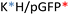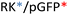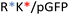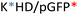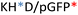Open Access Article
Open Access ArticleUnravelling the role of individual components in pBAE/polynucleotide polyplexes in the synthesis of tailored carriers for specific applications: on the road to rational formulations†
María
Navalón-López
a,
Aurora
Dols-Perez
 b,
Santiago
Grijalvo
b,
Santiago
Grijalvo
 c,
Cristina
Fornaguera
c,
Cristina
Fornaguera
 *a and
Salvador
Borrós
*a and
Salvador
Borrós
 a
a
aGrup d’Enginyeria de Materials (GEMAT) Institut Químic de Sarrià (IQS) Universitat Ramon Llull (URL), Via Augusta 390, 08017 Barcelona, Spain. E-mail: cristina.fornaguera@iqs.url.edu
bInstitut de Bioenginyeria de Catalunya (IBEC), The Barcelona Institute of Science and Technology (BIST), C/Baldiri i Reixac 11‐15, 08028 Barcelona, Spain
cInstitute for Advanced Chemistry of Catalonia (IQAC-CSIC), Networking Center on Bioengineer-ing, Biomaterials and Nanomedicine (CIBER-BBN), C/ Jordi Girona 18-26, 08034 Barcelona, Spain
First published on 6th February 2023
Abstract
Oligopeptide end–modified poly(β-amino ester)s (OM-pBAEs) offer a means for the effective implementation of gene therapeutics in the near future. A fine-tuning of OM-pBAEs to meet application requirements is achieved by the proportional balance of oligopeptides used and provide gene carriers with high transfection efficacy, low toxicity, precise targeting, biocompatibility, and biodegradability. Understanding the influence and conformation of each building block at molecular and biological levels is therefore pivotal for further development and improvement of these gene carriers. Herein, we unmask the role of individual OM-pBAE components and their conformation in OM-pBAE/polynucleotide nanoparticles using a combination of fluorescence resonance energy transfer, enhanced darkfield spectral microscopy, atomic force microscopy, and microscale thermophoresis. We found that modifying the pBAE backbone with three end-terminal amino acids produces unique mechanical and physical properties for each combination. Higher adhesion properties are seen with arginine and lysine-based hybrid nanoparticles, while histidine provides an advantage in terms of construct stability. Our results shed light on the high potential of OM-pBAEs as gene delivery vehicles and provide insights into the influence of the nature of surface charges and the chemical nature of the pBAE modifications on their paths towards endocytosis, endosomal escape, and transfection.
Introduction
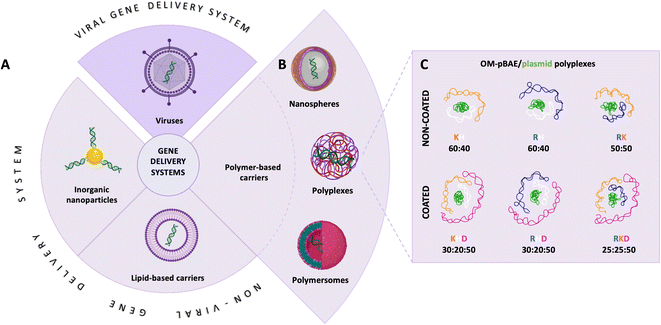
Polynucleotide-based therapies steadily grow into transforming the whole (bio) medicine field as they show tremendous potential for the prevention or treatment of a wide range of infectious diseases or genetic disorders.1 Currently there are more than 2400 clinical trials comprising gene therapies.2,3 Remarkably, the first treatment applied worldwide, SARS-CoV-2 prophylactic vaccines have only given a glimpse of involved intricacies and revealed the untapped potential of polynucleotide-based therapies.4–7 Despite their theoretical full potential, these therapies still find unsatisfactory clinical outcomes, which are mainly related to the low chemical stability and poor bioavailability of polynucleotides in vivo.8,9 Furthermore, the human body inherently protects itself from foreign nucleic acids; thus, they are prematurely cleared up from circulation before they arrive at their target cells.10Overcoming these drawbacks is accomplished by compartmentalizing the polynucleotides into designed nanocarriers (Fig. 1A) programmed to deliver them into the specific tissue and intracellular compartment.11 Genetically engineering viral vectors emerged as a natural carrier for gene therapies with ubiquitous abilities to infect and transport the therapeutic nucleic acids across the cellular barriers.12,13 However, viral vectors are concerned with different aspects, such as safety, immunogenicity, insertional mutagenesis, low loading capacity, and high production costs.14 These drawbacks triggered the development of nanotechnology based non-viral delivery systems (Fig. 1B), which on the one hand are modular to meet the application requirements and on the other hand, their production is cost-effective.15
Among non-viral carriers, poly(β-amino ester)s (pBAE) are an outstanding option as a carrier, due to their high building blocks versatility, high transfection efficacy, low toxicity, and excellent biocompatibility and biodegradation profile.16–18 Upon contact with polynucleotides, pBAEs condense into polymeric nanoparticles, (polyplexes), which protect the fragile cargo and transport it into the targeted tissue.15 Additionally, pBAE building blocks contain hexyl monomers (C6) that provide certain hydrophobicity to the polymer (see structure and characterization in Fig. S1†), which allows the freeze-drying of the polyplexes, a remarkable property for achieving long-term storage and ameliorated logistics of the distribution.15 Further functionalization of the pBAE vectors with short cationic oligopeptides (OM-pBAE), such as lysine (K), arginine (R), and histidine (H) (C6CK3, C6CR3, and C6CH3 respectively, their structures and characterization data are shown in Fig. S1†) are designed not only to increase their buffering capacity and ability to compact nucleic acids due to electrostatic interactions but also to promote cellular internalization and nucleic acid transfection.15,19,20 As we have previously described,11 C6CK3 promotes the plasmatic membrane crossing, C6CH3 assists in the endosomal scape development, and C6CR3 gives an advantage to nuclear subcellular localization. Cationic peptides, however, increase the polyplex-positive surface charge resulting in transfection promiscuity.11 Consequently, these pBAE-based polyplexes have low selectivity. Decreasing the surface charge and thus promiscuity is achieved by coating the polyplexes with aspartic acid (D)-functionalized pBAEs. In this sense, it is clear that a careful combination of different OM-pBAEs is thus crucial for achieving optimal functionality.11,15,21
Indeed, various OM-pBAEs combinations have achieved optimal results in vitro and in vivo.15 Despite these outstanding outcomes, different structures of pBAEs in the polyplexes, and therefore their influence on the final properties, are still unclear. These unknown characteristics involve time-consuming experiments without specific and clear application due to the uncertainty of their target and uptake specificity, which are needful characteristics to achieve medical needs.
Recently, we demonstrated the microstructure of C6CR3/pDNA polyplexes and an oligopeptide dependence of the time evolution of the cationic polyplexes using high-resolution optical microscopy.22,23 In this work, we aimed to understand the microstructure of polyplexes composed of mixtures of OM-pBAE and determine the role of individual components in the final formulation and create a library of OM-pBAE carriers based on a rational selection in the function of the chased properties for a specific application. For this purpose, we studied six polyplex constructions with different compositions: KH, KHD, RH, RHD, RK, and RKD, combining different OM-pBAEs (Fig. 1C). We revealed the microstructure and physicochemical properties of the different polyplexes using a combination of fluorescence resonance energy transfer (FRET), enhanced darkfield spectral microscopy, atomic force microscopy (AFM), and microscale thermophoresis (MST). Our results allow the rational engineering of carriers for any required application and allow for the further development of pBAEs building blocks for the optimal delivery of polynucleotides.
Results and discussion
Different OM-pBAE combinations (structures are shown in Fig. 1C, and structure and characterization in Fig. S1, ESI†) were tested, encapsulating pGFP as a model reporter plasmid. Physicochemical NPs' properties depend on the OM-pBAEs end-terminal peptides.Materials at the nanometric scale have distinct physicochemical characteristics in terms of size, shape, surface properties, composition, and stability, which is crucial in determining their physiological behavior.24 A rigorous approach aimed to characterize nanomaterials is essential to ensure quality and safety and will contribute to the construction of a competent delivery system. With this idea in mind, we studied six different NPs: KH, RH, and RK as cationic polyplexes (uncoated) and KHD, RHD, and RKD referred to as the anionic-complexed ones (Fig. 1C).
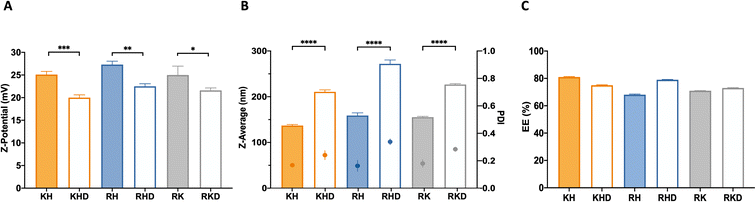
Once the different NPs were freshly synthesized, the first step was to analyze the zeta potential in order to corroborate that the OM-pBAEs with the different terminal peptides shifted the surface potential of the nanoparticle. Results demonstrated that, as expected, the surface charge of cationic combinations was in the 25–27 mV range while the anionic-coating produced a significant 20% surface charge decrease,24,25 confirming the interaction of the two polymers (cationic and anionic) in the polyplex formation (Fig. 2A).
Regarding their hydrodynamic diameters, cationic polyplexes (KH, RH, and RK) showed a hydrodynamic size in a range of 140 and 160 nm, while the anionic-complexed NPs (RHD, KHD, RKD) were slightly bigger (200–250 nm). These differences cannot be explained from the C6CD3 coating. Moreover, due to the similarities in the polymer backbone (see Fig. S1†), no noticeable differences in the hydrodynamic diameters from the group of cationic and that of anionic-coated NPs are expected, as can be seen in the data shown in Fig. 2B. Accordingly, the polydispersity index (PDI) was slightly higher in the case of the negatively-coated NPs, which is coherent with the introduction of an extra component to the formulation, which increases sample heterogeneity. Nevertheless, all formulations have an agreeable dimension and size distribution for their parenteral administration.24 In terms of size, we can tune nanoparticles at our convenience for different applications. For example, coated formulations could be an interesting option for therapies requiring nasal delivery, where a higher size is required for nanoparticle deposition in the mucosa.26 This route, in addition, requires lower cationic surface potentials, making KHD and RHD formulations potential candidates.
The next step was the plasmid encapsulation efficiency (EE) quantification, a key parameter for the efficient production of drug delivery systems (DDS). The measured %EE of the fresh NPs was over 65% for all NP formulations (Fig. 2C). It is remarkable that for both groups (coated and naked), the NP combinations that have smaller sizes present slightly higher encapsulation ratios. As explained above, this can be attributed to the stronger interactions between the cationic polymers and the plasmid, allowing a higher plasmid packing capacity. Again, we could use this idea for optimizing the encapsulation of different nucleic acids. For example, if larger nucleic acids need to be encapsulated, KH may be the best option, for example, for vaccination purposes, where long sequences are needed to deliver antigen mRNAs.
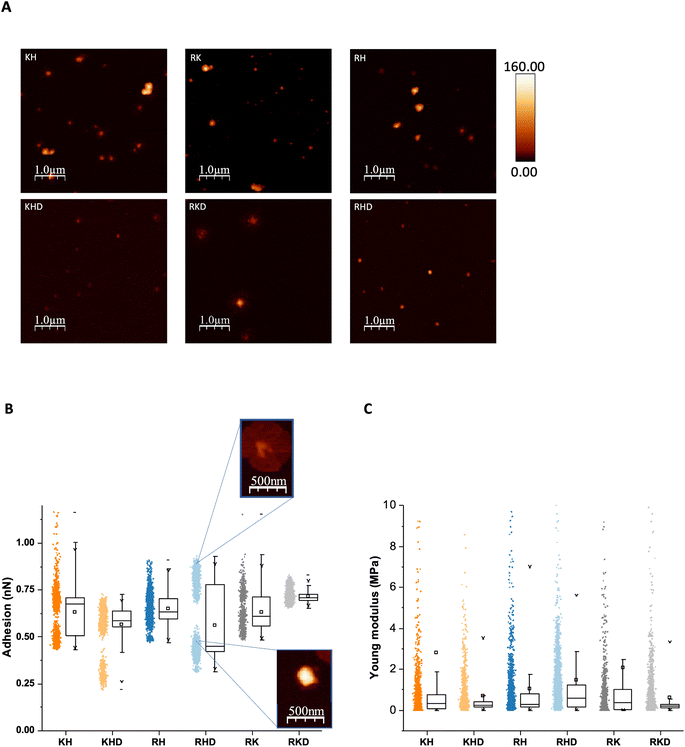
Nanomechanical/adhesion properties that are usually forgotten can affect transport pathways, cell targeting, and degradation.27 Thus, their characterization is important. Atomic force microscopy (AFM) experiments were performed to visualize the nanoparticle complexation state, shape, complex organization, and adhesion properties.
First, we collected topography images (Fig. 3A), confirming the spherical shape, as expected, due to their lowest energy of formation,28 for all NP formulations.
Adhesion values for uncoated and coated NPs are presented in Fig. 3B. The polyplexes containing C6CR3 polymer (RH and RHD, and RK and RKD) presented higher adhesion values, as expected, due to the highest cationic peptide that arginine represents. At the same time, histidine-containing polymers (KH and KHD, and RH and RHD), follow a pattern suggesting two differentiated populations with NP. We hypothesized that in some fraction of the polyplexes, the C6CH3 polymer was slightly disassembled,23 decreasing the adhesion values, due to the polymer loss. This trend was also observed in the stability, as discussed in the section below. Comparing the cationic polyplexes with anionic-coated ones, it can be observed that cationic constructions show a clear major population around 0.6 nN, and a smaller one around or below 0.5 nN, confirming our hypothesis of the C6CH3 loss, as described above. The RHD formulation is the only one that presents 3 populations. One could be attributed to the conserved polyplex (containing C6CH3). The second to the C6CH3 disassembled polyplex, previously described for the cationic constructions but with lower adhesion values, below 0.5 nN, as explained by the anionic coating. The third one, with values above 0.75 nN, suggests a loss of the negative coating, which leads to a higher adhesion value (Fig. 2B). It is worth noticing that some RHD polyplexes eventually showed a flower-like structure (Fig. 3B). These can be assigned to the third population that loses the anionic coating. In this sense, KHD ultimately showed some flower-like structure also, but in less quantity. It can also be seen that, as expected, RKD is the most compacted construct. As commented before, R and K are strongly cationic peptides, having high affinity to the anionic C6CD3 polymer. Additionally, NPs size was measured and also corroborated with the DLS measurements, as described previously (ESI Fig. S2†).
Furthermore, Young's modulus was also studied as part of their mechanical properties, but no significant differences were observed between the different formulations, which is reasonable since they all have the same backbone structure (Fig. 3C).
In summary, it can be confirmed that the H polymer is the one showing the lowest interaction within the NP. Also, the R-containing formulations are the ones presenting higher adhesion values. These adhesion properties are interesting for delivery system applications, such as muscle regeneration in which the nanoparticles may have higher adhesion values as scaffold-like structures for assisting cells in the reconstruction processes.29,30
OM-pBAEs physicochemical properties influence the nanoparticle structuration
The affinity between components of the polyplex is the key to understanding the intramolecular interactions driving the structure function. Affinity is also monitored as a part of the drug discovery process to help design drugs that bind their targets selectively and specifically.31Binding affinity is the strength of the interaction between a single biomolecule (pGFP, in this case) to its binding partner (OM-pBAEs). It is an essential and basic concept but poorly examined in the drug delivery systems area. This concept can be studied and quantified by the dissociation constant (KD), which, for the nanoparticles must be low enough to enable the condensation of the plasmid inside the polymer forming the nanoparticle and, at the same time, high enough to be able to deliver the cargo efficiently once interacted with the cells. To study this parameter, MicroScale Thermophoresis was carried out.
The obtained values for cationic NPs (KH, RH, and RK; Fig. 4A) show higher affinity (lower KD) that is achieved with KH NPs, followed by RH and RK. Referring to the coated nanoparticles (Fig. 4B), the same dissociation curve was followed without significant KD differences. Comparing both groups, cationic KD is nearly doubling the value of the coated polyplexes, which can be explained due to the combination of cationic and anionic peptides in coated NPs. Both cationic and anionic OM-pBAEs present a strong interaction between them, reducing the interaction with plasmid. Specifically, it is interesting to comment on the KD of KH NPs, which is comparable to that of the coated group. This is in agreement with the size, surface potential, and EE that these NPs as shown in their physicochemical characterization (see Section 1). Moreover, we have demonstrated that we can tailor the KD of our NPs combining different polymers opening the door for new applications. If a sustained release is needed, such as in the cosmetics field to retard aging, drug delivery systems (DDS) based on the coated constructions or KH would be a good option.
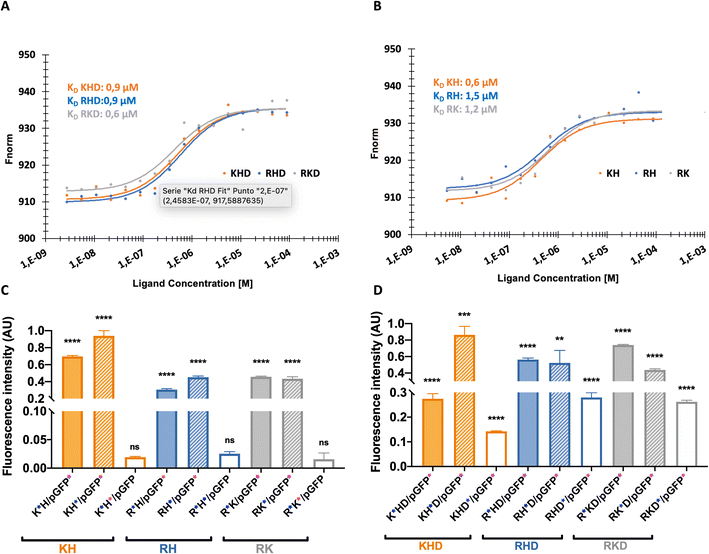 | ||
Fig. 4 Binding affinity and FRET intensities profiles. (A) and (B) dose response charts showing binding affinity curve and (C) and (D) fluorescent energy transference for: (A) and (C): cationic and (B) and (D) coated constructs. The“ ” at the x axis in the nanoparticle's names mean the nanoparticle elements that was labelled with Cy5 and “ ” at the x axis in the nanoparticle's names mean the nanoparticle elements that was labelled with Cy5 and “ ”the one labelled with Cy3. Results are the mean ± SD values of a triplicate. P value < 0.0001 (****), p value < 0.001 (***), p value < 0.01(**), p value < 0.05 (*) and p value > 0.05 (ns). More details in FRET experiments are given in Fig. S4, from ESI.† ”the one labelled with Cy3. Results are the mean ± SD values of a triplicate. P value < 0.0001 (****), p value < 0.001 (***), p value < 0.01(**), p value < 0.05 (*) and p value > 0.05 (ns). More details in FRET experiments are given in Fig. S4, from ESI.† | ||
The next step was to define the complexation and structuration to quantitatively study the distribution of the components within the polyplex by Förster resonance energy transfer (FRET). In FRET, a higher transference of energy means that the fluorophores are closer, thus, indicating that the smallest NPs size is obtained. As shown in Fig. 4C, all cationic polyplexes are sufficiently small within the optimum range for the parenteral transport.24 KH polyplexes present higher values than RK and RH NPs. This can be explained by the fact that KH polyplexes are the ones with smaller sizes (Fig. 2B). Additionally, it was observed that the energy transference between polymers is negligible (see Fig. S3, ESI†). This is reasonable due to the repulsion between both (positively charged) polymers causing spatial distancing and as a consequence, the lowering of the signal. The obtained values also indicate which polymer is on the inner layer closer to the plasmid and which is on the external layer. The FRET intensity of the C6CH3 polymer with pGFP is slightly higher than that of other polymers, meaning that C6CH3 is relatively closer to the plasmid once the nanoparticle is formed. This could be attributed not only to the charge effects of protonated histidine but also to the geometry of the binding between π systems of both the biomolecules (DNA nucleobases and His cyclopentane).32 Besides, in the RK polyplex, C6CR3 is slightly nearer to the nucleic acid than the C6CK3 polymer. Therefore, the NPs insight conformations, with the C6CR3 surrounding the C6CK3 polymer can be deduced (see illustration in Fig. 1C).
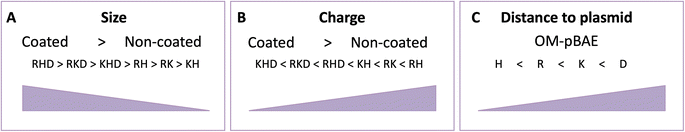
As can be seen in Fig. 4D, the coated constructs, KHD, RKD, and RHD follow the same trend as the cationic nanoparticle but with reduced energy-transferred values when measuring D interaction. The decrease indicates that the negative polymer coating shields the FRET between the other polymers and the plasmid. Additionally, the energy transferred between the polymer C6CD3 and plasmid is smaller; hence, this confirms that the negative polymer is conforming to the outer layer of the nanoparticle, as we have hypothesized. This result is highly important, defining these kinds of constructs, which is the external polymer where the targeting moiety should be attached. In summary, the FRET experiments are in agreement with the hydrodynamic diameter obtained with DLS; coated NPs show higher hydrodynamic diameter and lower surface potential in comparison to the cationic complexes (Fig. 5A and B). Furthermore, the transference of energy has allowed us to understand the distribution of the components inside the nanoparticle, where it was seen that H is the nearest polymer to the genetic material followed by arginine, lysine, and aspartic acid OM-pBAEs (Fig. 5C).
Finally, darkfield hyperspectral microscopy was performed to qualitatively confirm the morphology and unveil the polymer internal composition of the particles. The 6 different polyplexes were freshly prepared and the optical images of the constructions were obtained as can be observed in Fig. 6A. Polyplexes do not have a core–shell structure but a plasmid core surrounded by a cloud polymer (Fig. 6B) resulting in a nanosphere shape, as represented in the sketches of Fig. 1C and preserving the order deduced in the previous section (C6CH3, C6CR3, C6CK3, and C6CD3 from the inner to the outer layer). In fact, this cloud structure was previously described by our group by only using the C6CR3 polymer, and here we confirmed that it also appeared for polymer combinations.15,33 To remark that, thermodynamically, the inner plasmid core, completely made up of anionic molecules, surrounded by the cloud of the cationic polymer layer is not the most stable conformation. Nevertheless, we have given pieces of evidence here and in our previous works that it is the conformation that experimentally results. It could be hypothesized that it can be maintained stable thanks to the close position between the core and the shell layer of the polymer that avoids the expected electrostatic repulsions.
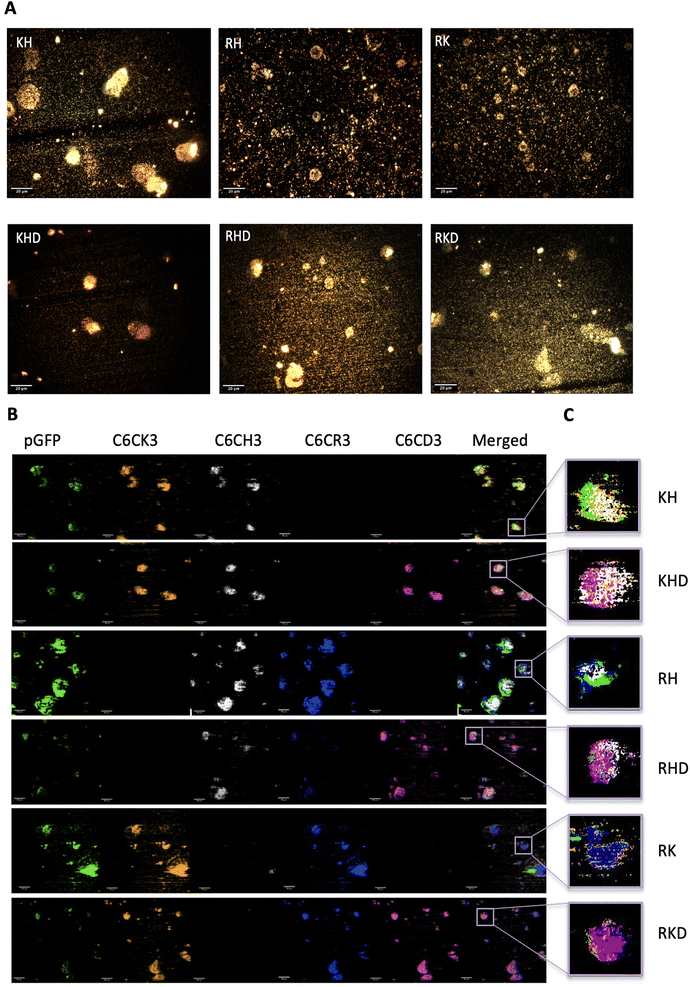 | ||
| Fig. 6 Polyplex optical and hyperspectral matching images. (A) Optical images of the different nanoparticles' samples. Images were obtained with a 40× objective. Scale bars = 20 μm. More optical images can be found in Fig. S6, from ESI.† (B) Hyperspectral images attributing each nanoparticles component to a channel (hyperspectral profiles of each biomaterial in Fig. S7†), in where pGFP channel is shown in green, C6CK3 in orange, C6CH3 in white, C6CR3 in blue, CHCD3 in magenta and last channel illustrates merging. Scale bars = 10 μm. (C) Zoom in of the merged channels of a single nanoparticle for each construct type, following the same colour code. | ||
The presented particles in Fig. 6B are shown in different planes. For this reason, some of the samples show the outer layer while others the inner. Additionally, as shown in Fig. 6C, there is a zoom-in of the merged spectral reconstructions in which a nanosphere can be appreciated together with the polymer and plasmid distribution. Again, this is in accordance with and confirms our previous results. In conclusion, with this qualitative technique, we have been able to visualize and corroborate the previous results obtaining a more concise idea of how the NP components are distributed inside the nanosphere. In addition, the possibility to determine which OM-pBAE is localized on the outer layer of the NP may enable us to obtain an insight into the interactions between the polymer and biological media more closely. In fact, defining the internal structuration of the NPs is not the only important factor, but also selecting the right polymer where the targeting moiety needs to be attached to achieve the best surface exposure and thus the highest interaction with the corresponding receptor, required for directing the particles to the target tissues.
OM-pBAEs structuration goes together with nanoparticle stability
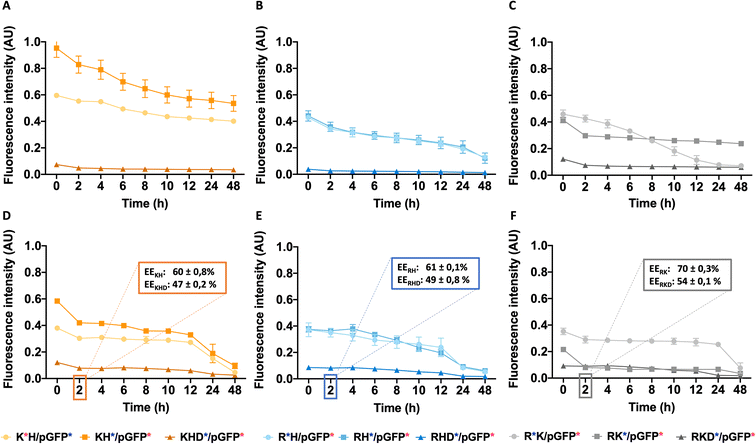
To design the effective polymeric NPs for drug delivery, their stability has to be thoroughly assessed not only the stock solutions but also in simulated biological fluids conditions. To remark, the importance of safety evaluation that, although out of the scope of the present work, must be taken into account. For parenterally administered formulations, both, cell compatibility and blood compatibility must be ensured. We have reported in previous works the non-cytotoxic character of all formulations15,21,34 (see summarized data in Fig. S5†), thus, they can be safely administered.Temperature study is an essential aspect to determine the stability, 25 °C a key parameter for particle shelf storage and synthesis while 37 °C is the homeostasis body temperature. For this purpose, the stability of different polyplexes was studied in phosphate-buffered saline (PBS) at different times by the FRET technique (for more information see Table S2 from ESI†). The same procedure was also performed in fetal bovine serum (FBS) to simulate the human body fluid environment, as a model of human plasma. Once the stability study was performed, a more precise application could be assigned to each nanoparticle as a function of their lifetime. Cationic and anionically-coated NPs were incubated with FBS, used to model the NP behavior in plasma In Fig. 7, cationic and anionic NPs incubated with FBS of each construct type are represented. The transferred fluorescence between polymers was not significant, so only FRET between the polymer and plasmid was represented in all cases (as shown in the previous results from Fig. 4C and D and Table S2 from ESI†). In general, all the combinations follow the same decay pattern. After 2 h, the decay is more accentuated, suggesting that there is a percentage of NPs dissociated during this period,23 which was clearly corroborated by the obtained AFM values. Getting into details shown in Fig. 7A–C, we can confirm that each polymer type follows a similar tendency with slight differences at 25 °C. On the one hand, for KH (Fig. 7A), the energy transferred is notably higher than that for the other constructions, as we have seen before. On the other hand, in the RH construct (Fig. 7B) both polymers follow a comparable disassembly process and are similar to the fluorescence energy-transferred values of RK NPs (Fig. 7C). In this last construction, RK, the FRET results of the C6CR3 polymer show a signal being gradually reduced over time due to its destabilization over time. Despite this, C6CK3 of this RK polyplex has an intense polymer drop at 2 h of incubation. This phenomenon could indicate that C6CK3 is the first polymer that tends to decomplex from the nanoparticle structure. Whereas, for the other two combinations (KH and RH) the first polymer in decomplex is C6CH3, for KH this disaggregation is more abrupt than in RH whose C6CR3 polymer follows a similar pattern degradation to that of C6CH3.
Concerning the coated constructions, no significant variations were observed along time at 25 °C with the exception of the mentioned 2 h period decay. The polymer C6CD3 remains on the outer layer but it does not conform to a core structure; it is more likely to be a woolen ball allowing the release of the inner polymers.
The same experiment was performed at 37 °C to study the effect of temperature on the stability of NPs (Fig. 7D–F). Coated NPs had slightly higher values in comparison to those described previously at 25 °C but were still far from the values obtained with the cationic polymers. This is expected since C6CD3 was added a posteriori once the polyplex was already formed. Concerning the cationic NPs, FRET values were remarkably decreased and in addition to the 2 h decrease, a prominent fluorescence drop occurred between the 12–24 h was seen, thus, indicating they are temperature sensitive. The same behavior was observed when NPs were incubated at 37 °C except for the C6CR3 polymer in RK NPs combination that displayed stability values over 24 h incubation, without the 12 h drop. All this indicates that our NPs are able to perform a partial release of the cargo between a 2 h period, which is more than enough time for the NPs to reach the target organ. Moreover, as expected, the presence of the enzymes in the physiological media will also accelerate the decomplexation process. To complete and corroborate the 2 h-cargo release hypothesis, an encapsulation efficiency experiment was performed, obtaining the results shown in Fig. 7D–F. Results showed a decrease in the %EE (compared to initial values shown in Fig. 2) for all the combinations after this time, meaning that there are small percentages of the release for all the constructions. Specifically, the delivery of the cationic combinations is delayed in comparison with the coated ones. For instance, the KH construction delivers 25% of the pGFP cargo, while its respective-coated KHD nanoparticles liberated 40% of the pGFP cargo. This can be justified since there is a reduction of the cationic polymer over time (as seen from FRET experiments and AFM results), so the overall charge of the nanoparticle may decrease giving rise to a non-stable system with pGFP, which is negatively charged, is more exposed to the C6CD3 coating, which is also anionic, leading to a repulsion process and thereby cargo release.
In this stability section, it will be illustrated that each polymer of NP constructs has its own biodegradability properties; thus, NP stability is under the aegis of the terminal peptides used. The C6CH3 polymer was first disassembled, as deduced from the AFM results, and proceeded by C6CK3, C6CR3, and C6CD3 when the temperature was increased. This property together with the binding affinity are interesting in order to select a proper nanoparticle in which the release kinetics is important, for example, higher binding affinity and stability will promote sustained release therapies, making RKD an attractive option for long-term release. All these studied, and now, well-described properties have allowed us to tailor the OM-pBAE carriers by selecting the most accurate oligopeptide combination, based on the needs or interests requested for the specific application goal; all these properties are summarized in Table 1.
| NPs formulation | Z – potential | Size | EE | Adhesion | Binding affinity | Stability |
|---|---|---|---|---|---|---|
| KH | *** | * | *** | ** | *** | * |
| KHD | * | ** | ** | ** | ** | ** |
| RH | *** | * | * | *** | * | ** |
| RHD | * | *** | * | *** | ** | ** |
| RK | ** | * | * | ** | * | ** |
| RKD | * | ** | ** | *** | *** | ** |
Conclusions
The aim of this study was to mechanically and physiochemically decipher the NP properties as a function of the end peptide polymer combinations. The comprehension of these features fulfills the lack of basic science studies, always taken for granted, but essential for engineering carriers and, hence, boosts a rational selection for the most suitable carrier for a specific application and its further commercialization success. These constructions have been deeply analyzed and have demonstrated different structures as a function of the OM-pBAE combination used. Cationic structures have smaller sizes but higher Z-potential whilst it is contrary for the C6CD3 coated NPs. Despite this, nucleic acid encapsulation efficiency is preserved for all the constructions with elevated values, headed by KH polyplexes. Additionally, this wide study allowed the understanding of a new concept: the in-sight NP components distribution and therefore the outer exposed polymer allowing the addition of specific targets, providing an efficient delivery. Alluding to the mechanical approach, the C6CH3 polymer is the first in leaving the nanoparticle probably facilitating the cargo release. This affirmation has been reinforced by the stability studies that demonstrated that depending on the OM-pBAE used, the nanoparticle construction has higher sensitivity to temperature and media variations, and the coated constructions are less susceptible than the cationic ones. This provides different timings for the cargo release in the function of the end-terminal peptide used and, therefore, sustained or rapid release can be achieved with different constructions. Thus, for the first time, we revealed properties that will allow tailoring the carriers by selecting the most accurate oligopeptide combination based on the needs or interests requested for the specific application goal. Consequently, our results will boost future research on OM-pBAE to facilitate their transfer to clinical applications.Experimental section
Materials
Various reagents have been used in this project. Sucrose, Sodium Acetate, Hepes, DEPC-treated water, and PBS were purchased from Sigma-Aldrich®. Cyanine dyes were purchased from Lumiprobe for polymer labeling but for plasmid labeling Label IT® TrackerTM Intracellular Nucleic Acid Localization Kit from Mirus was purchased. Fetal Bovine Serum (FBS) was obtained from Lab Clinics. Plasmid GFP (3486 bp) was produced and purified from E. coli. Quant-iT PicoGreen Kit from Invitrogen was used for DNA quantification. Arginine, histidine, lysine, and aspartic acid end-modified poly(β-amino ester) (PBAE) polymers were synthesized by a group from the materials Engineering team, following a two-step procedure previously described in the literature.15Methods
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 ratio and RK in a 50
40 ratio and RK in a 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 ratio. The nucleic acid was added to the polymer solution, and mixed by pipetting, followed by 30 min of incubation at room temperature (V1). For the preparation of the coated pBAE/pGFP NPs, the C6CD3 (aspartic acid, named D) polymer in sodium acetate solution (12.5 mM pH 5.2), was added to the formed polyplex during the incubation (at 15 min) in the same quantity as the cationic polymer. For the formation of the discrete structures after incubation, the mixture was nanoprecipitate in the same volume of MiliQ water. Thereafter, the same initial volume of Hepes 20 mM + 4 wt% sucrose (pH 7.4) solution was also added as cryo and lyoprotectors ending with sample 1/3 diluted compared with the first incubation.
50 ratio. The nucleic acid was added to the polymer solution, and mixed by pipetting, followed by 30 min of incubation at room temperature (V1). For the preparation of the coated pBAE/pGFP NPs, the C6CD3 (aspartic acid, named D) polymer in sodium acetate solution (12.5 mM pH 5.2), was added to the formed polyplex during the incubation (at 15 min) in the same quantity as the cationic polymer. For the formation of the discrete structures after incubation, the mixture was nanoprecipitate in the same volume of MiliQ water. Thereafter, the same initial volume of Hepes 20 mM + 4 wt% sucrose (pH 7.4) solution was also added as cryo and lyoprotectors ending with sample 1/3 diluted compared with the first incubation.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 v/v mixture of diethyl ether: acetone and posteriorly dried overnight and dissolved in DMSO to obtain a solution of 100 mg ml−1.
3 v/v mixture of diethyl ether: acetone and posteriorly dried overnight and dissolved in DMSO to obtain a solution of 100 mg ml−1.
 ” indicates the component labelled with Cy5 and the “
” indicates the component labelled with Cy5 and the “ ” the component labelled with Cy3 in each case
” the component labelled with Cy3 in each case
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 (v/w) and reaction time was increased to 2 h, in order to reach a labeling density of 10 dye
10 (v/w) and reaction time was increased to 2 h, in order to reach a labeling density of 10 dye![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DNA molecule.22 Afterwards, the sample of Cy3/5-DNA was quantified by means of fluorescence and absorbance, respectively, with an Infinite M Plex microplate reader from TECAN.
DNA molecule.22 Afterwards, the sample of Cy3/5-DNA was quantified by means of fluorescence and absorbance, respectively, with an Infinite M Plex microplate reader from TECAN.
The target, pGFP-Cy5, was loaded into the capillaries at a 5 nM concentration. Excitation power was set to 40% and MST power to a medium. Then, it was excited with pico-RED with a 15% excitation power at a thermostat setpoint of 25 °C. Serial dilutions of the ligands (KH, RH, RK, KHD, RHD, and RKD) were added starting from 0.2 μM to 5 nM. A timeframe of 0.5–1.5 s was chosen to analyze the data by the MO Affinity Analysis software (NanoTemper) by Hill fit.
Author contributions
Conceptualization: SBG, CFP data curation: ADP, SGR, SBG, CFP formal analysis: MNL, ADP, SGR, CFP funding acquisition: ADP, SGR, SBG investigation: MNL, CFP methodology: MNL, ADP, SGR, CFP project administration: CFP, SBG resources: SBG, CFP software: MNL, ADP, SGR supervision: SBG, CFP validation: ADP, SGR, SBG, CFP visualization: SBG, CFP writing – original draft: MNL writing – review & editing: ADP, SGR, CFP, SBG.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge the financial support received from MINECO (grant RTI2018-094734-B-C22 and BIGDATASPM PID2019-110210GB-I00), MICIN (grant RTC2019-007260-1), MCIN/AEI /10.13039/501100011033/FEDER, UE (grant PID2021-125910OB-I00) and Generalitat de Catalunya (2017 SGR 1559). A. D-P. received funding from the postdoctoral fellowships program Beatriu de Pinós, funded by the Secretary of Universities and Research (Government of Catalonia), and from the Horizon 2020 program of research and innovation of the European Union under the Marie Skłodowska-Curie grant agreement No 801370. S. G. thanks the Instituto de Salud Carlos III (ISCIII) (CB06/01/1058). CIBER BBN is an initiative funded by the VI National R + D + I Plan 2008–2011, Iniciativa Ingenio 2010, Consolider Program, CIBER actions and financed by ISCIII with assistance from the European Regional Development Fund. The authors want to thank NANBIOSIS ICTS and the Nanostructured Liquid Characterization Unit (U12) for the characterization of materials by high-resolution optical microscopy with spectral analysis. Laura Olmo and Maria Stampa Lopez-Pinto are acknowledged for polymer synthesis. Maria Stampa Lopez-Pinto and Nil González-Rios are acknowledged for the NMR characterization of the polymers. The Nanothemper team is also acknowledged for their complimentary sample processing data of the NPs binding affinity results.References
- B. A. Bunnell and R. A. Morgan, Gene Therapy for Infectious Diseases, 1998, vol. 11 Search PubMed.
- B. Thapa and R. Narain, Mechanism, Current Challenges and New Approaches for Non Viral Gene Delivery, Elsevier Ltd, 2016, DOI:10.1016/B978-0-08-100520-0.00001-1.
- K. I. Papadopoulos, P. Wattanaarsakit, W. Prasongchean and R. Narain, Gene Therapies in Clinical Trials, Elsevier Ltd, 2016, DOI:10.1016/B978-0-08-100520-0.00010-2.
- M. J. Mulligan, K. E. Lyke, N. Kitchin, J. Absalon, A. Gurtman, S. Lockhart, K. Neuzil, V. Raabe, R. Bailey, K. A. Swanson, P. Li, K. Koury, W. Kalina, D. Cooper, C. Fontes-Garfias, P. Y. Shi, Ö. Türeci, K. R. Tompkins, E. E. Walsh, R. Frenck, A. R. Falsey, P. R. Dormitzer, W. C. Gruber, U. Şahin and K. U. Jansen, Phase I/II Study of COVID-19 RNA Vaccine BNT162b1 in Adults, Nature, 2020, 586(7830), 589–593, DOI:10.1038/s41586-020-2639-4.
- U. Sahin, M. Alexander, E. Derhovanessian, I. Vogler, L. M. Kranz, M. Vormehr, A. Baum, K. Pascal, D. M. Jasmin Quandt, S. Brachtendorf, V. Lörks, S. Julian, H. Rolf, D. Becker, A.-K. Eller, J. Grützner, C. Boesler, C. Rosenbaum, M.-C. Kühnle, U. Luxemburger, A. Kemmer-Brück, D. Langer, K. U. J. Martin and O. T. Bexon, COVID-19 Vaccine BNT162b1 Elicits Human Antibody and TH1 T Cell Responses, Nature, 2021, 590, E17, DOI:10.1038/s41586-020-03102-w.
- L. R. Baden, H. M. El Sahly, B. Essink, K. Kotloff, S. Frey, R. Novak, D. Diemert, S. A. Spector, N. Rouphael, C. B. Creech, J. McGettigan, S. Khetan, N. Segall, J. Solis, A. Brosz, C. Fierro, H. Schwartz, K. Neuzil, L. Corey, P. Gilbert, H. Janes, D. Follmann, M. Marovich, J. Mascola, L. Polakowski, J. Ledgerwood, B. S. Graham, H. Bennett, R. Pajon, C. Knightly, B. Leav, W. Deng, H. Zhou, S. Han, M. Ivarsson, J. Miller and T. Zaks, Efficacy and Safety of the MRNA-1273 SARS-CoV-2 Vaccine, N. Engl. J. Med., 2021, 384(5), 403–416, DOI:10.1056/nejmoa2035389.
- J. Mateus, J. M. Dan, Z. Zhang, C. R. Moderbacher, M. Lammers, B. Goodwin, A. Sette, S. Crotty and D. Weiskopf, Low-Dose MRNA-1273 COVID-19 Vaccine Generates Durable Memory Enhanced by Cross-Reactive T Cells, Science, 2021, 374(6566), 1–27, DOI:10.1126/science.abj9853.
- M. A. Kay, State-of-the-Art Gene-Based Therapies: The Road Ahead, Nat. Rev. Genet., 2011, 12(5), 316–328, DOI:10.1038/nrg2971.
- H. Herweijer and J. A. Wolff, Progress and Prospects: Naked DNA Gene Transfer and Therapy, Gene Ther., 2003, 10(6), 453–458, DOI:10.1038/sj.gt.3301983.
- B. B. Mendes, J. Conniot, A. Avital, D. Yao, X. Jiang, X. Zhou, N. Sharf-Pauker, Y. Xiao, O. Adir, H. Liang, J. Shi, A. Schroeder and J. Conde, Nanodelivery of Nucleic Acids, Nat. Rev. Methods Primers, 2022, 2(1), 24, DOI:10.1038/S43586-022-00104-Y.
- P. Dosta, N. Segovia, A. Cascante, V. Ramos and S. Borrós, Surface Charge Tunability as a Powerful Strategy to Control Electrostatic Interaction for High Efficiency Silencing, Using Tailored Oligopeptide-Modified Poly(Beta-Amino Ester)s (PBAEs), Acta Biomater., 2015, 20, 82–93, DOI:10.1016/j.actbio.2015.03.029.
- S. E. YlÃ, Glybera Finally Recommended for Approval as the First Gene Therapy Drug in the European Union, 2012, DOI:10.1038/mt.2012.194.
- R. P. Khemariya and P. S. Khemariya, New-Fangled Approach in the Management of Alzheimer by Formulation of Polysorbate 80 Coated Chitosan Nanoparticles of Rivastigmine for Brain Delivery and Their In Vivo Evaluation, Int. J. Sci. Eng. Invent., 2016, 02, 18–29 Search PubMed.
- M. Ramamoorth and A. Narvekar, Non Viral Vectors in Gene Therapy – An Overview, J. Clin. Diagn. Res., 2015, 9(1), GE01–6, DOI:10.7860/JCDR/2015/10443.5394.
- C. Fornaguera, M. Guerra-Rebollo, M. Ángel Lázaro, C. Castells-Sala, O. Meca-Cortés, V. Ramos-Pérez, A. Cascante, N. Rubio, J. Blanco and S. Borrós, mRNA Delivery System for Targeting Antigen-Presenting Cells In Vivo, Adv. Healthcare Mater., 2018, 7(17), e1800335, DOI:10.1002/adhm.201800335.
- Y. Liu, Y. Li, D. Keskin and L. Shi, Poly(β-Amino Esters): Synthesis, Formulations, and Their Biomedical Applications, Adv. Healthcare Mater., 2019, 8(2), 1–24, DOI:10.1002/adhm.201801359.
- J. Karlsson, K. R. Rhodes, J. J. Green and S. Y. Tzeng, Poly(beta-amino ester)s as gene delivery vehicles: challenges and opportunities, Expert Opin. Drug Delivery, 2020, 17(10), 1395–1410, DOI:10.1080/17425247.2020.1796628.
- R. A. Cordeiro, A. Serra, J. F. J. Coelho and H. Faneca, Poly(β-Amino Ester)-Based Gene Delivery Systems: From Discovery to Therapeutic Applications, J. Controlled Release, 2019, 310, 155–187, DOI:10.1016/j.jconrel.2019.08.024.
- G. T. Zugates, N. C. Tedford, A. Zumbuehl, S. Jhunjhunwala, C. S. Kang, L. G. Griffith, D. A. Lauffenburger, R. Langer and D. G. Anderson, Gene Delivery Properties of End-Modified Poly(β-Amino Ester)S, Bioconjugate Chem., 2007, 18(6), 1887–1896, DOI:10.1021/bc7002082.
- J. C. Sunshine, S. B. Sunshine, I. Bhutto, J. T. Handa and J. J. Green, Poly(β-Amino Ester)-Nanoparticle Mediated Transfection of Retinal Pigment Epithelial Cells in Vitro and in Vivo, PLoS One, 2012, 7(5), e37543, DOI:10.1371/journal.pone.0037543.
- N. Segovia, P. Dosta, A. Cascante, V. Ramos and S. Borrós, Oligopeptide-terminated poly(β-amino ester)s for highly efficient gene delivery and intracellular localization, Acta Biomater., 2014, 10(5), 2147–2158, DOI:10.1016/j.actbio.2013.12.054.
- R. Riera, N. Feiner-Gracia, C. Fornaguera, A. Cascante, S. Borrós and L. Albertazzi, Tracking the DNA Complexation State of PBAE Polyplexes in Cells with Super Resolution Microscopy, Nanoscale, 2019, 11(38), 17869–17877, 10.1039/c9nr02858g.
- R. Riera, J. Tauler, N. Feiner-Gracia, S. Borrós, C. Fornaguera and L. Albertazzi, Complex PBAE Nanoparticle Cell Trafficking: Tracking Both Position and Composition Using Super Resolution Microscopy, 2022, DOI:10.1002/cmdc.202100633.
- M. A. Dobrovolskaia and S. F. McNeil, Frontiers in Nanobiomedical Research and Key Considerations for Nanoparticle Characterization Prior to Immunotoxicity Studies, Handbook of Immunological Properties of Engineered Nanomaterials, World Scientific, 2nd edn, 2013 Search PubMed.
- P. Dosta, C. Demos, V. Ramos, D. W. Kang, S. Kumar, H. Jo and S. Borrós, Delivery of siRNA to Endothelial Cells In Vivo Using Lysine/Histidine Oligopeptide-Modified Poly(β-amino ester) Nanoparticles, Cardiovasc. Eng. Technol., 2021, 12(1), 114–125, DOI:10.1007/s13239-021-00518-x.
- A. P. Singh, A. Biswas, A. Shukla and P. Maiti, Targeted Therapy in Chronic Diseases Using Nanomaterial-Based Drug Delivery Vehicles, Signal Transduction Targeted Ther., 2019, 4(1), 1–21, DOI:10.1038/s41392-019-0068-3.
- J. R. Smith, T. O. B. Olusanya and D. A. Lamprou, Characterization of Drug Delivery Vehicles Using Atomic Force Microscopy: Current Status, Taylor & Francis, 2018, vol. 15, DOI:10.1080/17425247.2018.1546693.
- N. Kapate, J. R. Clegg and S. Mitragotri, Non-Spherical Micro- and Nanoparticles for Drug Delivery: Progress over 15 Years, Adv. Drug Delivery Rev., 2021, 177, 113807, DOI:10.1016/J.ADDR.2021.05.017.
- V. Ogay, E. A. Mun, G. Kudaibergen, M. Baidarbekov, K. Kassymbek, Z. Zharkinbekov and A. Saparov, Progress and Prospects of Polymer-Based Drug Delivery Systems for Bone Tissue Regeneration, Polymers, 2020, 12(12), 1–25, DOI:10.3390/polym12122881.
- M. Filippi, G. Born, M. Chaaban and A. Scherberich, Natural Polymeric Scaffolds in Bone Regeneration, Front. Bioeng. Biotechnol., 2020, 8, 474, DOI:10.3389/fbioe.2020.00474.
- Binding Affinity | Dissociation Constant | Malvern Panalytical https://www.malvernpanalytical.com/en/products/measurement-type/binding-affinity, accessed 2022 -05 -13 Search PubMed.
- C. D. M. Churchill and S. D. Wetmore, Noncovalent Interactions Involving Histidine: The Effect of Charge on π–π Stacking and T-Shaped Interactions with the DNA Nucleobases, J. Phys. Chem. B, 2009, 113(49), 16046–16058, DOI:10.1021/jp907887y.
- C. Fornaguera, M. Díaz-Caballero, C. García-Fernandez, L. Olmo, M. S. L. Pinto, M. Navalón-López, M. Guerra-Rebollo and S. Borrós, Synthesis and Characterization of Mrna-Loaded Poly(Beta Aminoesters) Nanoparticles for Vaccination Purposes, J. Visualized Exp., 2021, 2021(174) DOI:10.3791/62889.
- WSXM: A Software for Scanning Probe Microscopy and a Tool for Nanotechnology, 2007, DOI:10.1063/1.2432410.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2na00800a |
| This journal is © The Royal Society of Chemistry 2023 |