2312 CrossRef CAS PubMed.
- D. Larsen, A. M. Kietrys, S. A. Clark, H. S. Park, A. Ekebergh and E. T. Kool, Chem. Sci., 2018, 9, 5252 RSC.
- M. Suchý, C. Lazurko, A. Kirby, T. Dang, G. Liu and A. J. Shuhendler, Org. Biomol. Chem., 2019, 17, 1843 RSC.
- Y. Wang, H. Yu, Y. Zhang, C. Jia and M. Ji, Dyes Pigm., 2021, 190, 109284 CrossRef CAS.
- V. Ntziachristos, C. Bremer and R. Weissleder, Eur. Radiol., 2003, 13(1), 195–208 CrossRef PubMed.
- Z. Guo, S. Park, J. Yoon and I. Shin, Chem. Soc. Rev., 2014, 43, 16–29 RSC.
- C. Ding and T. Ren, Coord. Chem. Rev., 2023, 482, 215080 CrossRef CAS.
- S. A. Hilderbrand and R. Weissleder, Curr. Opin. Chem. Biol., 2010, 14(1), 71–79 CrossRef CAS PubMed.
- Q. Wang, Z. Li, Y. Hao, Y. Zhang and C. Zhang, Anal. Chem., 2022, 94, 17328 CrossRef CAS PubMed.
- M. Oe, K. Miki, Y. Ueda, Y. Mori, A. Okamoto, Y. Funakoshi, H. Minami and K. Ohe, ACS Sens., 2021, 6, 3320 CrossRef CAS PubMed.
- W. Quan, G. Zhang, Y. Li, W. Song, J. Zhan and W. Lin, Anal. Chem., 2023, 95, 2925 CrossRef CAS PubMed.
- Z. Min, M. Zhang, H. Sun, L. Xu, X. Wang and Z. Liu, Dyes Pigm., 2023, 218, 111446 CrossRef CAS.
- F. Liang, W. Huang, L. Wu, Y. Wu, T. Zhang, X. He, Z. Wang, X. Yu, Y. Li and S. Qian, Analyst, 2023, 148(7), 1437 RSC.
- N. Ding, Z. Li, Y. Hao and C. Zhang, Anal. Chem., 2022, 94, 12120 CrossRef CAS PubMed.
- R. Wills, J. Farhi, P. Czabala, S. Shahin, J. M. Spangle and M. Raj, Chem. Sci., 2023, 14, 8305 RSC.
- H. W. Goedde, D. P. Agarwal, G. Fritze, D. Meier-Tackmann, S. Singh, G. Beckmann, K. Bhatia, L. Z. Chen, B. Fang, R. Lisker, Y. K. Paik, F. Rothhammer, N. Saha, B. Segal, L. M. Srivastava and A. Czeizel, Hum. Genet., 1992, 88, 344 CrossRef CAS PubMed.
- T. Bura, P. Retailleau, G. Ulrich and R. Ziessel, J. Org. Chem., 2011, 76, 1109 CrossRef CAS PubMed.
- J. López-García, M. Lehocký, P. Humpolíček and P. Sáha, J. Funct. Biomater., 2014, 5, 43 CrossRef PubMed.
- R. Edmondson, J. J. Broglie, A. F. Adcock and L. Yang, Assay Drug Dev. Technol., 2014, 12, 207 CrossRef CAS PubMed.
- M. Huch, J. A. Knoblich, M. P. Lutolf and A. Martinez-Arias, Development, 2017, 144, 938 CrossRef CAS PubMed.
- M. Yoon, M. C. Madden and H. A. Barton, Toxicol. Sci., 2006, 89, 386 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: These include reaction procedures for the synthesis of tunable thiolamine-BODIPY probes from visible to IR range, characterization of probes by 1H, 13C NMR, and MS, procedure for carrying out chemoselectivity studies with probes, procedure for calculating quantum yields, procedure for determining the cell viability, quantification of aldehyde concentrations inside and outside cells by measuring turn on fluorescence, procedure for growing and analyzing aldehydes in organoids and procedure for analysis of aldehydes in mice organs. The authors have cited additional references within the supporting information. See DOI: https://doi.org/10.1039/d4sc00391h |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2024 |

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
: