Open Access Article
Open Access ArticleCritical assessment of interactions between ct-DNA and choline-based magnetic ionic liquids: evidences of compaction†
Kiran Devi
Tulsiyan
ab,
Saroj Kumar
Panda
c,
Malay Kumar
Rana
 *c and
Himansu S.
Biswal
*c and
Himansu S.
Biswal
 *ab
*ab
aSchool of Chemical Sciences, National Institute of Science Education and Research (NISER), PO – Bhimpur-Padanpur, Via-Jatni, District – Khurda, PIN – 752050, Bhubaneswar, India. E-mail: himansu@niser.ac.in
bHomi Bhabha National Institute, Training School Complex, Anushakti Nagar, Mumbai 400094, India
cDepartment of Chemical Sciences, Indian Institute of Science Education and Research (IISER) Berhampur, Odisha-760010, India
First published on 15th March 2024
Abstract
Ionic liquids (ILs) have become an alternative green solvent for storage and for stability of DNA. However, an in-depth understanding of binding and molecular interactions between ILs and DNA is needed. In this respect, magnetic ILs (MILs) are promising due to their tunable physicochemical properties. Various spectroscopic techniques and molecular simulations have been employed to unravel the critical factors of the strength and binding mechanism of MILs with DNA. UV-vis spectra unravel the multimodal binding of MILs with DNA, and the intrusion of IL molecules into the minor groove of DNA has been observed from dye displacement studies. Fluorescence correlation spectroscopic studies and scanning electron microscopy confirm the compaction of the DNA. ITC and molecular docking studies estimate the binding affinity of DNA with MILs, of ∼7 kcal mol−1. The 1 μs long-MD simulations give insight into the structural changes in the DNA in the MIL environment. Due to strong interaction with choline ions in the close vicinity, DNA helixes bend or squeeze in length and dilate in diameter (elliptical → spherical), leading to compaction. The post-MD parameters suggest a stronger interaction with [Ch]2[Mn] IL than with [Ch][Fe] IL; hence, the former induces DNA compaction to a more significant extent. Furthermore, decompaction is observed with the addition of sodium salts and is characterized using spectroscopic methods.
Introduction
Ionic liquids (ILs) are organic salts with low melting points. They are alternatives to aqueous and organic solvents due to their diverse physio-chemical properties, such as low vapor pressure, low toxicity, high thermal and chemical stability, biocompatibility, and high conductivity.1 Owing to the diverse nature of ILs, their application is not only limited to electrochemical devices and synthetic chemistry,2–4 but they can also act as a green solvent for biological applications.5–9 The low cytotoxicity and biodegradability of ILs render them promising solvents for storing and extracting biomolecules.10 Several exciting studies on the interaction of various imidazolium and ammonium-based ILs with nucleic acids have been reported.11,12 However, very few reports on magnetic ILs (MILs) have attracted the attention of the scientific community, to look into favorable interactions between ILs and nucleic acids.13,14 Anderson and coworkers efficiently extracted DNA from aqueous solutions using phosphonium-based MILs.15–17 They also reported that ammonium- and phosphorous-based MILs enhance DNA stability at room temperature. Recently, Kumar and coworkers explored the stability of DNA in phosphonium-based MILs and its efficacy in acting as a contrast agent for MRI applications.18,19In addition, the past decade has witnessed bio-ILs, namely choline-based ILs (ChILs), as promising ILs based on the principles of green chemistry.20 Recently, Biswal and coworkers have provided spectroscopic evidence on long-term storage and extraction of DNA and RNA using choline amino acid-based ionic liquids (ChAAILs).21,22 They proposed that cations of ILs play an important role in the stability of biomolecules. Further, Frade et al. studied the toxicity of choline-based magnetic ILs (ChMILs) in two human cell lines. They reported [FeCl4]− and [MnCl4]2− as the most and least suitable magnetic anions, respectively.23 However, on the contrary, Sintra et al. found [MnCl4]2− to be less toxic to marine bacteria.24 In this regard, we aimed to understand the DNA–ChMIL interaction using various spectroscopic techniques, such as steady-state absorption and emission and time-resolved emission spectroscopy, circular dichroism (CD), scanning electron microscopy (SEM), and fluorescence correlation spectroscopy (FCS). Thermodynamic parameters involved in DNA–ChMIL interactions and the binding mode were evaluated using isothermal titration calorimetry (ITC) and molecular dynamics studies, respectively. A deliberate study of interactions between ILs and DNA has also been put together from the perspective of molecular dynamic simulations. We have also tried to show that counter-anions play a vital role in binding with biomolecules. We observed that the solution turns turbid on adding ChMIL to DNA. However, the transparency is resumed when NaCl is added to the precipitate (Fig. 1). This is the first indication of the compaction of DNA by ChMIL and decompaction by NaCl, which needs careful investigation.
The transition of DNA from its double helical structure to a compact and coiled form is known as compaction of DNA.25 The reverse process is termed decompaction, wherein the DNA resumes its double helical structure. The compaction and decompaction of DNA have potential applications in nanomaterial templating,26 stress protection, gene regulation,27 photocontrol of gene expression, and DNA integrity protection.28 Hence, enormous efforts have been made in the past decade to discover and explore several compacting and decompacting agents.29 Owing to its negative charge, DNA can be compacted using various cationic agents, such as polyamines, polymers, nanoparticles, vesicles, and surfactants. Some compacting agents include light divalent and trivalent ions, such as Co3+, Fe2+, Fe3+, etc., and small polyamines, such as spermine.30–33 On the contrary, monovalent ions, such as Na+ or divalent cations, such as Mg2+, act as decompacting agents.34–36 Thus, multivalent salts act as compacting or decompacting agents. Enhancement of compaction in DNA in the presence of negatively charged nanoparticles has also been reported. However, these multivalent ions and nanoparticles pose toxic effects on DNA, which encourages the exploration of ionic liquids (ILs) as alternative compacting agents for DNA.
Herein, we have tried to unravel interactions between DNA and ChMILs using several spectroscopic techniques and molecular dynamics (MD) simulation studies.
Results and discussion
MIL-induced conformational changes in ct-DNA
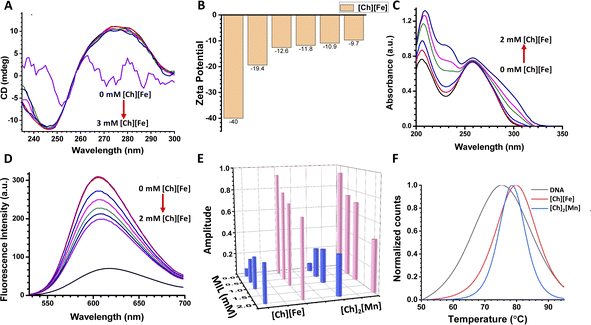
CD spectroscopy is a valuable technique to monitor the structural stability of nucleic acids and proteins.37 CD spectra have different characteristic peaks for different types of DNA. The A-form of DNA has a dominant positive band at 260 nm and a negative band at 210 nm. The B form of DNA (B-DNA) has a positive band at 276 nm due to π–π base stacking, and a negative band at 246 nm due to helicity. The Z-form of DNA has a negative band at 290 nm, a positive band around 260 nm, and another characteristic negative band at around 205 nm.38Fig. 2A shows the CD spectra of ct-DNA on increasing concentrations of [Ch][Fe] IL. Upon addition of [Ch][Fe] IL up to 2 mM, minimal conformational changes were observed in the positive and negative bands of B-DNA. In contrast, a significant change was observed with further addition of IL. The bands in the CD spectra disappeared on adding 3 mM [Ch][Fe] IL, and the solution turned turbid. Control experiments were also performed in the presence of FeCl3 and choline hydroxide, where we did not observe any turbidity. Therefore, the turbidity is attributed solely to the presence of [Ch][Fe] IL. The CD spectra of ct-DNA with increasing concentration of [Ch]2[Mn] IL were recorded to unravel the effect of anions on ct-DNA (Fig. S6A†). It was observed that [Ch]2[Mn] IL behaved similarly to [Ch][Fe] IL. No significant change was noticed up to 2 mM concentration of [Ch]2[Mn] IL. However, the solution turned turbid, and the band disappeared, after adding IL. These unaltered CD spectra indicate that the DNA is stable in ChMILs at low concentrations. However, compaction of DNA happens at higher concentrations, as revealed by the change in the CD spectra. This is in agreement with previous reports, according to which, at a high concentration of Fe and Mn, chelation leads to the compaction of DNA.31,39 In addition, there is a deviation from the well-established concept that cations play an essential role in maintaining the structural stability and integrity of DNA, as the [Ch] cation is known to maintain the structural integrity of DNA.22Further, the effect of MILs on the surface of ct-DNA was explored using zeta potential measurements. The effective surface charge of ct-DNA derived from the zeta potential (ζ) in the presence of different [Ch][Fe] IL concentrations is shown in Fig. 2B. The ct-DNA shows a ζ value of −40 mV due to phosphate groups on its surface. The negative value of ζ decreases to −13 mV after adding 2 mM [Ch][Fe] IL, indicating significant interaction of choline cations with ct-DNA.19 At higher concentrations (5 mM), negative counter ions of [Ch][Fe] IL also interact with the exposed surface charge of ct-DNA, and, subsequently, only a 3 mV change in the ζ value was observed. It is presumed that the overall negative surface of ct-DNA was neutralized through the positive counterion of ChMIL at lower concentrations, whereas the negative counterion interacts at higher concentrations. The zeta potentials of ct-DNA in the presence of [Ch]2[Mn] IL are given in Fig. S6B.†
The value of ζ on adding 2 mM [Ch]2[Mn] IL was reduced to −9 mV due to the higher interaction of choline cations with the phosphate group of ct-DNA. Akin to previous results, minimal change in ζ value at a higher concentration of [Ch]2[Mn] IL indicates the interaction between the anionic counterpart and ct-DNA.
Spectroscopic insight on the interaction between MIL and ct-DNA
On establishing the interaction between ChMIL and ct-DNA, the nature of the binding mode was investigated using steady-state absorption and emission measurements. A shift in absorption maxima for the characteristic peak of ct-DNA suggests interaction between ct-DNA and ChMILs. In addition, a dye displacement study was employed to characterize interactions between ChMILs and ct-DNA. On monitoring the absorbance at 260 nm for ct-DNA, the addition of [Ch][Fe] IL results in a hyperchromic shift (Fig. 2C). Broadening of the 260 nm peak was also observed on increasing the concentration of [Ch][Fe] IL. The change in peak shape confirms the binding interaction between the positive and negative counterions of the ILs and ct-DNA. Steady-state fluorescence studies were carried out using the well-known ct-DNA-binding probe ethidium bromide (EB). EB binds to intercalating sites40,41 of ct-DNA,42 resulting in an increase in the fluorescence intensity of the ct-DNA–EB complex. However, quenching of the fluorescence intensity upon addition of [Ch][Fe] IL to the complex validates the displacement of EB from the ct-DNA–EB complex and binding of [Ch][Fe] IL at the intercalating sites of ct-DNA (Fig. 2D). The binding of choline cation to the intercalating site of DNA through electrostatic interactions has previously been reported by Khan et al.43 and Sahoo et al.22 Steady-state absorption and dye displacement studies for [Ch]2[Mn] IL with ct-DNA are depicted in Fig. S6C and D,† respectively. Fluorescence quenching was observed for the ct-DNA–EB complex in the presence of [Ch]2[Mn] IL. The quenching rate is higher in [Ch]2[Mn] IL, which is attributed to the higher interaction of the choline cation to ct-DNA, as validated by zeta potential studies.The mode of interaction between ChMILs and ct-DNA was also investigated using time-resolved fluorescence spectroscopy. As given below, a biexponential fluorescence decay model was used to obtain excited-state lifetimes.
I(t) = a1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) exp(−t/τ1) + a2 exp(−t/τ1) + a2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) exp(−t/τ2), exp(−t/τ2), |
The lifetime decay profile of EB follows a single exponential decay, with an excited-state lifetime of 1.62 ns. On formation of the complex with ct-DNA, the decay profile follows a biexponential decay. The excited-state lifetimes of a shorter component of 1.59 ns and a longer component of 18.73 ns, with corresponding amplitudes of 0.07 and 0.93 for EB and ct-DNA–EB complex, respectively, were observed. Similar excited lifetimes were also reported by Hellar et al.44 The lifetime decay profiles of the ct-DNA–EB complex on gradual addition of [Ch][Fe] IL are shown in Fig. S6E.† The lifetime decay profiles still follow a biexponential decay in the presence of [Ch][Fe] IL. However, the amplitude of the shorter component increases from 7% to 33% with an increase in IL concentration, and the amplitude of the longer component decreases from 93% to 67%. Similar observations were obtained for [Ch]2[Mn] IL, as shown in Fig. S6F of the ESI.† A similar increase in the shorter component was observed in [Ch]2[Mn] IL, i.e., 7 to 35%. The increase in the amplitude of the shorter component corroborates with the dye displacement study. It is attributed to the release of EB from the ct-DNA–EB complex on gradual addition of IL. The increase in the shorter component is attributed to the higher quenching in [Ch]2[Mn] IL. The fluorescence decay parameters of the ct-DNA–EB complex in the presence of ILs are provided in Fig. 2E and Table S2.†
Information on the thermal stability of ct-DNA was obtained from a UV melting study. Melting of DNA is accompanied by a hyperchromic effect; the DNA absorbs UV light at 260 nm, and this increases with cleavage of the hydrogen bonding in ct-DNA.45 The UV melting plots were analyzed by plotting the first derivative of the absorbance with respect to temperature against temperature. The first derivative of melting data is shown in Fig. 2F. The melting temperature (Tm) of ct-DNA in the absence of ChMILs was found to be 75 °C. The melting temperature increases by 3–4 °C in the presence of [Ch][Fe], while it increases by 4–5 °C in the presence of [Ch]2[Mn]. This change in melting point is attributed to the thermal stability of ct-DNA. Our findings are supported by a recent simulation study by Garai et al., which shows a significant increase in persistence length and stretch modulus of DNA with a higher concentration of IL.46 The increased rigidity of ct-DNA would result in a higher melting point, as observed here.
Thermodynamics of the interaction between ionic liquid and the ct-DNA system
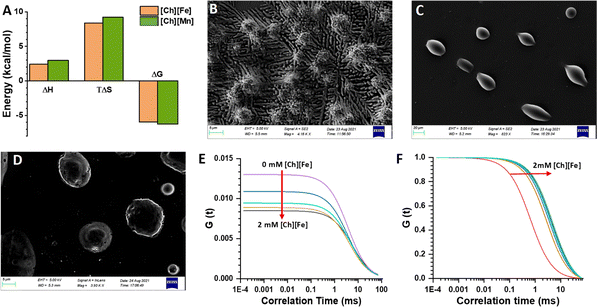
Isothermal titration calorimetry (ITC) is a technique that provides information about the thermodynamics of binding of ligands to proteins or nucleic acids. Thermodynamic parameters, such as binding affinity (kA), enthalpy (ΔH), entropy (ΔS), Gibbs free energy (ΔG), and stoichiometry (n), are evaluated based on the heat change due to ligand–DNA interactions.47 The ITC profile for the ct-DNA–EB complex is shown in Fig. S7A,† where individual peaks represent each injection. The heat change due to EB and DNA interactions was calculated by the integration method. The thermodynamic parameters, upon fitting, gave a negative ΔH and a positive ΔS value, indicating the process is enthalpy and entropy driven. The binding of [Ch][Fe] IL with ct-DNA was also characterized using an ITC enthalpogram. The integrated heat data of ct-DNA in the presence of [Ch][Fe] IL are shown in Fig. S7B.† The thermodynamic parameters reveal positive ΔH and TΔS values, and the reaction is entropically driven by the gradual addition of [Ch][Fe] IL (Fig. 3A). According to the literature, groove binding of ligands leads to an increase in the entropy of the system, with the binding being entropically driven. The ITC profiles for [Ch]2[Mn] IL are shown in Fig. S7C,† and the thermodynamic parameters confirm that the process is entropically driven. We propose that the anions of ionic liquids result in the aggregation of cations through electrostatic interactions, which promotes the compaction of ct-DNA.Compaction of DNA
Field emission scanning electron microscopy (FESEM) provides insight into the morphology of ct-DNA.48 As shown in Fig. 3B, ct-DNA forms a coiled structure nearly 50 nm in size in the buffer solution. The ct-DNA compaction occurred in the presence of [Ch][Fe] IL (see Fig. 3C), and the size of the globular form is nearly 5 μm. Similar compaction was observed in [Ch][Mn] IL (Fig. 3D); however, the size of the globular form was almost 7 μm. The FESEM image gives pictorial evidence of compaction on addition of MILs. The morphology of the ct-DNA globule is in good agreement with the previous literature. Earlier, Satpathi et al. reported that, as FESEM is conducted in a dried condition, the probability of aggregation increases, which explains the larger aggregation globule in our case.48Further, fluorescence correlation spectroscopic (FCS) measurements were carried out using the well-known DNA-binding probe 4′,6-diamidino-2-phenylindole (DAPI).49 DAPI is known to bind at the minor groove region of DNA.50Fig. 3E shows that with increase in concentration of [Ch][Fe] IL in the ct-DNA–DAPI complex, the G(τ) value at zero time delay, represented as G(0), decreases. G(0) is related to N as G(0) = 1/N(1 + A), where A is the ratio of the number of molecules (ct-DNA–DAPI complex) present in the native state. The decrease in the G(0) value suggests an increase in the effective number of molecules in the observation volume undergoing intense fluctuation. Simultaneously, on normalizing the G(τ) function, we observe that the diffusion coefficient increases with increasing concentration of [Ch][Fe] IL (Fig. 3F). Similar observations were made for [Ch]2[Mn] IL and are shown in Fig. S8A and B.† The FCS technique also provides information on the hydrodynamic radius (RH) of biomolecules. Herein, we found the RH of the ct-DNA–DAPI complex to be 35–40 nm. However, adding ChMILs increases the RH, which is estimated to be 115–130 nm. The increase in RH is attributed to the compaction of ct-DNA in the presence of ChMILs.
Decompaction of ct-DNA
Monovalent anions and cations act as decompacting agents. We have observed the decompaction of ct-DNA structure with the assistance of common salt NaCl. The excess NaCl (5 mM) was added to the compacted ct-DNA solution, which converted condensed ct-DNA into a soluble form, likely due to an efficient ion exchange process.Decompaction of ct-DNA was validated by UV-vis spectroscopy (Fig. S8C†) and CD spectroscopy (Fig. S8D†), which showed same bands as native ct-DNA without any degradation. The characteristic positive peak at 274 nm and negative peak at 245 nm, confirm the secondary structural stability of ct-DNA on decompaction. Along with this, the ratio of absorbance at 260 and 280 nm was found to be 1.8, which substantiates the purity of ct-DNA.
The experimental results were aided by molecular docking and molecular dynamics simulations.
Molecular dynamics (MD) simulations and post-MD parameters
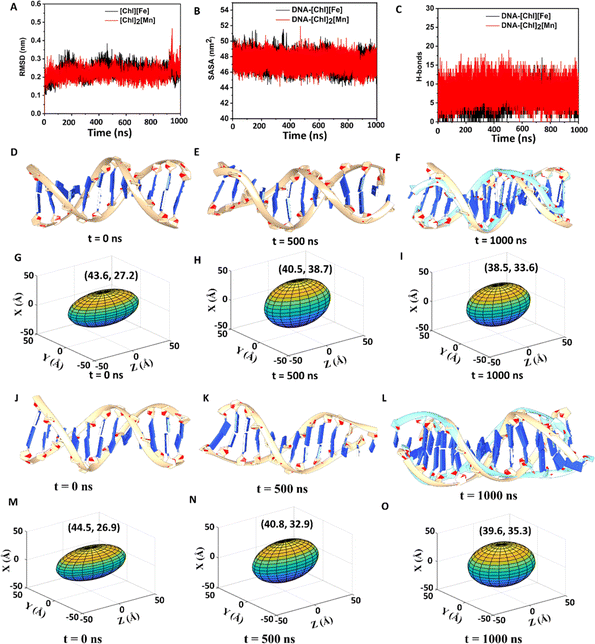
An initial idea of binding of ChMILs to ct-DNA was obtained using molecular docking.51,52 The most favorable binding pose along with the interactions of ct-DNA with both the ChMILs is shown in Fig. S9† and was used as the starting conformation for MD simulations. MD simulations were conducted to extract microscopic insight into interactions between ct-DNA and ChMILs over time. Classical MD helps to explore physical processes and conformational space of systems over a large span. The time evolution of systems can be characterized by post-MD parameters, such as root-mean-square-deviation (RMSD), solvent-accessible surface area (SASA), principal component analysis (PCA), and radial distribution function (RDF), using MD trajectories.RMSD is an essential parameter to judge conformational stability. A large variation of RMSD is a measure of large instability and weak interaction between DNA and IL. Simulations show that DNA–[Ch]2[Mn] has a lower RMSD for DNA than DNA–[Ch][Fe], indicating greater stability or stronger interaction between DNA and [Ch]2[Mn] IL (see Fig. 4A). While analyzing SASA for DNA, a similar trend is observed between [Ch]2[Mn] and [Ch][Fe]. Irrespective of the IL present, SASA, in Fig. 4B, reveals that the amount of DNA surface exposed to solvent steadily decreases during simulation. This indicates that ILs have closer contact with the DNA surface than water. To dispel any ambiguity, we computed the RDF for both systems (Fig. S10†), which unearths some crucial aspects of structural organization among DNA, [Ch]+, [FeCl4]−, [MnCl4]2−, and water. It is interesting to observe that [Ch]+ ions are closer to DNA in both systems, with the maximum likelihood peak at around 0.5 nm. Conversely, water, [FeCl4]−, and [MnCl4]2− ions are far from DNA. Due to coulombic forces, the [Ch]+ ions interact with DNA (specifically with the phosphate groups of DNA) in the closest proximity among other constituents of ILs or water.
This unequivocally demonstrates that the DNA surface has some residual negative charge, congruent with the zeta potential value. On the basis of RDF data, the decrease of ζ value from −40 mV to −13 mV in [Ch][Fe] IL or to −9 mV in [Ch]2[Mn] IL, is thus well justified by a charge-neutralizing combination between [Ch]+ and DNA. As evident in Fig. 4C, [Ch]2[Mn] establishes a more significant number of H-bond interactions with DNA than [Ch][Fe]. Consequently, the former should exert a much stronger attraction to DNA than the latter.
As the MD simulation progresses, the DNA structure in [Ch][Fe] IL changes gradually. Due to the interaction with [Ch]+/IL, the molecule begins to bend and dilate in diameter as major and minor grooves alter. An unsuccessful overlay of DNA structures between 0 ns and 1000 ns in Fig. 4D–F illustrates the degree of structural deformation. One of the DNA terminals at 1000 ns is seen to be aberrantly bent. In a similar way, but to a significant extent, [Ch]2[Mn] IL distorts the DNA structure (Fig. S11†), causing it to lose one primary and one minor groove. IL, particularly [Ch]+, pulls helixes/strands apart with strong coulombic and H-bond forces that markedly reduce the DNA length and increase its diameter, leading to DNA compaction. To track changes in DNA structure, we provide the lengths and diameters of DNA in Tables S3 and S4† at different time intervals. More conveniently, 3D elliptical images in Fig. 4G–I and S12† adequately depict the change in DNA volume during the simulation, to support FESEM data. The major and minor axes of ellipses represent DNA length and diameter, respectively. A change from an elliptical to a spherical shape (decrease of major axis and increase of minor axis) over time is evident, indicative of DNA compaction in ILs, which is more substantial in [Ch]2[Mn] IL.
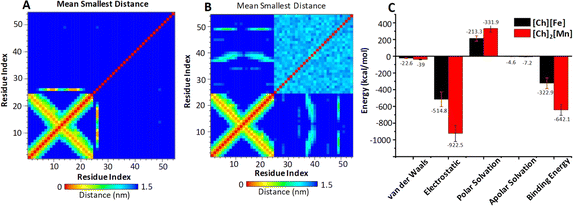
To quantify the affinity and binding strength between DNA and ILs, we estimated binding free energy (ΔGbind) by taking the final 10 ns of the MD trajectories. Fig. 5C displays the computed ΔGbind values with error bars and their contributory interactions for both systems. For the DNA–[Ch][Fe] system, overall ΔGbind and other energy components, such as van der Waals, electrostatic, polar, and apolar solvation energy components, are −322.9, −22.6, −514.8, 213.3, and −4.6 kcal mol−1, respectively. Similarly, respective values in the DNA–[Ch]2[Mn] system are −642.1, −39, −922.5, 331.9, and −7.2 kcal mol−1, respectively. As is clear, the electrostatic component accounts for most of the interaction between DNA and MILs, which is greater (by −407.7 kcal mol−1) in [Ch]2[Mn] IL. Consequently, [Ch]2[Mn] IL has a stronger affinity for DNA (twice that of [Ch][Fe] IL), which is consistent with earlier post-MD parameters. Qualitatively, we also see an excellent agreement between ΔGbind values and ITC Gibbs free energy changes of about −6 kcal mol−1 in [Ch][Fe] IL and about −9 kcal mol−1 in [Ch]2[Mn] IL. On the basis of the computational interaction energy analysis, we can thus explain experimental observations, including the higher quenching rate and melting temperature of DNA in [Ch]2[Mn] IL.
The entire MD trajectories were subjected to PC analysis. Taking the first two PCs into account, Fig. S13† displays the conformational space of DNA in two systems for the last 10 ns of the MD trajectories. Comparing between the two systems, DNA in [Ch]2[Mn] IL occupies a smaller subspace than that in [Ch][Fe] IL. A smaller subspace indicates a more pronounced interaction between DNA and [Ch]2[Mn] IL. In addition, the correlation contact map for residues generated on the final 10 ns MD trajectories for both systems provides more evidence. Fig. 5A and B reveals that [Ch]2[Mn] IL has more contact with DNA base pairs than [Ch][Fe] during the simulation period.
Conclusions
Various experimental techniques and molecular docking experiments have explored the binding characteristics and molecular mechanisms of ChMILs with ct-DNA. The turbidity in ct-DNA with increasing concentrations of ChMILs confirms the compaction of the DNA, and SEM and FCS analysis validated the compaction of ct-DNA. The molecular interactions of ILs and ct-DNA via electrostatic, hydrogen bonding, and hydrophobic interactions were revealed through UV-vis spectra, fluorescence dye displacement study, zeta potential, and molecular docking. Based on the experimental and theoretical evidence, it is concluded that MILs can bind ct-DNA through the choline cation irrespective of the anions used in this study. MD studies suggest a strong interaction between [Ch]2[Mn] and DNA. In comparison to [Ch][Fe], DNA suffers a more significant conformational shift in the presence of [Ch]2[Mn]. During the simulation, the surface of DNA exposed to solvent steadily decreases for both complexes. The structure of the DNA bends or becomes more spherical compared to its initial form due to interactions with the two magnetic ionic liquids. Due to a more pronounced interaction, DNA compaction is more prominent in [Ch]2[Mn]. Of the two ILs, the estimated binding free energy, ΔG, is −642 kcal mol−1, for [Ch]2[Mn], confirming its stronger DNA affinity.Data availability
The datasets supporting this article have been uploaded as part of the ESI.†Author contributions
The research problem was conceptualized by H. S. B. K. D. T. performed all the experiments through the supervision of H. S. B. The MD simulation was performed by S. K. P. and M. K. R. The manuscript was written through the contributions of all authors. All authors have given approval to the final version of the manuscript.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
K. D. T. and H. S. B. acknowledge the Centre for Interdisciplinary Sciences (CIS), NISER for experimental facilities. K. D. T. and H. S. B. acknowledge financial support from the Department of Atomic Energy, Department of Science and Technology (Project File No. CRG/2022/001096), Govt. of India.Notes and references
- B. C. Zhigang Lei, Y.-M. Koo and D. R. MacFarlane, Introduction: Ionic Liquids, Chem. Rev., 2017, 117, 6633–6635 CrossRef.
- S. S. Choudhury, S. Mahapatra, A. K. Sahu, P. Hembram, S. Jena and H. S. Biswal, Synthesis of α,β-Unsaturated Ketones in Water: The Claisen–Schmidt Condensation Revisited, ACS Sustainable Chem. Eng., 2022, 10, 14271–14279 CrossRef CAS.
- S. S. Choudhury, S. Mahapatra and H. S. Biswal, Hydrogen bond mediated conversion of benzenenitriles and arylacetonitriles to amides: an “on/in-water” reaction strategy, Green Chem., 2022, 24, 4981–4990 RSC.
- S. S. Choudhury, S. Jena, D. K. Sahoo, S. Shekh, R. K. Kar, A. Dhakad, K. H. Gowd and H. S. Biswal, Gram-Scale Synthesis of 1,8-Naphthyridines in Water: The Friedlander Reaction Revisited, ACS Omega, 2021, 6, 19304–19313 CrossRef CAS.
- M. Watanabe, M. L. Thomas, S. Zhang, K. Ueno, T. Yasuda and K. Dokko, Application of Ionic Liquids to Energy Storage and Conversion Materials and Devices, Chem. Rev., 2017, 117, 7190–7239 CrossRef CAS.
- A. Benedetto and H.-J. Galla, Editorial of the “ionic liquids and biomolecules” special issue, Biophys. Rev., 2018, 10, 687–690 CrossRef.
- D. K. Sahoo, A. Chand, S. Jena and H. S. Biswal, Hydrogen-bond-driven thiouracil dissolution in aqueous ionic liquid: A combined microscopic, spectroscopic and molecular dynamics study, J. Mol. Liq., 2020, 319, 114275 CrossRef CAS.
- D. K. Sahoo, V. R. Mundlapati, A. A. Gagrai and H. S. Biswal, Efficient SO2 Capture through Multiple Chalcogen Bonds, Sulfur-Centered Hydrogen Bonds and S⋯π Interactions: A Computational Study, ChemistrySelect, 2016, 1, 1688–1694 CrossRef CAS.
- D. K. Sahoo, S. Jena, K. D. Tulsiyan, J. Dutta, S. Chakrabarty and H. S. Biswal, Amino-Acid-Based Ionic Liquids for the Improvement in Stability and Activity of Cytochrome c: A Combined Experimental and Molecular Dynamics Study, J. Phys. Chem. B, 2019, 123, 10100–10109 CrossRef CAS PubMed.
- M. Sivapragasam, M. Moniruzzaman and M. Goto, Recent advances in exploiting ionic liquids for biomolecules: Solubility, stability and applications, Biotechnol. J., 2016, 11, 1000–1013 CrossRef CAS.
- A. Benedetto and P. Ballone, Room Temperature Ionic Liquids Meet Biomolecules: A Microscopic View of Structure and Dynamics, ACS Sustainable Chem. Eng., 2016, 4, 392–412 CrossRef CAS.
- S. K. Shukla and J.-P. Mikkola, Use of Ionic Liquids in Protein and DNA Chemistry, Phys. Chem. Chem. Phys., 2020, 8, 598662 CAS.
- X. Ding, K. D. Clark, M. Varona, M. N. Emaus and J. L. Anderson, Magnetic ionic liquid-enhanced isothermal nucleic acid amplification and its application to rapid visual DNA analysis, Anal. Chim. Acta, 2019, 1045, 132–140 CrossRef CAS PubMed.
- M. N. Emaus, K. D. Clark, P. Hinners and J. L. Anderson, Preconcentration of DNA using magnetic ionic liquids that are compatible with real-time PCR for rapid nucleic acid quantification, Anal. Bioanal. Chem., 2018, 410, 4135–4144 CrossRef CAS PubMed.
- A. N. Bowers, M. J. Trujillo-Rodríguez, M. Q. Farooq and J. L. Anderson, Extraction of DNA with magnetic ionic liquids using in situ dispersive liquid–liquid microextraction, Anal. Bioanal. Chem., 2019, 411, 7375–7385 CrossRef CAS PubMed.
- K. D. Clark, M. M. Yamsek, O. Nacham and J. L. Anderson, Magnetic ionic liquids as PCR-compatible solvents for DNA extraction from biological samples, Chem. Commun., 2015, 51, 16771–16773 RSC.
- K. D. Clark, O. Nacham, H. Yu, T. Li, M. M. Yamsek, D. R. Ronning and J. L. Anderson, Extraction of DNA by Magnetic Ionic Liquids: Tunable Solvents for Rapid and Selective DNA Analysis, Anal. Chem., 2015, 87, 1552–1559 CrossRef CAS PubMed.
- P. S. Gehlot, H. Gupta and A. Kumar, Paramagnetic Surface Active Ionic Liquids: Interaction with DNA and MRI Application, Colloid and Interface Science Communications, 2018, 26, 14–23 CrossRef CAS.
- P. S. Gehlot, H. Gupta, M. S. Rathore, K. Khatri and A. Kumar, Intrinsic MRI contrast from amino acid-based paramagnetic ionic liquids, Materials Advances, 2020, 1, 1980–1987 RSC.
- R. Caparica, A. Júlio, A. R. Baby, M. E. M. Araújo, A. S. Fernandes, J. G. Costa and T. Santos de Almeida, Choline-Amino Acid Ionic Liquids as Green Functional Excipients to Enhance Drug Solubility, Pharmaceutics, 2018, 10, 288 CrossRef CAS.
- K. D. Tulsiyan, S. Jena, M. González-Viegas, R. K. Kar and H. S. Biswal, Structural Dynamics of RNA in the Presence of Choline Amino Acid Based Ionic Liquid: A Spectroscopic and Computational Outlook, ACS Cent. Sci., 2021, 7, 1688–1697 CrossRef CAS PubMed.
- D. K. Sahoo, S. Jena, J. Dutta, S. Chakrabarty and H. S. Biswal, Critical Assessment of the Interaction between DNA and Choline Amino Acid Ionic Liquids: Evidences of Multimodal Binding and Stability Enhancement, ACS Cent. Sci., 2018, 4, 1642–1651 CrossRef CAS PubMed.
- R. F. M. Frade, S. Simeonov, A. A. Rosatella, F. Siopa and C. A. M. Afonso, Toxicological evaluation of magnetic ionic liquids in human cell lines, Chemosphere, 2013, 92, 100–105 CrossRef CAS PubMed.
- T. E. Sintra, M. Nasirpour, F. Siopa, A. A. Rosatella, F. Gonçalves, J. A. P. Coutinho, C. A. M. Afonso and S. P. M. Ventura, Ecotoxicological evaluation of magnetic ionic liquids, Ecotoxicol. Environ. Saf., 2017, 143, 315–321 CrossRef CAS PubMed.
- A. Estévez-Torres and D. Baigl, DNA compaction: fundamentals and applications, Soft Matter, 2011, 7, 6746–6756 RSC.
- H. A. Becerril and A. T. Woolley, DNA-templated nanofabrication, Chem. Soc. Rev., 2009, 38, 329–337 RSC.
- B. Demeneix, Z. Hassani and J.-P. Behr, Towards Multifunctional Synthetic Vectors, Curr. Gene Ther., 2004, 4, 445–455 CrossRef CAS PubMed.
- Y.-Z. You, Z.-Q. Yu, M.-M. Cui and C.-Y. Hong, Preparation of Photoluminescent Nanorings with Controllable Bioreducibility and Stimuli-Responsiveness, Angew. Chem., Int. Ed., 2010, 49, 1099–1102 CrossRef CAS PubMed.
- A. González-Pérez, J. Carlstedt, R. S. Dias and B. Lindman, Cyclodextrins in DNA decompaction, Colloids Surf., B, 2010, 76, 20–27 CrossRef PubMed.
- A. A. Zinchenko, V. G. Sergeyev, K. Yamabe, S. Murata and K. Yoshikawa, DNA Compaction by Divalent Cations: Structural Specificity Revealed by the Potentiality of Designed Quaternary Diammonium Salts, ChemBioChem, 2004, 5, 360–368 CrossRef CAS PubMed.
- S. Gawęda, M. C. Morán, A. A. C. C. Pais, R. S. Dias, K. Schillén, B. Lindman and M. G. Miguel, Cationic agents for DNA compaction, J. Colloid Interface Sci., 2008, 323, 75–83 CrossRef PubMed.
- T. Kikuchi, S. Sato, D. Fujita and M. Fujita, Stepwise DNA condensation by a histone-mimic peptide-coated M12L24 spherical complex, Chem. Sci., 2014, 5, 3257–3260 RSC.
- M. Deiana, Z. Pokladek, K. Matczyszyn, P. Mlynarz, M. Buckle and M. Samoc, Effective control of the intrinsic DNA morphology by photosensitive polyamines, J. Mater. Chem. B, 2017, 5, 1028–1038 RSC.
- R. S. Dias, B. Lindman and M. G. Miguel, Compaction and Decompaction of DNA in the Presence of Catanionic Amphiphile Mixtures, J. Phys. Chem. B, 2002, 106, 12608–12612 CrossRef CAS.
- E. Grueso, C. Cerrillos, J. Hidalgo and P. Lopez-Cornejo, Compaction and Decompaction of DNA Induced by the Cationic Surfactant CTAB, Langmuir, 2012, 28, 10968–10979 CrossRef CAS PubMed.
- X. Guo, B. Cui, Y. Li and J. Ding, Effects of salt and temperature on single-chained cationic surfactant/oligodeoxynucleotide vesicle formation, J. Polym. Sci., Part A: Polym. Chem., 2012, 50, 1740–1745 CrossRef CAS.
- R. L. Webb, Circular Dichroism and the Conformational Analysis of Biomolecules Edited by Gerald D. Fasman. Plenum Press, New York and London. 1996. ix + 738 pp. 17 × 25.5 cm. ISBN 0-306-45152-5. $125.00, J. Med. Chem., 1996, 39, 5285–5286 CrossRef CAS.
- J. Kypr, I. Kejnovská, D. Renčiuk and M. Vorlíčková, Circular dichroism and conformational polymorphism of DNA, Nucleic Acids Res., 2009, 37, 1713–1725 CrossRef CAS PubMed.
- D. Roncarati, S. Pelliciari, N. Doniselli, S. Maggi, A. Vannini, L. Valzania, L. Mazzei, B. Zambelli, C. Rivetti and A. Danielli, Metal-responsive promoter DNA compaction by the ferric uptake regulator, Nat. Commun., 2016, 7, 12593 CrossRef CAS PubMed.
- C. C. Tsai, S. C. Jain and H. M. Sobell, X-ray crystallographic visualization of drug-nucleic acid intercalative binding: structure of an ethidium-dinucleoside monophosphate crystalline complex, Ethidium: 5-iodouridylyl (3'-5') adenosine, Proc. Natl. Acad. Sci. U. S. A., 1975, 72, 628–632 CrossRef CAS PubMed.
- R. Galindo-Murillo and T. E. Cheatham III, Ethidium bromide interactions with DNA: an exploration of a classic DNA–ligand complex with unbiased molecular dynamics simulations, Nucleic Acids Res., 2021, 49, 3735–3747 CrossRef CAS PubMed.
- J. Kapuscinski, DAPI: a DNA-Specific Fluorescent Probe, Biotech. Histochem., 1995, 70, 220–233 CrossRef CAS PubMed.
- A. Haque, I. Khan, S. I. Hassan and M. S. Khan, Interaction studies of cholinium-based ionic liquids with calf thymus DNA: Spectrophotometric and computational methods, J. Mol. Liq., 2017, 237, 201–207 CrossRef CAS.
- D. P. Heller and C. L. Greenstock, Fluorescence lifetime analysis of DNA intercalated ethidium bromide and quenching by free dye, Biophys. Chem., 1994, 50, 305–312 CrossRef CAS.
- S. U. Rehman, T. Sarwar, M. A. Husain, H. M. Ishqi and M. Tabish, Studying non-covalent drug–DNA interactions, Arch. Biochem., 2015, 576, 49–60 CrossRef.
- A. Garai, D. Ghoshdastidar, S. Senapati and P. K. Maiti, Ionic liquids make DNA rigid, J. Chem. Phys., 2018, 149, 45104 CrossRef PubMed.
- M. W. Freyer and E. A. Lewis, in Methods in Cell Biology, Academic Press, 2008, vol. 84, pp. 79–113 Search PubMed.
- S. Satpathi, A. Sengupta, V. M. Hridya, K. Gavvala, R. K. Koninti, B. Roy and P. Hazra, A Green Solvent Induced DNA Package, Sci. Rep., 2015, 5, 9137 CrossRef.
- L. Yu, Y. Lei, Y. Ma, M. Liu, J. Zheng, D. Dan and P. Gao, A Comprehensive Review of Fluorescence Correlation Spectroscopy, Frontiers in Physics, 2021, 9 DOI:10.3389/fphy.2021.644450.
- A. Pabbathi and A. Samanta, Spectroscopic and Molecular Docking Study of the Interaction of DNA with a Morpholinium Ionic Liquid, J. Phys. Chem. B, 2015, 119, 11099–11105 CrossRef CAS.
- S. S. Mati, S. S. Roy, S. Chall, S. Bhattacharya and S. C. Bhattacharya, Unveiling the Groove Binding Mechanism of a Biocompatible Naphthalimide-Based Organoselenocyanate with Calf Thymus DNA: An “Ex Vivo” Fluorescence Imaging Application Appended by Biophysical Experiments and Molecular Docking Simulations, J. Phys. Chem. B, 2013, 117, 14655–14665 CrossRef CAS PubMed.
- J. Eberhardt, D. Santos-Martins, A. F. Tillack and S. Forli, AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings, J. Chem. Inf. Model., 2021, 61, 3891–3898 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sc00004h |
| This journal is © The Royal Society of Chemistry 2024 |