Open Access Article
Open Access ArticleArtificial intelligence performance in testing microfluidics for point-of-care†
Mert Tunca
Doganay
a,
Purbali
Chakraborty
a,
Sri Moukthika
Bommakanti
a,
Soujanya
Jammalamadaka
a,
Dheerendranath
Battalapalli
a,
Anant
Madabhushi
bc and
Mohamed S.
Draz
 *ade
*ade
aDepartment of Medicine, Case Western Reserve University School of Medicine, Cleveland, OH 44106, USA. E-mail: mohamed.draz@case.edu
bDepartment of Biomedical Engineering, Emory University, Atlanta, GA, USA
cAtlanta Veterans Administration Medical Center, Atlanta, GA, USA
dDepartment of Biomedical Engineering, Case Western Reserve University, Cleveland, OH, USA
eDepartment of Biomedical Engineering, Cleveland Clinic, Cleveland, OH 44106, USA
First published on 20th September 2024
Abstract
Artificial intelligence (AI) is revolutionizing medicine by automating tasks like image segmentation and pattern recognition. These AI approaches support seamless integration with existing platforms, enhancing diagnostics, treatment, and patient care. While recent advancements have demonstrated AI superiority in advancing microfluidics for point of care (POC) diagnostics, a gap remains in comparative evaluations of AI algorithms in testing microfluidics. We conducted a comparative evaluation of AI models specifically for the two-class classification problem of identifying the presence or absence of bubbles in microfluidic channels under various imaging conditions. Using a model microfluidic system with a single channel loaded with 3D transparent objects (bubbles), we challenged each of the tested machine learning (ML) (n = 6) and deep learning (DL) (n = 9) models across different background settings. Evaluation revealed that the random forest ML model achieved 95.52% sensitivity, 82.57% specificity, and 97% AUC, outperforming other ML algorithms. Among DL models suitable for mobile integration, DenseNet169 demonstrated superior performance, achieving 92.63% sensitivity, 92.22% specificity, and 92% AUC. Remarkably, DenseNet169 integration into a mobile POC system demonstrated exceptional accuracy (>0.84) in testing microfluidics at under challenging imaging settings. Our study confirms the transformative potential of AI in healthcare, emphasizing its capacity to revolutionize precision medicine through accurate and accessible diagnostics. The integration of AI into healthcare systems holds promise for enhancing patient outcomes and streamlining healthcare delivery.
Tribute to George WhitesidesI joined George at a challenging time in my scientific career, searching for guidance and inspiration. George became that mentor, and working in his lab was both life-saving and life-changing for me. His approach to science—always thinking from unexpected angles—left a lasting impression. What stood out most was not just his unique perspective but the system he developed to manage both the lab and the science, which was unlike anything I had encountered before.I never left a meeting with George without feeling more inspired. We shared many enriching discussions, particularly around innovation and technology. His probing questions pushed me to think more deeply, especially at the intersections of biology and engineering. Most significantly, George took the time to personally guide and support my career development—an experience his administrative staff often described as exceptional. This personal investment, combined with the support of his outstanding lab management team, made my time in his lab one of the most inspiring periods of my career. Mohamed Draz |
Introduction
The convergence of artificial intelligence (AI) and healthcare has opened up a new era of possibilities, particularly in detection diagnostics and treatment. With AI algorithms continuously advancing, the integration of these approaches into healthcare systems holds immense promise for transforming traditional practices and addressing longstanding challenges in healthcare delivery.1–3 Healthcare applications driven by sophisticated machine learning (ML) and deep learning (DL) algorithms stand at the forefront of modern healthcare innovation.4–6 These algorithms empower machines to obtain insights from vast datasets, predict clinical outcomes, and assist healthcare providers in making informed decisions.6 From medical imaging analysis to personalized treatment strategies, AI-driven approaches have demonstrated significant efficacy in improving diagnostic precision and ultimately enhancing patient outcomes.7–10POC diagnostics represent a cornerstone of modern healthcare, offering timely and accessible testing solutions, particularly in resource-limited settings.11–13 The integration of AI into microfluidic systems presents a promising avenue for enhancing the accessibility and efficiency of POC testing.14,15 By harnessing advanced ML and DL algorithms, AI enhances the sensitivity, specificity, and multiplexing capabilities of microfluidic devices, enabling rapid and accurate detection of a wide range of diseases and biomarkers directly at the POC.16–18 An important approach where AI is utilized to enhance microfluidic systems is in image processing. ML and DL learning models excel at image classification and pattern recognition tasks and can support microfluidic devices to perform rapid and multiplex assays, allowing for comprehensive screening or testing using minimal resources.19–21 This integration addresses critical gaps in healthcare access and empowers a new level of POC diagnostics, equipping frontline providers with actionable insights and revolutionizing the delivery of healthcare services.
Recent advancements have demonstrated superior performance in identifying disease biomarkers, detecting cancer,22 viruses,23 bacteria,24 and other pathogens,25 underscoring the robustness and clinical relevance of AI-integrated microfluidic platforms in modern healthcare settings. However, despite these advancements, there remains a gap in the comparative evaluations of different AI algorithms in testing microfluidics, and the optimal approach for maximizing their performance in this context remains unclear, particularly in the POC diagnostics.26–31 In POC settings, practical constraints such as cost, power consumption, memory limitations, and computational efficiency are crucial, making the choice of algorithm highly impactful. For instance, logistic regression is relatively simple, with a complexity of O(n × m), where n is the number of samples and m the number of features. It requires moderate computational power and memory, making it a good fit for POC settings that have limited central processing unit (CPU) power and memory.32 Decision trees, with complexity O(n × m × log(n)),33 and random forests, which add an additional factor for the number of trees (O(k × n × m × log(n)),34 where k is the number of trees), require moderate resources. They build tree structures that evaluate multiple features at once. While computationally more demanding than logistic regression, they can still be feasible in many POC setups, especially with fewer trees. Naive Bayes classifiers are computationally efficient due to their independence assumption for features, with complexity O(n × m). This makes them ideal in resource-limited environments. However, this simplification can sometimes reduce predictive performance if feature independence is not a valid assumption.35 On the other hand, support vector machines (SVMs), especially with non-linear kernels, can have significantly higher complexities (O(n2) to O(n3)), making them less suitable for constrained environments without powerful CPUs or graphics processing units (GPUs). However, using linear kernels or approximation methods (e.g., linear SVM or fast SVM) can reduce the computational load, making SVMs a more viable option for POC.36K-Nearest neighbors (K-NN), while simple in terms of training complexity (O(n × m)), can become computationally intensive during inference due to distance calculations between all data points. Optimization techniques like KD-trees (K-dimensional trees) or Ball-trees can speed up inference, making K-NN more feasible for real-time POC applications.37 Neural networks and deep learning models (e.g., convolutional neural networks (CNNs)) typically have a higher complexity of O(n × m × d), where d is the depth of the network. These models require substantial memory and processing power, particularly using GPU/TPU resources (where TPU stands for tensor processing units), which are not commonly available in POC devices. However, methods like dropout, batch normalization, weight pruning, and model distillation can help reduce the computational burden, allowing for more lightweight versions of these models to be deployed on smaller devices.38 Foundation models, like large-scale AI models (e.g., generative pre-trained transformers (GPT), bidirectional encoder representations from transformers (BERT)), present an even bigger challenge due to their high computational demands during both training and inference. These models often require substantial GPU clusters or high-performance computing (HPC) environments, making them impractical for resource-constrained POC settings. In such cases, pre-trained models fine-tuned for specific tasks or more compact versions of these models (e.g., TinyBERT, DistilBERT) might be used instead.39 This trade-off between computational demands and resource availability emphasizes the importance of balancing model performance with resource constraints in POC settings.
We employed a model microfluidic system, featuring a single microfluidic channel loaded with 3D transparent objects of bubbles. This model is designed to rigorously challenge the performance of commonly used AI models and provide insights into their effectiveness in real-world diagnostic scenarios. We integrated various ML and DL algorithms into our study, including CNNs like MobileNetV2, ResNet101V2, and DenseNet169, alongside commonly used ML models in healthcare applications such as Naive Bayes, logistic regression, KNN, SVM, and random forest.40–44 Among the six evaluated ML algorithms, the random forest model performed best, achieving 95.52% sensitivity, 82.57% specificity, and 97% AUC. Similarly, among the nine DL models, DenseNet169 stood out, achieving 92.63% sensitivity, 92.22% specificity, and 92% AUC. Such a comparative study is critical in gaining a comprehensive understanding of the strengths and weaknesses of different algorithms, informing algorithm selection, optimization, and deployment decisions across diverse domains and applications.45–48
Results and discussion
The integration of AI in medicine is driven by its remarkable ability to analyze and classify images and datasets. This computational capability of AI algorithms is foundational across diverse domains, prominently within diagnostics and medical testing, where AI-driven image analysis stands as a transformative force, providing rapid data processing and precise assessment devoid of infrastructure constraints or specialized human oversight.3,49,50 This technological paradigm bears profound implications, particularly on POC diagnostics, through its role in facilitating the integration of microfluidics into POC applications.51 By harnessing sophisticated ML and DL algorithms, AI streamlines the imaging and analysis of microfluidic devices, such as smartphone-captured assays, reducing the total testing cost and time, enhancing accuracy, and expanding utility.19,52,53 This convergence of AI and microfluidics within POC holds immense potential to democratize healthcare access, particularly in underserved regions, by providing affordable, accurate, and accessible diagnostic solutions.14,19,54,55In our study, we investigated the efficacy of AI algorithms, including both ML and DL, to facilitate the process of testing microfluidics within POC settings. We employed a microfluidic system comprising a single microfluidic channel to rigorously assess a set of 15 AI models recognized for data analysis and image classification across biomedical and diagnostic domains. Our experimental setup incorporated testing configurations featuring varying densities of bubbles. Bubbles as a readout was selected to probe the imaging and analytical performance of the examined algorithms. Despite bubbles being less prevalent than conventional color-based or fluorescence-based readouts, their inherent 3D transparency poses challenges, as they may be mistaken for non-targeted constituents within the sample matrix, microfluidic system or the testing environment and background. In addition, transparent bubbles can introduce challenges such as refraction and variable light scattering, which may impact imaging accuracy and algorithm performance. By using these bubbles, we aimed to simulate complex real-world imaging conditions and evaluate how well the AI models could handle such complexities. Colorimetric readouts, though linear and would allow comparatively easier workflow, fail to sufficiently encapsulate the intricacies necessary for discerning strengths and weaknesses of the tested algorithms. Meanwhile, fluorescence, although known to support high specificity and sensitivity testing, remains impractical for widespread POC adoption due to the need for bulky equipment and specialized setup to achieve the required sensitivity and specificity in most analyses.
Our set of AI algorithms included ML models, such as Naive Bayes, logistic regression, k-nearest neighbors (KNN), support vector machines (SVM), and random forest, alongside DL CNNs such as MobileNetV2, ResNet101V2, and DenseNet169. By combining traditional ML algorithms with state-of-the-art CNN architectures, we created a diverse ensemble of models that can collectively leverage different aspects of the data. This ensemble approach is essential to enhance robustness and generalization performance, particularly in scenarios where the dataset may be limited or the target features are challenging to discern (i.e., bubbles). The incorporation of traditional ML algorithms stemmed from their robustness in handling various types of features, including those extracted from images, and their suitability for the often constrained datasets characteristic of microfluidic diagnostics at POC settings. The CNN architectures like MobileNetV2, ResNet101V2, and DenseNet169 have unparalleled ability to capture intricate spatial relationships within images, which is crucial for discerning subtle patterns like challenging signals such as bubbles. This aligns with the evolving field of diagnostics, which is moving towards inventing and incorporating more versatile readouts like bubbles to allow for more sensitive and unique detection capabilities, distinct from common ones like color and fluorescence. These CNN architectures offer distinct trade-offs in terms of model size, computational efficiency, and classification accuracy, offering flexibility in addressing the specific nuances of the dataset.
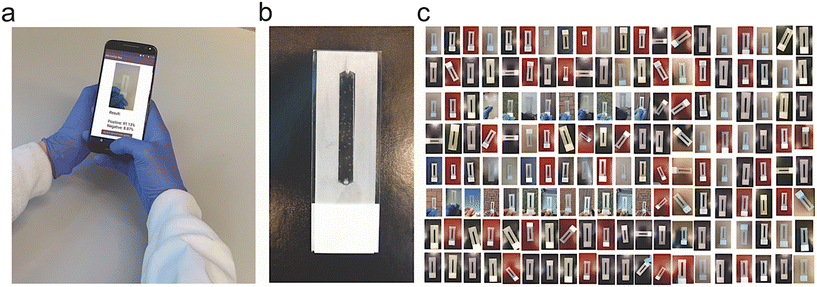
To investigate the capabilities of the selected set of ML and DL algorithms in testing microfluidics, we captured 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 097 images of our microfluidic model with bubbles in various settings, including different environments, lighting conditions, times of the day, and backgrounds (Fig. 1). We labeled the captured images either positive or negative, based on the number of bubbles, around a threshold value of 10 bubbles per microchip, to train our ML and DL models (Fig. 1a). Out of the 19
097 images of our microfluidic model with bubbles in various settings, including different environments, lighting conditions, times of the day, and backgrounds (Fig. 1). We labeled the captured images either positive or negative, based on the number of bubbles, around a threshold value of 10 bubbles per microchip, to train our ML and DL models (Fig. 1a). Out of the 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 097 labelled images (Fig. 1b), 15
097 labelled images (Fig. 1b), 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 530 images were utilized for training using Python running on Lambda Vector GPU Workstation (Intel i9-10900x CPU, NVIDIA RTX A6000 GPU) system.
530 images were utilized for training using Python running on Lambda Vector GPU Workstation (Intel i9-10900x CPU, NVIDIA RTX A6000 GPU) system.
To test the performance of ML models, we used 1595 randomly selected images, excluding those used for training, to evaluate their classification accuracy. We employed standard performance metrics, including accuracy, precision, recall (i.e., sensitivity), specificity, F1 score, and Matthews's correlation coefficient (MCC) (Table S1†), obtained from each model to determine their effectiveness.56 We conducted all statistical analyses and data visualizations using TensorFlow and TensorBoard tools with necessary Python libraries as matplotlib, NumPy, Keras, Sklearn, pandas, torch.57,58 The comparison primarily centered around specificity and sensitivity values, which are metrics influencing overall performance and gives information about other metrics.
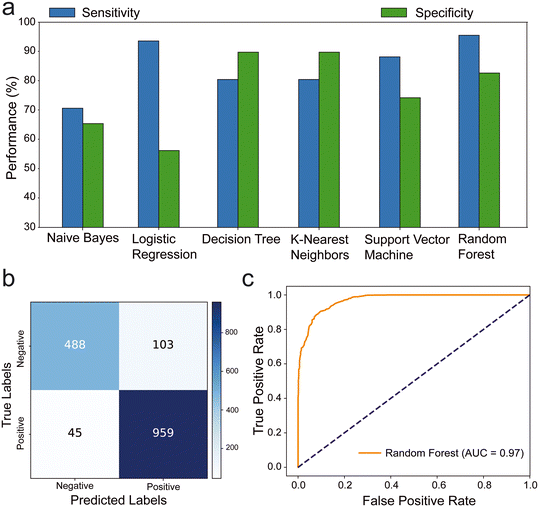
Our analysis of the ML models revealed that logistic regression and random forest models exhibited exceptional sensitivity (>90%), while K-nearest neighbors and random forest models demonstrated high specificity (>80%) (Fig. 2a). The results showed that the highest sensitivity value was obtained from the random forests (95.52%) and the highest specificity value was obtained from K-nearest neighbors (89.68%) ML models. we assessed the confusion matrix to better understand the positive and negative predictions. Out of 1595 images, 1447 were classified correctly, with 45 false negatives and 103 false positives. The model primarily made errors in the classification of negative samples (Fig. 2b and S1†). The ROC analysis of the trained models indicated that the random forest (AUC: 97%) (Fig. 2c) and K-nearest neighbors (AUC: 90%) have highest area under the ROC, which represents the diagnostic ability of the model (Fig. S2†). Additionally, the random forest model outperformed others in terms of F1 score (92.8%) and accuracy (90.72%). This shows that the random forest provides most balanced results between precision and sensitivity with highest accuracy. Consequently, the most effective model was observed as random forest with notable metrics as 95.52% sensitivity, 82.57% specificity, 90.72% accuracy, 90.3% precision, 92.8% F1 score, 79.95% MCC, and 97% AUC (Table S1†).
To test the performance of DL models, we continued by evaluating the performance of the selected CNNs architectures using the same dataset of 1595 images. The performance evaluation step was conducted using developed Python algorithms with the help of Pandas, NumPy, Sklearn, Matplotlib, Keras and Tensorflow libraries.57 The deep learning models utilized for this evaluation included MobileNetV2, EfficientNetV2B0, EfficientNetV2B2, DenseNet169, DenseNet201, InceptionV3, ResNet50V2, EfficientNetB5, and ResNet101V2. In selecting these deep learning models, we prioritized those that does not require significant computing power and thus ensure compatibility for evaluation and testing microfluidics at POC. We also ensured that the chosen models were commonly employed for computer vision tasks, prioritizing ease of integration and robust performance on POC compatible mobile devices.19
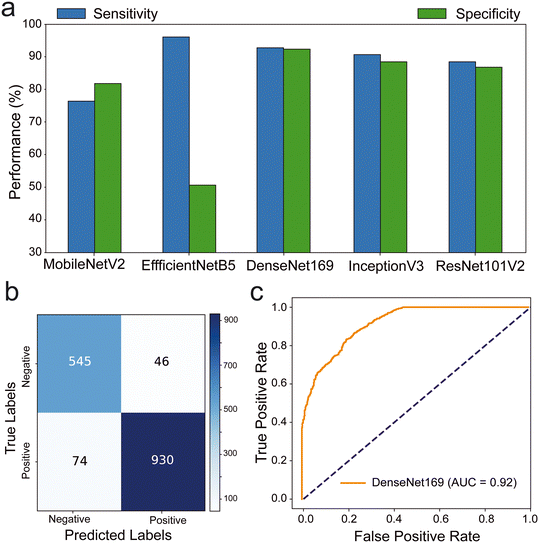
Our results indicated that DenseNet169, EfficientNetB5, and EfficientNetV2B0 exhibited outstanding sensitivity values of 92.63%, 95.82%, and 91.93%, respectively (Fig. 3a and S3–S5†). ResNet50V2 (89.17%) and InceptionV3 (88.49%) demonstrated high specificity values, while DenseNet169 displayed an exceptional specificity of 92.22% (Table S2†). The confusion matrix revealed further insights into the performance of these algorithms. DenseNet169 algorithm excelled in detecting negative samples, accurately classifying 545 out of 591, while also achieving the second-highest performance in positive classification with 930 out of 1004, resulting in the highest overall performance at 92% (Fig. 3b). Other algorithms including EfficientNetB5 correctly identified 962 out of the tested 1004 positive samples. However, it misclassified 293 negative samples as positive, resulting in a 50.4% performance rate for negative samples and an overall performance rate of 79%. EfficientNetV2B0 exhibited similar performance, albeit with a 7% overall performance rate downgrade, reflecting a 4% difference in true positive performance rate and an 11% decrease in true negative performance rate. The results of MobileNetV2, EfficientNetV2B2, DenseNet201, InceptionV3, ResNet50V2, and ResNet101V2 algorithms are shown in Fig. S4 and S5† with misclassification rates <38%. The ROC analysis of the trained DL models, ResNet50V2 (AUC: 96%), ResNet101V2 (AUC: 96%), InceptionV3 (AUC: 95%) and DenseNet169 (AUC: 92%) and DenseNet201 (AUC: 90%) had the highest area under the ROC (Fig. S6 and S7†). Additionally, the DenseNet169 model outperformed other models in terms of F1 score (93.94%) and accuracy (92.48%) (Table S2†). Overall, DenseNet169 outperformed other models with the performance metrics and gives the applicable model with 0.92 AUC (Fig. 3c).
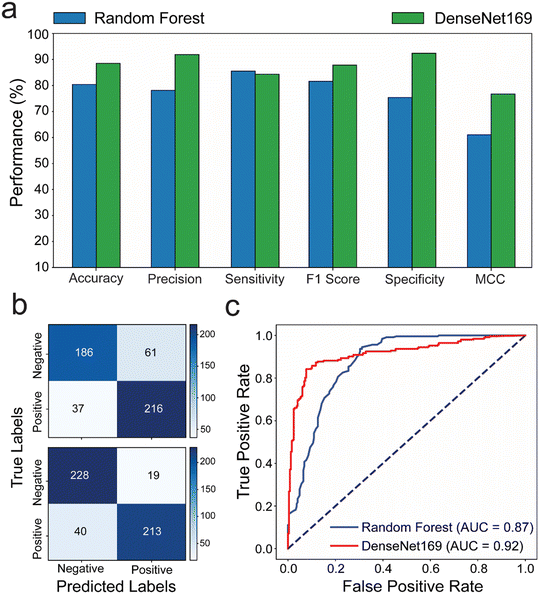
We compared the performance of random forest and DenseNet169, as these models had outperformed others in our evaluations. To challenge them further, we used a set of 184 microchips prepared with varying numbers of bubbles. A new test set of images was created under different environmental conditions than those used during training. This test set included images taken against different backgrounds (including black, red, brown, metallic grey, and dark blue), rotation, and brightness. This approach allowed us to assess user experience in suboptimal conditions, ensuring a thorough and comprehensive evaluation of the models' performance in real-world microchip testing scenarios. The generated positive and negative prediction rates were analyzed against the ground truth values of bubbles per chip to evaluate the performance of each model. The results revealed that the DenseNet169 DL model achieves prediction rates with better performance compared to the random forest ML model with 80.4% and 88.2% accuracy; 77.98% and 91.81% precision; 81.51% and 87.84% F1 score; 75.3% and 92.31% specificity; and 61.03% and 76.69% MCC for random forest and DenseNet169, respectively. The confusion matrix and ROC analyses, on the other hand, confirmed that the DenseNet169 DL algorithm is the optimal prediction model for testing our microfluidic model, outperforming the random forest ML algorithm by 87% in AUC and 92% in accuracy classifying true positive and true negative (Fig. 4b and c).
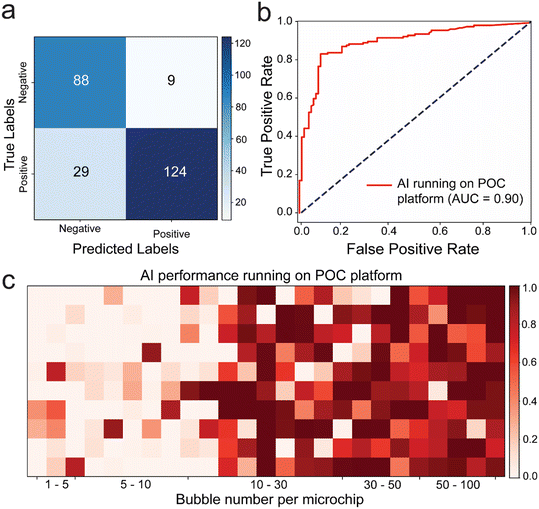
To demonstrate the effectiveness of incorporating AI in real-world sample testing scenarios using POC-compatible systems, a mobile application capable of running the DenseNet169 model seamlessly was developed, without the need for further optimization. The application features a simple interface for initiating model evaluation and presents results in terms of positive and negative prediction rates, along with images of the tested microfluidic chips (Fig. S8†). Out of 250 images, 212 were classified correctly, 29 were classified as false negatives, and 9 were classified as false positives. The model primarily made errors in classifying positive samples. The performance metrics were as follows: accuracy: 84.8%, precision: 93.23%, sensitivity/recall: 81.05%, F1 score: 86.71%, specificity: 90.72%, and MCC: 70.09. The deep learning model achieved an AUC value of 0.90, highlighting its superiority in testing our microfluidic model with bubbles (Fig. 5b). Furthermore, upon examining the confusion matrix alongside sensitivity and specificity values. Results showed that the DenseNet169 deep learning model achieved 81.05% sensitivity and 90.72% specificity (Fig. 5a). Heatmap analysis was conducted using images with bubble counts ranging from 0 to 100. The results indicated a higher margin of error around the threshold of 10 bubbles, particularly chips with around 20 to 30 bubbles are ∼30% misclassified as negative.
Our study provides a comprehensive evaluation of both ML and deep learning DL algorithms in the context of microfluidics testing under POC settings. Among the ML models, random forest emerged as the top performer with a sensitivity of 95.52%, specificity of 82.57%, and an AUC of 97%, showcasing its strong capability in accurately classifying microfluidic device images. The high sensitivity and specificity values underscore random forest's effectiveness in distinguishing positive from negative samples even in challenging imaging conditions. However, the higher rate of false positives indicates a potential area for improvement. In contrast, DL models, particularly DenseNet169, exhibited outstanding performance with sensitivity and specificity values of 92.63% and 92.22%, respectively. DenseNet169's consistent high performance across different testing conditions, including variations in background and lighting, highlights its robustness and adaptability, making it highly suitable for real-world POC diagnostics where consistent and reliable performance is crucial.
Despite the promising results, several challenges must be addressed to facilitate the widespread adoption of AI in microfluidic POC diagnostics. One key issue is the misclassification of samples with a marginal number of bubbles, especially around the threshold of 10 bubbles, which was evident in the heatmap analysis. Further refinement of the AI models and incorporating additional features or training data will be necessary to enhance accuracy in borderline cases. Combining multiple algorithms can also help overcome these challenges. For example, employing ensemble techniques that integrate models like U-Net for image segmentation and Canny edge detection for edge detection could improve precision in detecting subtle features. Additionally, integrating algorithms such as YOLO (You Only Look Once) for real-time object detection and HOG (histogram of oriented gradients) for robust feature extraction can further enhance the accuracy and reliability of microfluidic POC diagnostics. Such hybrid approaches can leverage the strengths of different algorithms, providing a more comprehensive and accurate analysis.
Moreover, integrating AI models into mobile applications for POC testing will necessitate ensuring seamless operation across a wide range of devices and environmental conditions, with a strong emphasis on user-friendliness and reliability. This integration is pivotal for achieving the robustness required for practical deployment in diverse healthcare settings. The successful implementation of AI in microfluidic POC diagnostics has far-reaching implications for the healthcare industry, especially in resource-limited settings where access to sophisticated medical infrastructure is often constrained. By enabling rapid, accurate, and on-site testing, AI-driven POC systems address one of the most pressing challenges in modern medicine: the need for timely and precise diagnostics. By democratizing access to high-quality diagnostic tools, AI-integrated POC systems empower frontline healthcare providers with actionable insights, fostering a more equitable distribution of medical resources. This shift supports personalized medicine approaches, tailoring treatment plans to individual patient profiles based on accurate and immediate diagnostic data. Ultimately, the widespread adoption of AI-enhanced microfluidic POC diagnostics can transform healthcare delivery, making it more accessible, efficient, and responsive to the needs of diverse populations worldwide.
Conclusion
The transformative impact of AI on healthcare is rapidly increasing, particularly in advancing precision medicine through accurate and accessible diagnostics. By conducting a comprehensive comparative evaluation of AI models in testing microfluidics, we have demonstrated the superiority of AI-driven approaches over traditional methods, particularly in the context of POC diagnostics. Through the integration of ML and DL algorithms, we created a diverse ensemble of models capable of leveraging various aspects of the data, thereby enhancing robustness and generalization performance. Our results revealed that the random forest ML model and the DenseNet169 DL model exhibited exceptional performance, surpassing other algorithms in terms of sensitivity, specificity, and AUC values. DenseNet169 integration into a mobile POC system demonstrated exceptional accuracy, outperforming traditional visual interpretation by a significant margin. This confirms the potential of AI to revolutionize diagnostics, offering more accurate and efficient testing solutions in resource-limited settings. Moreover, our findings highlight the significant role that AI can play into healthcare systems, as it holds promise for enhancing patient outcomes, streamlining healthcare delivery, and ultimately, democratizing access to high-quality diagnostic services. Moving forward, further research and development efforts are warranted to optimize AI algorithms for real-world deployment, ensuring their seamless integration into clinical practice and maximizing their impact on global health outcomes.Material and methods
Microfluidic chip model design and fabrication
We developed a microfluid chip system that features a single microfluidic channel. The microchip was designed using the vector graphics editor CorelDRAW Graphics suite software, and fabricated of polymethyl methacrylate (PMMA) (3.125 mm thick), DSA film (100 μm thick, 3 M, USA), and glass slides (25 mm × 75 mm). The fabrication process starts by cutting PMMA and DSA film using a laser cutter (Boss Laser LS-1416, USA). The PMMA was prepared to contain the microfluidic channel inlet and outlet, while DSA film included the main testing channel. All materials were precleaned with 70% ethanol, and deionized water using lint-free tissue. The surface of the cleaned glass slides was treated and cleaned using oxygen plasma (PE-25, 100 mW, 15% oxygen; Plasma Etch Inc.) for 10 minutes. Then PMMA and DSA film were assembled on the modified glass slide, forming the model microfluidic chip system. Each system was loaded with platinum nanoparticle-seeded bubbles. PtNPs synthesized using our previously published protocol were mixed with a peroxide-containing solution (5% hydrogen peroxide and 20% glycerol) and loaded on chip system. The concentration of added PtNPs was controlled to prepare systems with variable numbers of bubbles (0–>200 bubbles per chip), randomly distributed within the microfluidic channel.AI models selection, training and performance testing
We selected a set of 15 models that encompass a number of machine learning and deep learning models, widely reported to have high performance in image classification and pattern recognition. The machine learning models included Naive Bayes, logistic regression, decision tree, K-nearest neighbors, support vector machine and random forest, while the deep learning models of MobileNetV2, EfficientNetV2B0, EfficientNetV2B2, DenseNet169, DenseNet201, InceptionV3, ResNet50V2, EfficientNetB5 and ResNet101V2, were selected to support workflow running on mobile devices and systems. We generated a dataset of 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 097 images of the model microfluidic system captured using Moto XT1575, iPhone X and Vivo smartphones. The dataset comprises two groups, i.e., positive (with >10 bubbles per microchip) and negative (in range of <10 bubbles per microchip) sample images. The microfluidic system imaging was performed at different angles (0–360°) and backgrounds and environments to maximize the variations, and make our dataset more robust and comprehensive. We used 15
097 images of the model microfluidic system captured using Moto XT1575, iPhone X and Vivo smartphones. The dataset comprises two groups, i.e., positive (with >10 bubbles per microchip) and negative (in range of <10 bubbles per microchip) sample images. The microfluidic system imaging was performed at different angles (0–360°) and backgrounds and environments to maximize the variations, and make our dataset more robust and comprehensive. We used 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 530 images for training, 1788 images for validation and 1012 images for testing the performance of the selected ML and DL models in testing the model microfluidic system and classifying samples into positive and negative based on bubble signal. We started the process by importing pre-trained models available from Scikit-learn and Keras libraries to develop the selected ML and DL models, respectively. In the pre-processing step, the images of our training dataset were resized to the input dimensions of the selected models, leveraging the features learned by ImageNet pretrained network. We performed the batch normalization then used Adam optimizer to fine-tune the network using a global learning rate of 0.001. In addition, we employed a varied number of epochs to test the algorithms optimal performance and we set the number to 50 epochs. Then we performed the transfer learning by removing the final classification layer from the chosen networks and trained with our dataset. All the algorithms were trained on Vector Workstation (Intel i9-10900x CPU and NVIDIA RTX A6000 GPU, Lambda) and after training, we tested the performance of the best-performing ML and DL algorithms individually using a challenging dataset of 400 images. This testing dataset included rotated images, images with various colored backgrounds (matte, bright, reflective), and images with lens distortion and brightness variations. The ML algorithms were evaluated using the sklearn and torch libraries, while the DL algorithm was evaluated using the TensorFlow library. Performance metrics such as accuracy, precision, sensitivity, and F1-score were employed to quantitatively measure classification accuracy and the ability of each model to correctly identify the tested microchip.
530 images for training, 1788 images for validation and 1012 images for testing the performance of the selected ML and DL models in testing the model microfluidic system and classifying samples into positive and negative based on bubble signal. We started the process by importing pre-trained models available from Scikit-learn and Keras libraries to develop the selected ML and DL models, respectively. In the pre-processing step, the images of our training dataset were resized to the input dimensions of the selected models, leveraging the features learned by ImageNet pretrained network. We performed the batch normalization then used Adam optimizer to fine-tune the network using a global learning rate of 0.001. In addition, we employed a varied number of epochs to test the algorithms optimal performance and we set the number to 50 epochs. Then we performed the transfer learning by removing the final classification layer from the chosen networks and trained with our dataset. All the algorithms were trained on Vector Workstation (Intel i9-10900x CPU and NVIDIA RTX A6000 GPU, Lambda) and after training, we tested the performance of the best-performing ML and DL algorithms individually using a challenging dataset of 400 images. This testing dataset included rotated images, images with various colored backgrounds (matte, bright, reflective), and images with lens distortion and brightness variations. The ML algorithms were evaluated using the sklearn and torch libraries, while the DL algorithm was evaluated using the TensorFlow library. Performance metrics such as accuracy, precision, sensitivity, and F1-score were employed to quantitatively measure classification accuracy and the ability of each model to correctly identify the tested microchip.
AI testing on a POC compatible system
We utilized the open-source platform Android Studio (version Giraffe 2022.3.1) to develop an AI-enabled mobile application. Android Studio offers an integrated development environment (IDE) tailored for Android application development. The application facilitates the capture of sensor images through the smartphone's built-in camera or from images stored in the device's memory. A trained DL model, DenseNet169, was converted to TensorFlow Lite and integrated into the application, which was developed for Android 6.0 (API level 23). This application was installed on a Moto XT1575 and used as a proof-of-concept system for testing microfluidics with images simulating real-world conditions. We evaluated the performance of the AI model using a testing set of 250 images, each featuring 0–100 bubbles per chip. This testing set included images with challenging backgrounds and imaging conditions, such as noise, blur, hand interaction, daylight, artificial light, natural and artificial occlusion, resolution variability, and the presence of small bubbles. The classification results, displayed on the user interface, indicate the probability of a sample being positive (>50%) or negative (<50%). The correlation between AI-generated classification results and the number of bubbles per chip was analyzed, and prediction accuracy rates were employed to generate performance metrics.Data availability
The authors confirm that all data supporting the findings of this study are included within the article and its ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
Research reported in this publication was partially supported by the National Institutes of Health under award numbers U54CS254566 and P30DA054557. The authors would like to thank Dr. Steven J. Eppell, Dr. Michael Jenkins, and Dr. James Basilion for their valuable discussions, as well as Dr. Cyril R. A. John Chelliah, Ethan Roman, Ebenesh Chandrakumar, Dheeksha Devaraj, and Aravinth Kumar for their assistance with the initial trials. We are also grateful to Dr. Robert Bonomo for his insightful feedback and edits to the final manuscript.References
- P. Rajpurkar, E. Chen, O. Banerjee and E. J. Topol, Nat. Med., 2022, 28, 31–38 CrossRef CAS PubMed.
- A. Esteva, A. Robicquet, B. Ramsundar, V. Kuleshov, M. DePristo, K. Chou, C. Cui, G. Corrado, S. Thrun and J. Dean, Nat. Med., 2019, 25, 24–29 CrossRef CAS PubMed.
- E. J. Topol, Nat. Med., 2019, 25, 44–56 CrossRef CAS PubMed.
- J. N. Acosta, G. J. Falcone, P. Rajpurkar and E. J. Topol, Nat. Med., 2022, 28, 1773–1784 CrossRef CAS PubMed.
- A. Hosny, C. Parmar, J. Quackenbush, L. H. Schwartz and H. J. W. L. Aerts, Nat. Rev. Cancer, 2018, 18, 500–510 CrossRef CAS PubMed.
- D. S. Kermany, M. Goldbaum, W. Cai, C. C. Valentim, H. Liang, S. L. Baxter, A. McKeown, G. Yang, X. Wu and F. Yan, Cell, 2018, 172, 1122–1131.e1129 CrossRef CAS PubMed.
- R. Aggarwal, V. Sounderajah, G. Martin, D. S. W. Ting, A. Karthikesalingam, D. King, H. Ashrafian and A. Darzi, NPJ Digit. Med., 2021, 4, 65 CrossRef PubMed.
- P. Lambin, R. T. H. Leijenaar, T. M. Deist, J. Peerlings, E. E. C. de Jong, J. van Timmeren, S. Sanduleanu, R. T. H. M. Larue, A. J. G. Even, A. Jochems, Y. van Wijk, H. Woodruff, J. van Soest, T. Lustberg, E. Roelofs, W. van Elmpt, A. Dekker, F. M. Mottaghy, J. E. Wildberger and S. Walsh, Nat. Rev. Clin. Oncol., 2017, 14, 749–762 CrossRef PubMed.
- M. Sermesant, H. Delingette, H. Cochet, P. Jaïs and N. Ayache, Nat. Rev. Cardiol., 2021, 18, 600–609 CrossRef PubMed.
- O. Oren, B. J. Gersh and D. L. Bhatt, Lancet Digital Health, 2020, 2, e486–e488 CrossRef PubMed.
- P. Yager, G. J. Domingo and J. Gerdes, Annu. Rev. Biomed. Eng., 2008, 10, 107–144 CrossRef CAS PubMed.
- C. P. Y. Chan, W. C. Mak, K. Y. Cheung, K. K. Sin, C. M. Yu, T. H. Rainer and R. Renneberg, Annu. Rev. Anal. Chem., 2013, 6, 191–211 CrossRef CAS.
- C. Wang, M. Liu, Z. Wang, S. Li, Y. Deng and N. He, Nano Today, 2021, 37, 101092 CrossRef CAS.
- J. Riordon, D. Sovilj, S. Sanner, D. Sinton and E. W. Young, Trends Biotechnol., 2019, 37, 310–324 CrossRef CAS PubMed.
- S. Chen, Z. Qiao, Y. Niu, J. C. Yeo, Y. Liu, J. Qi, S. Fan, X. Liu, J. Y. Lee and C. T. Lim, Nat. Rev. Bioeng., 2023, 1, 950–971 CrossRef.
- J. Zhou, J. Dong, H. Hou, L. Huang and J. Li, Lab Chip, 2024, 24(5), 1307–1326 RSC.
- W. Zhao, Y. Zhou, Y.-Z. Feng, X. Niu, Y. Zhao, J. Zhao, Y. Dong, M. Tan, Y. Xianyu and Y. Chen, ACS Nano, 2023, 17, 13700–13714 CrossRef CAS PubMed.
- Z. Ao, H. Cai, Z. Wu, L. Hu, A. Nunez, Z. Zhou, H. Liu, M. Bondesson, X. Lu and X. Lu, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2214569119 CrossRef CAS PubMed.
- B. Wang, Y. Li, M. Zhou, Y. Han, M. Zhang, Z. Gao, Z. Liu, P. Chen, W. Du and X. Zhang, Nat. Commun., 2023, 14, 1–18 CAS.
- H. Liu, L. Nan, F. Chen, Y. Zhao and Y. Zhao, Lab Chip, 2023, 23, 2497–2513 RSC.
- J. Zheng, T. Cole, Y. Zhang, J. Kim and S.-Y. Tang, Biosens. Bioelectron., 2021, 194, 113666 CrossRef CAS PubMed.
- W. L. Bi, A. Hosny, M. B. Schabath, M. L. Giger, N. J. Birkbak, A. Mehrtash, T. Allison, O. Arnaout, C. Abbosh and I. F. Dunn, Ca-Cancer J. Clin., 2019, 69, 127–157 CrossRef PubMed.
- N. Arora, A. K. Banerjee and M. L. Narasu, Future Virol., 2020, 15(11), 717–724 CrossRef CAS.
- K. P. Smith, H. Wang, T. J. Durant, B. A. Mathison, S. E. Sharp, J. E. Kirby, S. W. Long and D. D. Rhoads, Clin. Microbiol. Newsl., 2020, 42, 61–70 CrossRef.
- K. P. Smith and J. E. Kirby, Clin. Microbiol. Infect., 2020, 26, 1318–1323 CrossRef CAS.
- A. J. London, Hastings Cent. Rep., 2019, 49, 15–21 CrossRef PubMed.
- C. Gilvary, N. Madhukar, J. Elkhader and O. Elemento, Trends Pharmacol. Sci., 2019, 40, 555–564 CrossRef CAS PubMed.
- J. E. Dayhoff and J. M. DeLeo, Cancer, 2001, 91, 1615–1635 CrossRef CAS.
- O. Koteluk, A. Wartecki, S. Mazurek, I. Kołodziejczak and A. Mackiewicz, J. Pers. Med., 2021, 11, 32 CrossRef PubMed.
- H. Bhaskar, D. C. Hoyle and S. Singh, Comput. Biol. Med., 2006, 36, 1104–1125 CrossRef PubMed.
- A. A. de Hond, A. M. Leeuwenberg, L. Hooft, I. M. Kant, S. W. Nijman, H. J. van Os, J. J. Aardoom, T. P. Debray, E. Schuit and M. van Smeden, NPJ Digit. Med., 2022, 5, 2 CrossRef PubMed.
- M. Sumner, E. Frank and M. Hall, Speeding up logistic model tree induction, in European conference on principles of data mining and knowledge discovery, Springer Berlin Heidelberg, Berlin, Heidelberg, 2005, pp. 675–683 Search PubMed.
- H. M. Sani, C. Lei and D. Neagu, Computational complexity analysis of decision tree algorithms, in Artificial Intelligence XXXV: 38th SGAI International Conference on Artificial Intelligence, AI 2018, Cambridge, UK, December 11–13, 2018, Proceedings, Springer International Publishing, 2018, pp. 191–197 Search PubMed.
- X. Zheng, J. Jia, S. Guo, J. Chen, L. Sun, Y. Xiong and W. Xu, IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens., 2021, 14, 2222–2235 Search PubMed.
- Z. Zheng, Naive Bayesian classifier committees, in European Conference on Machine Learning, Springer Berlin Heidelberg, Berlin, Heidelberg, 1998, pp. 196–207 Search PubMed.
- L. Bottou and C. J. Lin, Support vector machine solvers, 2007 Search PubMed.
- K. Hajebi, Y. Abbasi-Yadkori, H. Shahbazi and H. Zhang, Fast approximate nearest-neighbor search with k-nearest neighbor graph, in Twenty-Second International Joint Conference on Artificial Intelligence, 2011 Search PubMed.
- Y. Zhang, L. Wang, J. Zhao, X. Han, H. Wu, M. Li and M. Deveci, Inf. Sci., 2024, 670, 120644 CrossRef.
- R. Bommasani, D. A. Hudson, E. Adeli, R. Altman, S. Arora, S. von Arx, M. S. Bernstein, J. Bohg, A. Bosselut and E. Brunskill, arXiv, 2021, preprint, arXiv:2108.07258, DOI:10.48550/arXiv.2108.07258.
- R. C. Deo, Circulation, 2015, 132, 1920–1930 CrossRef PubMed.
- A. Alanazi, Informatics in Medicine Unlocked, 2022, 30, 100924 CrossRef.
- M. A. Morid, A. Borjali and G. Del Fiol, Comput. Biol. Med., 2021, 128, 104115 CrossRef PubMed.
- A. W. Salehi, S. Khan, G. Gupta, B. I. Alabduallah, A. Almjally, H. Alsolai, T. Siddiqui and A. Mellit, Sustainability, 2023, 15, 5930 CrossRef.
- M. M. Rahaman, C. Li, Y. Yao, F. Kulwa, M. A. Rahman, Q. Wang, S. Qi, F. Kong, X. Zhu and X. Zhao, J. X-Ray Sci. Technol., 2020, 28, 821–839 CAS.
- F. Zhuang, Z. Qi, K. Duan, D. Xi, Y. Zhu, H. Zhu, H. Xiong and Q. He, Proc. IEEE, 2020, 109, 43–76 Search PubMed.
- S. J. Pan and Q. Yang, IEEE Trans. Knowl. Data Eng., 2009, 22, 1345–1359 Search PubMed.
- J. Ooge, G. Stiglic and K. Verbert, Wiley Interdiscip. Rev.: Data Min. Knowl. Discov., 2022, 12, e1427 Search PubMed.
- K. Moulaei, A. Yadegari, M. Baharestani, S. Farzanbakhsh, B. Sabet and M. R. Afrash, Int. J. Med. Inform., 2024, 105474 CrossRef PubMed.
- X. Tang, BJR|Open, 2019, 2, 20190031 CrossRef PubMed.
- N. Hasani, M. A. Morris, A. Rahmim, R. M. Summers, E. Jones, E. Siegel and B. Saboury, PET Clin., 2022, 17, 1–12 CrossRef PubMed.
- Z. Angehrn, L. Haldna, A. S. Zandvliet, E. Gil Berglund, J. Zeeuw, B. Amzal, S. A. Cheung, T. M. Polasek, M. Pfister and T. Kerbusch, Front. Pharmacol., 2020, 11, 759 CrossRef PubMed.
- Y. Yang, F. Xu, J. Chen, C. Tao, Y. Li, Q. Chen, S. Tang, H. K. Lee and W. Shen, Biosens. Bioelectron., 2023, 115233 CrossRef CAS PubMed.
- D. Xu, X. Huang, J. Guo and X. Ma, Biosens. Bioelectron., 2018, 110, 78–88 CrossRef CAS PubMed.
- D. McIntyre, A. Lashkaripour, P. Fordyce and D. Densmore, Lab Chip, 2022, 22, 2925–2937 RSC.
- I. Hernández-Neuta, F. Neumann, J. Brightmeyer, T. Ba Tis, N. Madaboosi, Q. Wei, A. Ozcan and M. Nilsson, J. Intern. Med., 2019, 285, 19–39 CrossRef PubMed.
- Y. Jiao and P. Du, Quant. Biol., 2016, 4, 320–330 CrossRef.
- I. Stančin and A. Jović, An overview and comparison of free Python libraries for data mining and big data analysis, in 2019 42nd International convention on information and communication technology, electronics and microelectronics (MIPRO), IEEE, 2019, pp. 977–982 Search PubMed.
- B. J. Erickson, P. Korfiatis, Z. Akkus, T. Kline and K. Philbrick, J. Digit. Imaging, 2017, 30, 400–405 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4lc00671b |
| This journal is © The Royal Society of Chemistry 2024 |