Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
App-based quantification of crystal phases and amorphous content in ZIF biocomposites†
Michael R.
Hafner
a,
Laura
Villanova
 *b and
Francesco
Carraro
*b and
Francesco
Carraro
 *a
*a
aInstitute of Physical and Theoretical Chemistry, Graz University of Technology, 8010 Graz, Austria. E-mail: francesco.carraro@tugraz.at
bFaculty of Technical Chemistry, Chemical and Process Engineering, Biotechnology, Graz University of Technology, 8010 Graz, Austria. E-mail: laura.villanova@tugraz.at
First published on 4th March 2022
Abstract
The performance of zeolitic imidazolate frameworks (ZIFs) as protective hosts for proteins in drug delivery or biocatalysis strongly depends on the type of crystalline phase used for the encapsulation of the biomacromolecule (biomacromolecule@ZIF). Therefore, quantifying the different crystal phases and the amount of amorphous content of ZIFs is becoming increasingly important for a better understanding of the structure–property relationship. Typically, crystalline ZIF phases are qualitatively identified from diffraction patterns. However, accurate phase examinations are time-consuming and require specialized expertise. Here, we propose a calibration procedure (internal standard ZrO2) for the rapid and quantitative analysis of crystalline and amorphous ZIF phases from diffraction patterns. We integrated the procedure into a user-friendly web application, named ZIF Phase Analysis, which facilitates ZIF-based data analysis. As a result, it is now possible to quantify i) the relative amount of various common crystal phases (sodalite, diamondoid, ZIF-CO3-1, ZIF-EC-1, U12 and ZIF-L) in biomacromolecule@ZIF biocomposites based on Zn2+ and 2-methylimidazole (HmIM) and ii) the crystalline-to-amorphous ratio. This new analysis tool will advance the research on ZIF biocomposites for drug delivery and biocatalysis.
Introduction
Metal–organic frameworks (MOFs) are a class of coordination polymers that are formed via the assembly of inorganic nodes (metal clusters or ions) and organic linkers.1,2 Recent progress on composites based on MOFs and biomacromolecules, namely, MOF biocomposites, has been generating significant interest in the field of biotechnology and biomedicine.3–6 In particular, zeolitic imidazolate frameworks (ZIFs) prepared from Zn2+ and 2-methylimidazole (HmIM) are widely studied because their facile synthesis is compatible with several different classes of biomacromolecules (e.g., proteins, carbohydrates, lipids, nucleic acids, viruses, and cells).3–5,7–11 By mixing biomacromolecules to an aqueous solution of Zn2+ cations and HmIM, it is possible to find conditions inducing the spontaneous formation of the ZIF material around biomacromolecules (i.e., biomimetic mineralization).3 Once a ZIF biocomposite (biomacromolecule@ZIF) is formed, the encapsulated biomacromolecule can be protected against harsh environments,3,12 can be stored without the need for refrigeration for long periods5,9,12 and can be released in a controlled fashion by exposing the biocomposite to chelating agents (e.g. EDTA),13 acidic environments (e.g., pH ≤ 6.5)13,14 and buffer solutions (e.g., PBS).13–15During the preparation of biomacromolecule@ZIF systems, reaction parameters (metal-to-ligand ratio, concentration, stirring or static conditions) and post-synthetic treatments (washing with water or ethanol) strongly influence the crystal structure of Zn(mIM)2-based frameworks.3,16,17 The type of crystalline phase strongly influences the physical–chemical properties of the final biocomposite.16–20 For example, diamondoid (dia) ZIF-8 is a MOF non-porous to N2 with a Brunauer–Emmett–Teller (BET) surface area of 40 m2 g−1,21 whereas sodalite (sod) ZIF-8 possesses permanent microporosity and has a BET surface area of 1200 m2 g−1.21 Consequently, different ZIF structures react differently towards external stimuli like pH changes (e.g., the release time of Bovine Serum Albumin (BSA) from dia ZIF-8 is 5 times longer than that from sod ZIF-8 at pH 5.5).16 More peculiar properties were shown by amorphous ZIF biocomposites: in drug delivery, pH changes can induce the instantaneous ZIF degradation and release of the biotherapeutics;22,23 in biocatalysis, an enhancement of the substrate diffusion and high enzymatic activities in enzyme@ZIF-8 biocatalysts were measured.22 We note that the quantification of the amorphous content is a challenging step that could require the combination of different characterization techniques (e.g. X-ray diffraction (XRD), N2 physisorption, thermogravimetric analysis, infrared spectroscopy (IR), and X-Ray photoemission spectroscopy).22 In general, a careful investigation of the obtained biocomposite typically shows a mixture of phases with a dominant component. As these phases have different functional properties, an inaccurate phase identification results in erroneous assessments of their structure–property–function relationships.16 Thus, a tool that enables a rapid and accurate phase assessment in ZIFs will help the progress of biomacromolecule@ZIF composites in biomedicine and biocatalysis.16,22
Currently, a straightforward tool for the identification and quantification of both the crystalline phases and the amorphous fraction of ZIF biocomposites is missing. Furthermore, powder X-Ray diffraction (PXRD) analysis is among the most time-consuming steps in the development cycle of novel materials.24 To have a time-efficient and quantitative analysis of ZIF biocomposites, we developed a web-based platform (ZIF Phase Analysis) that combines the customizable analytical capacities of the R environment25 and the intuitiveness of the user-friendly Shiny interface.26 The platform is freely accessible via a dedicated website (https://rapps.tugraz.at/apps/porousbiotech/start/) or directly at https://rapps.tugraz.at/apps/porousbiotech/ZIFphaseanalysis/. It includes a novel XRD calibration procedure that we developed to quantify the phases which are commonly found when synthesising ZIF biocomposites from Zn2+ and HmIM (i.e., sodalite and diamondoid ZIF-8, ZIF-CO3-1, ZIF-EC-1, U12, and ZIF-L,16,19,27–29 here referred to as sod, dia, ZIF-C, ZIF-EC-1, U12 and ZIF-L) and quantify the amorphous content via the analysis of XRD patterns. This tool will facilitate the data analysis and accelerate the research in the emerging field of ZIF biocomposites, allowing researchers from different fields to precisely correlate the functional properties of the ZIF biocomposites (e.g., drug release profiles and biocatalytic activity) to the crystal phase(s) of the biocomposite.
Calibration procedure
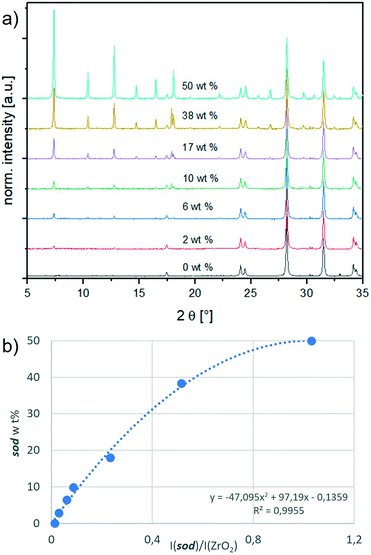
We designed a calibration method with an internal standard (ZrO2) for PXRD data analysis. ZrO2 was chosen as it is insoluble in water (thus it does not form a MOF with 2-methylimidazole) and its PXRD diffraction peaks do not overlap with ZIF diffraction peaks. Firstly, by optimizing the synthesis conditions, we synthesised each phase – sod, dia, ZIF-C, ZIF-EC-1, U12 and ZIF-L – in a pure form, excluding the formation of phase mixtures. These reaction conditions can be found in section S2.1.† In Fig. S1,† the PXRD patterns of the different phases are compared to the available simulated patterns to ascertain the type and purity of the obtained phases.16,17,19,27 Secondly, increasing amounts of a phase were mixed with the internal standard in different weight fractions (ranging from 2 to 50 wt%; to ensure the reproducibility of the results, we suggest not to use a ZIF wt% lower than 2% when preparing the samples for the calibration curve; see S7†). The detailed procedure is given in section S3.1, and the resulting calibration curves are shown in Fig. S3.†Fig. 1 depicts the calibration curve for sod. The relative sod weight percent (i.e., sod wt%) in the sod/ZrO2 mixture is plotted in the y-axis, whereas the sod/ZrO2 integrated intensities ratio (i.e., I(sod)/I(ZrO2)) is plotted in the x-axis. Each integrated intensity is calculated on the quantification peak (QP), namely, the reflection in the diffractogram with the highest intensity. The sod QP is at 7.36°, and the ZrO2 QP is at 28.20° (2θ, Cu Kα radiation). The QPs for all phases are listed in Table S1.† Thirdly, for each crystal phase, we determined the ratio between the phase integrated intensity and weight %. We then compared this intensity per weight ratio (phase/ZrO2) to a common reference (Al2O3, corundum) to experimentally derive an instrument-independent constant referred to as the reference intensity ratio (RIR, see Table S1† and section S5).30–32Crystal phase quantification based on experimental RIRs
In this work, a calibration procedure was used to generate experimentally derived RIRs subsequently exploited for the investigation of ZIF phases (i.e., sod, dia, ZIF-C, ZIF-EC-1, U12 and ZIF-L). Such experimentally derived RIRs were not previously reported in the literature. Instead, the literature reports the ZIF crystal structure from which a RIR can be calculated. However, a calculated RIR does not account for the experimental conditions (e.g., the absence of gas/liquid molecules in the pores) and could drastically differ from an experimentally derived RIR. In our study, the material is dried at atmospheric pressure and room temperature (ESI† for details). Consequently, the calculated RIR differs from the experimentally derived RIR. This difference is more prominent for the porous sod (the calculated RIR is 10.67,33 and the experimental RIR is 4.5) and less prominent for the dense phases (e.g., dia: the calculated RIR is 1.614,34 and the experimental RIR is 2.8; ZIF-EC-1: the calculated RIR is 1.403, and the experimental RIR is 2.5). For U12, the experimental RIR is 1.7 (since the crystal structure of U12 is not yet reported, there is no calculated RIR available). In Table S1† the calculated and experimental RIRs of the ZIF phases are listed.We used the experimentally derived RIRs to enhance the accuracy of a web application that was suitable for the identification of the main crystalline phase by using the calculated or estimated RIRs. Additionally, we revisited the phase selection criteria (section S6†), which now account for additional diffraction peaks and new ZIF phases. These changes resulted in improved identification and quantification of crystalline phases, from traditional (sod ZIF-8) to recently discovered phases (ZIF-EC-1).
Amorphous content quantification (bi-phasic system)
To quantify the amorphous content mixed with a pure ZIF phase (e.g., the amorphous content in a sample that shows only sod diffraction peaks) we estimated the phase wt% of the pure ZIF phase by using XRD data and its specific calibration curve (ESI† S3.3). Then, we compared this value with the expected phase wt% (i.e., considering the ZIF to be 100% pure and crystalline). If these two values differ, we hypothesise that a part of the material does not contribute to the intensities of the reflection peaks and postulate that the sample is a bi-phasic mixture where a certain proportion of the material is amorphous (e.g., amorphous/pure sod).32,35To verify this hypothesis, we prepared selected protein@ZIF biocomposites that are already reported in the literature.16 BSA is used as a model protein, and it is embedded in a ZIF matrix consisting of sod, dia, ZIF-C or U12. The synthesis protocols are listed in Table S2.†
With FT-IR spectroscopy we verified the formation of the BSA@ZIF composite materials (Fig. S4†).16
For structural analysis of the BSA@ZIFs composites, we measured the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
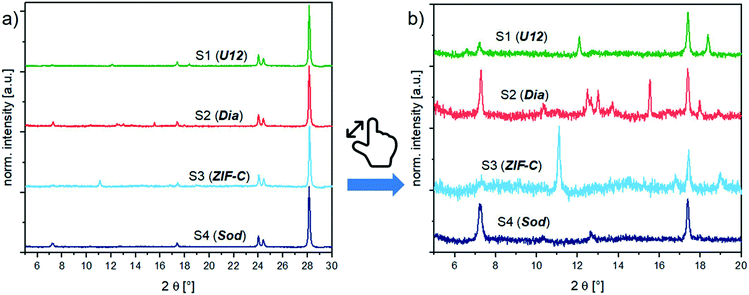
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 weight mixtures of BSA@ZIFs composites and the ZrO2 internal standard (prepared as in the calibration series, section S3.1†) with PXRD. Fig. 2 shows the diffractograms of the biocomposites. Depending on the reaction conditions (i.e., the metal
1 weight mixtures of BSA@ZIFs composites and the ZrO2 internal standard (prepared as in the calibration series, section S3.1†) with PXRD. Fig. 2 shows the diffractograms of the biocomposites. Depending on the reaction conditions (i.e., the metal![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ligand
ligand![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) BSA ratio) and the washing procedure (i.e., water or ethanol), different phases like sod (peaks at 7.36° (110), 10.45° (200) and 12.75° (211)), dia (peaks at 12.5° (002), 13.05° (011), 13.76° (20–2) and 15.57° (21–1)), ZIF-C (a peak at 11.05° (110)) and U12 (peaks at 12.13° and 18.43°) can be obtained.16
BSA ratio) and the washing procedure (i.e., water or ethanol), different phases like sod (peaks at 7.36° (110), 10.45° (200) and 12.75° (211)), dia (peaks at 12.5° (002), 13.05° (011), 13.76° (20–2) and 15.57° (21–1)), ZIF-C (a peak at 11.05° (110)) and U12 (peaks at 12.13° and 18.43°) can be obtained.16
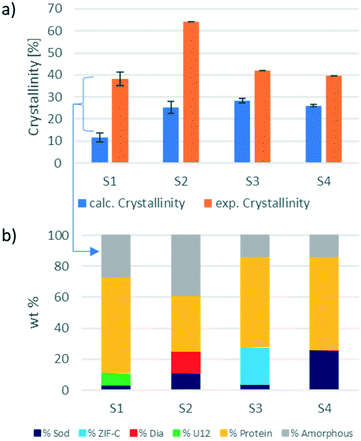
However, if the integrated intensity of the QP of the identified ZIF phase is inserted in the calibration curve, the calculated phase wt% is lower than expected. This confirms the hypothesis that only a portion of the weighed material contributes to the diffraction, and thus a certain amount is amorphous. In biocomposites, the formation of an amorphous content seems to be facilitated by the heterogeneous nucleation from the protein.3,4,36–38 The amount of encapsulated protein can be calculated via the analysis of the protein encapsulation efficiency (EE% determined via the Bradford assay; the details are listed in section S4.3†).16 The EE% and measured gravimetric yield (Fig. S5 and S6,† respectively) were used to calculate the weight of the encapsulated protein in the investigated sample. By subtracting the weight of the protein from the weight of the total biocomposite material, we determine the weight of the ZIF component for each sample. This theoretical ZIF wt% is then compared to the wt% calculated using the web app. The results of this comparison are shown in Fig. 3. For the analysed samples, the amorphous content ranges from 10% (sample 3) up to 35% (sample 2). Consequently, the amorphous phase has a substantial amount of the ZIF biocomposite, and this should not be overlooked as it can alter the expected properties of the material.16,22,39,40
Additionally, this procedure enables the precise evaluation of phase transitions during the processing of the same sample. For example, protein@ZIF-C biocomposites are often synthesized in water; then, when washed with ethanol, a phase transition from ZIF-C to sod can be observed.16 Here, we examined the conversion of BSA@ZIF-C (sample 3) to BSA@sod (sample 4) via ethanol washing. The quantification of the amorphous content in the two samples (15% in both cases) proves that the proportion of the crystalline MOF is maintained during the transition from ZIF-C to sod. In fact, the ZIF-C to sod transition does not involve a sensitive loss of crystalline material (S3: ZIF-C wt% = 24%, sod wt% = 4%; S4: sod wt% = 26%) and supports the hypothesis that ZIF-C can be fully converted to sod.
Web app implementation
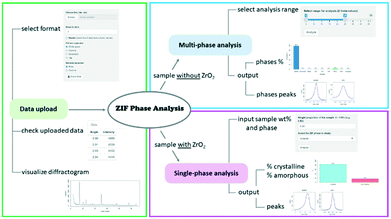
The ZIF Phase Analysis app consists of three sections, which are shown in Fig. 4. The data upload section is used to import the diffraction data (see the video in the ESI†). In this section, it is possible to select multiple data file options such as the number of rows to skip and the column/decimal separators. Once the data are uploaded, it is possible to see the first few rows of the data file and the diffractogram.The multi-phase analysis section is used to analyse samples prepared without the ZrO2 internal standard. In this section, it is possible to select the data analysis range by moving the extremes of the slider input to the desired 2θ values. We recommend using the 6–28° range as it includes all the reference peaks of the detectable crystal phases. The entire 6–39° range should be used if ZrO2 and/or ZnO and/or Zn(OH)2 are of interest as well. The analysis returns the identified phases and their relative proportions in the sample, along with the identified quantification peaks.
The single-phase analysis section is used to analyse the samples prepared with the ZrO2 internal standard. The user is required to input first the crystal phase to be investigated and then the phase wt% of the studied mixture (e.g., phase wt% = 3%; this implies that ZrO2 wt% = 97%). The 2θ values are automatically selected based on the crystal phase selected for analysis. The analysis returns the relative proportion of the crystalline and amorphous material in the sample, as well as the identified quantification peaks for both the crystal phase and internal standard. Researchers are encouraged to contact the authors for suggestions on the implementation of additional ZIF biocomposites to the existing web app portfolio.
Conclusion
We built XRD calibration curves and experimentally determined the reference intensity ratio, RIR, of ZIF phases (sod, dia, ZIF-C, ZIF-L, U12 and ZIF-EC-1) that are commonly obtained when preparing ZIF biocomposites. The XRD calibration curves and experimental RIRs were obtained by mixing the different Zn(mIM)2-based ZIF phases with an internal standard (ZrO2) with different wt% values. The measured experimental RIRs were then used for the crystal phase identification and multiphase analyses. The calibration curves were employed to quantify the amorphous content in the protein@ZIF biocomposite samples. For accurate quantification, an internal standard (ZrO2) was used, and the related diffraction patterns were analysed by a customized web application (ZIF Phase Analysis). By examining different protein@ZIF samples with our ZIF Phase Analysis app we estimated an amorphous content of up to 35%. Thus, 1/3 of the entire amount could significantly contribute to the essential functional properties for biocatalysis or drug delivery (enzymatic activity and release profile, respectively). Here we propose a procedure that requires the preparation of protein@ZIF samples and the related standards (controlled mixtures of phases with a ZrO2 internal standard). The XRD patterns can then be uploaded in our ZIF Phase Analysis app. The freely available ZIF Phase Analysis app will promptly analyse the data providing an accurate evaluation of the different ZIF phase components, including the amount of the amorphous phase. As protein@ZIF materials is a burgeoning research field approached by scientists with diverse expertise (e.g., biology, enzymology, and biomedicine), this app will especially support researchers with limited knowledge in crystallography. Thus, the here reported procedure combined with the ZIF phase analysis web application will progress research of ZIF biocomposites for multi-disciplinary applications including drug delivery or biocatalysis.16,22,40Author contributions
M. R. Hafner: design of methodology, experiment implementation, data collection and sorting, discussion of the results and writing of the manuscript; L. Villanova: programming, software development, design and testing of web app, design and launch of website, and revision of the manuscript; F. Carraro: conceptualisation, design of methodology, supervision of the project, discussion of the results and writing of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Prof. Paolo Falcaro for supporting this project. We acknowledge the European Union’s Horizon 2020 Programme (FP/2014–2020)/ERC Grant Agreement no. 771834 POPCRYSTAL and the TU Graz for the Lead Project (LP-03).References
- H.-C. Zhou, J. R. Long and O. M. Yaghi, Chem. Rev., 2012, 112, 673–674 CrossRef CAS PubMed.
- H. Furukawa, K. E. Cordova, M. O'Keeffe and O. M. Yaghi, Science, 2013, 341, 1230444 CrossRef PubMed.
- K. Liang, R. Ricco, C. M. Doherty, M. J. Styles, S. Bell, N. Kirby, S. Mudie, D. Haylock, A. J. Hill, C. J. Doonan and P. Falcaro, Nat. Commun., 2015, 6, 7240 CrossRef CAS PubMed.
- E. Astria, M. Thonhofer, R. Ricco, W. Liang, A. Chemelli, A. Tarzia, K. Alt, C. E. Hagemeyer, J. Rattenberger, H. Schroettner, T. Wrodnigg, H. Amenitsch, D. M. Huang, C. J. Doonan and P. Falcaro, Mater. Horiz., 2019, 6, 969–977 RSC.
- F. C. Herbert, S. S. Abeyrathna, N. S. Abeyrathna, Y. H. Wijesundara, O. R. Brohlin, F. Carraro, H. Amenitsch, P. Falcaro, M. A. Luzuriaga, A. Durand-Silva, S. D. Diwakara, R. A. Smaldone, G. Meloni and J. J. Gassensmith, Nat. Commun., 2021, 12, 2202 CrossRef CAS PubMed.
- Q. Qiu, H. Chen, Y. Wang and Y. Ying, Coord. Chem. Rev., 2019, 387, 60–78 CrossRef CAS.
- A. Poddar, J. J. Conesa, K. Liang, S. Dhakal, P. Reineck, G. Bryant, E. Pereiro, R. Ricco, H. Amenitsch, C. Doonan, X. Mulet, C. M. Doherty, P. Falcaro and R. Shukla, Small, 2019, 15, 1902268 CrossRef PubMed.
- Y. Zhang, F. Wang, E. Ju, Z. Liu, Z. Chen, J. Ren and X. Qu, Adv. Funct. Mater., 2016, 26, 6454–6461 CrossRef CAS.
- R. Riccò, W. Liang, S. Li, J. J. Gassensmith, F. Caruso, C. Doonan and P. Falcaro, ACS Nano, 2018, 12, 13–23 CrossRef PubMed.
- S. Li, M. Dharmarwardana, R. P. Welch, C. E. Benjamin, A. M. Shamir, S. O. Nielsen and J. J. Gassensmith, ACS Appl. Mater. Interfaces, 2018, 10, 18161–18169 CrossRef CAS PubMed.
- A. Poddar, S. Pyreddy, F. Carraro, S. Dhakal, A. Rassell, M. R. Field, T. S. Reddy, P. Falcaro, C. M. Doherty and R. Shukla, Chem. Commun., 2020, 56, 15406–15409 RSC.
- Y. Feng, H. Wang, S. Zhang, Y. Zhao, J. Gao, Y. Zheng, P. Zhao, Z. Zhang, M. J. Zaworotko, P. Cheng, S. Ma and Y. Chen, Adv. Mater., 2019, 31, 1805148 CrossRef PubMed.
- M. A. Luzuriaga, C. E. Benjamin, M. W. Gaertner, H. Lee, F. C. Herbert, S. Mallick and J. J. Gassensmith, Supramol. Chem., 2019, 0, 1–6 CAS.
- M. D. J. Velásquez-Hernández, R. Ricco, F. Carraro, F. T. Limpoco, M. Linares-Moreau, E. Leitner, H. Wiltsche, J. Rattenberger, H. Schröttner, P. Frühwirt, E. M. Stadler, G. Gescheidt, H. Amenitsch, C. J. Doonan and P. Falcaro, CrystEngComm, 2019, 21, 4538–4544 RSC.
- N. K. Maddigan, O. M. Linder-Patton, P. Falcaro, C. J. Sumby, S. G. Bell and C. J. Doonan, ACS Appl. Mater. Interfaces, 2021, 13, 51867–51875 CrossRef CAS PubMed.
- F. Carraro, M. D. J. Velásquez-Hernández, E. Astria, W. Liang, L. Twight, C. Parise, M. Ge, Z. Huang, R. Ricco, X. Zou, L. Villanova, C. O. Kappe, C. Doonan and P. Falcaro, Chem. Sci., 2020, 11, 3397–3404 RSC.
- W. Liang, R. Ricco, N. K. Maddigan, R. P. Dickinson, H. Xu, Q. Li, C. J. Sumby, S. G. Bell, P. Falcaro and C. J. Doonan, Chem. Mater., 2018, 30, 1069–1077 CrossRef CAS.
- M. Jian, B. Liu, R. Liu, J. Qu, H. Wang and X. Zhang, RSC Adv., 2015, 5, 48433–48441 RSC.
- Y. Lo, C. H. Lam, C.-W. Chang, A.-C. Yang and D.-Y. Kang, RSC Adv., 2016, 6, 89148–89156 RSC.
- Z.-X. Low, J. Yao, Q. Liu, M. He, Z. Wang, A. K. Suresh, J. Bellare and H. Wang, Cryst. Growth Des., 2014, 14, 6589–6598 CrossRef CAS.
- P. F. Rosen, J. J. Calvin, M. S. Dickson, A. D. Katsenis, T. Friščić, A. Navrotsky, N. L. Ross, A. I. Kolesnikov and B. F. Woodfield, J. Chem. Thermodyn., 2019, 136, 160–169 CrossRef CAS.
- X. Wu, H. Yue, Y. Zhang, X. Gao, X. Li, L. Wang, Y. Cao, M. Hou, H. An, L. Zhang, S. Li, J. Ma, H. Lin, Y. Fu, H. Gu, W. Lou, W. Wei, R. N. Zare and J. Ge, Nat. Commun., 2019, 10, 5165 CrossRef PubMed.
- A. Poddar, S. Pyreddy, F. Carraro, S. Dhakal, A. Rassell, M. R. Field, T. S. Reddy, P. Falcaro, C. M. Doherty and R. Shukla, Chem. Commun., 2020, 56, 15406–15409 RSC.
- F. Oviedo, Z. Ren, S. Sun, C. Settens, Z. Liu, N. T. P. Hartono, S. Ramasamy, B. L. DeCost, S. I. P. Tian, G. Romano, A. Gilad Kusne and T. Buonassisi, npj Comput. Mater., 2019, 5, 1–9 CrossRef CAS.
- R Core Team, R: A language and environment for statistical computing, R Foundation for Statistical Computing, Vienna, Austria, 2019 Search PubMed.
- W. Chang, J. Cheng, J. J. Allaire, Y. Xie and J. McPherson, shiny: Web Application Framework for R, 2020 Search PubMed.
- M. Ge, Y. Wang, F. Carraro, W. Liang, M. Roostaeinia, S. Siahrostami, D. M. Proserpio, C. Doonan, P. Falcaro, H. Zheng, X. Zou and Z. Huang, Angew. Chem., 2021, 60, 11391–11397 CrossRef CAS PubMed.
- R. Chen, J. Yao, Q. Gu, S. Smeets, C. Baerlocher, H. Gu, D. Zhu, W. Morris, O. M. Yaghi and H. Wang, Chem. Commun., 2013, 49, 9500 RSC.
- S. A. Basnayake, J. Su, X. Zou and K. J. Balkus, Inorg. Chem., 2015, 54, 1816–1821 CrossRef CAS PubMed.
- F. H. Chung, J. Appl. Crystallogr., 2018, 51, 789–795 CrossRef CAS.
- X. Zhou, D. Liu, H. Bu, L. Deng, H. Liu, P. Yuan, P. Du and H. Song, Solid Earth Sci., 2018, 3, 16–29 CrossRef.
- C. R. Hubbard and R. L. Snyder, Powder Diffr., 1988, 3, 74–77 CrossRef CAS.
- O. Shekhah, R. Swaidan, Y. Belmabkhout, M. du Plessis, T. Jacobs, L. J. Barbour, I. Pinnau and M. Eddaoudi, Chem. Commun., 2014, 50, 2089 RSC.
- Q. Shi, Z. Chen, Z. Song, J. Li and J. Dong, Angew. Chem., Int. Ed., 2011, 50, 672–675 CrossRef CAS PubMed.
- C. R. Hubbard, E. H. Evans and D. K. Smith, J. Appl. Crystallogr., 1976, 9, 169–174 CrossRef.
- K. Liang, C. Carbonell, M. J. Styles, R. Ricco, J. Cui, J. J. Richardson, D. Maspoch, F. Caruso and P. Falcaro, Adv. Mater., 2015, 27, 7293–7298 CrossRef CAS PubMed.
- X. Lian, Y. Fang, E. Joseph, Q. Wang, J. Li, S. Banerjee, C. Lollar, X. Wang and H.-C. Zhou, Chem. Soc. Rev., 2017, 46, 3386–3401 RSC.
- A. F. Ogata, A. M. Rakowski, B. P. Carpenter, D. A. Fishman, J. G. Merham, P. J. Hurst and J. P. Patterson, J. Am. Chem. Soc., 2020, 142, 1433–1442 CrossRef CAS PubMed.
- T. D. Bennett, A. L. Goodwin, M. T. Dove, D. A. Keen, M. G. Tucker, E. R. Barney, A. K. Soper, E. G. Bithell, J.-C. Tan and A. K. Cheetham, Phys. Rev. Lett., 2010, 104, 115503 CrossRef PubMed.
- C. Liu, J. Wang, J. Wan, Y. Cheng, R. Huang, C. Zhang, W. Hu, G. Wei and C. Yu, Angew. Chem., Int. Ed., 2020, 59, 3630–3637 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d2ce00073c |
| This journal is © The Royal Society of Chemistry 2022 |