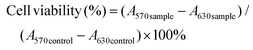Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Development of curcumin-loaded zein nanoparticles for transport across the blood–brain barrier and inhibition of glioblastoma cell growth†
Huaiying
Zhang
a,
Winant L.
van Os
b,
Xiaobo
Tian
c,
Guangyue
Zu
a,
Laís
Ribovski
 a,
Reinier
Bron
a,
Jeroen
Bussmann
d,
Alexander
Kros
a,
Reinier
Bron
a,
Jeroen
Bussmann
d,
Alexander
Kros
 b,
Yong
Liu
a and
Inge S.
Zuhorn
b,
Yong
Liu
a and
Inge S.
Zuhorn
 *a
*a
aDepartment of Biomedical Engineering, University of Groningen, University Medical Center Groningen, Groningen, The Netherlands. E-mail: i.zuhorn@umcg.nl
bDepartment of Supramolecular and Biomaterials Chemistry, Leiden Institute of Chemistry, Leiden University, Leiden, The Netherlands
cDepartment of Analytical Biochemistry and Interfaculty Mass Spectrometry Center, Groningen Research Institute of Pharmacy, University of Groningen, Groningen, The Netherlands
dDivision of BioTherapeutics, Leiden Academic Centre for Drug Research, Leiden University, Leiden, The Netherlands
First published on 28th January 2021
Abstract
Glioblastoma (GBM) is a devastating primary brain tumor resistant to conventional therapies. A major obstacle to GBM treatment is the blood–brain barrier (BBB), or blood–glioma barrier, which prevents the transport of systemically administered (chemotherapeutic) drugs into the tumor. This study reports the design of dodecamer peptide (G23)-functionalized polydopamine (pD)-coated curcumin-loaded zein nanoparticles (CUR-ZpD-G23 NPs) that efficiently traversed the BBB, and delivered curcumin to glioblastoma cells. The NPs enhanced the cellular uptake of curcumin by C6 glioma cells compared to free curcumin, and showed high penetration into 3D tumor spheroids. Functionalization of the NPs with G23 stimulated BBB crossing and tumor spheroid penetration. Moreover, the NPs markedly inhibited proliferation and migration and induced cell death in liquid and soft agar models of C6 glioma cell growth. Fluorescence microscopy and flow cytometry studies showed that the CUR-ZpD-G23 NPs increased cellular ROS production and induced apoptosis of C6 glioma cells. Following in vivo intravenous injection in zebrafish, ZpD-G23 NPs demonstrated the ability to circulate, which is a first prerequisite for their use in targeted drug delivery. In conclusion, zein-polydopamine-G23 NPs show potential as a drug delivery platform for therapy of GBM, which requires further validation in in vivo glioblastoma models.
Introduction
Limited uptake of chemotherapeutic drugs and chemo-resistance of glioma cells result in poor treatment options for glioblastoma (GBM) patients.1,2 The inadequate delivery of chemotherapeutics to GBM is largely caused by the presence of the blood–brain barrier (BBB).3 The BBB is a physiological barrier at the vascular-central nervous system interface, which is formed by a continuous layer of tight junction-expressing endothelial cells, supported by pericytes and astrocytes.4 It restricts the paracellular diffusion of macromolecules across the BBB. Temporary disruption of tight junction integrity to deliver macromolecules into the brain or intracranial injection for site-specific drug delivery into the brain are possible routes for drug delivery to the brain.5 However, these methods can be difficult to control spatiotemporally, significantly enhance the risk of medical complications and may be problematic for chronic use.At the BBB the process of selective transcellular transport of target molecules via carrier-mediated transport, cation-induced absorptive transcytosis, or receptor-mediated transcytosis, provide gateways for the delivery of nanoparticles into the brain.6,7
Many kinds of nanoparticles (NPs) have recently been employed to enhance the delivery of existing and novel therapeutics across the BBB.6,8–10 Among them, biodegradable NPs from natural polymers, such as protein-based polymers, have attracted remarkable attention as potential drug delivery carriers for their low cost and low toxicity.11 Zein, an alcohol-soluble protein, extracted from corn, has emerged as an ideal drug delivery system because of its excellent biocompatibility and biodegradability. Zein has been extensively investigated for the encapsulation of bioactive compounds (e.g. vitamin D3, resveratrol, quercetin and antimicrobials)12–16 because of its easy self-assembly into NPs, and sustained drug release capability.17 However, zein nanoparticles have poor physical stability at neutral pH or high salinity, and poor redispersibility after centrifugation or lyophilization. In addition, weak chemical reactivity appears to be a major problem that limits their further applications in the food and pharmaceutical field. Recently, it was proposed that surface modification of zein NPs with a layer of polydopamine (PDA) enhances their hydrophilicity, colloidal stability, biocompatibility and provides a conjugation site for bioactive functional groups.18,19 Moreover, polydopamine (pD) coating through oxidative self-polymerization of dopamine at neutral or a weak alkaline condition can function as pH-sensitive gatekeeper to control the release of drug molecules from the NPs in response to a pH-stimulus.20,21
Curcumin (CUR) is a polyphenol that is widely used in medicine for its pleiotropic anti-inflammatory, antimicrobial and anticancer activities.22–26 However, the use of curcumin is greatly hindered by its low bioavailability in both plasma and tissue because of its poor water solubility, poor absorption, rapid metabolism and rapid system elimination.27,28 Here, we developed self-assembled zein nanoparticles as a delivery vehicle for curcumin. The zein–curcumin NPs were colloidally stabilized by coating with a polydopamine (PDA) layer, which also improved their hydrophilicity, and reactivity with nucleophilic compounds allowing for further modification via Michael addition or Schiff base reactions. The PDA-coated zein-curcumin (CUR-ZpD) NPs were functionalized with the G23 dodecamer peptide. The ganglioside GM1-binding G23 peptide has been shown to mediate BBB crossing of polymersomes, RNA-binding proteins, and iron oxide nanoparticles.29–34 Moreover, effective delivery of BACE1-siRNA and doxorubicin to the brain has been reported for G23-functionalized nanoparticles, resulting in a reduced plaque load and improvement of cognitive function in an Alzheimer's disease model and a reduction in tumor volume in a glioblastoma model, respectively.31,32 The CUR-ZpD NPs were optimized to display a size <100 nm, low PDI, efficient curcumin loading and pH-dependent release. The G23-functionalized PDA-coated zein-curcumin (CUR-ZpD-G23) NPs showed great capacity to cross an in vitro BBB model. Using coumarin-6 as the fluorescent probe, the NPs showed high cellular uptake by C6 glioma cells and significant penetration into 3D tumor spheroids. In addition, we evaluated the effect of curcumin-loaded NPs on the proliferation, apoptosis, migration and cellular ROS production of C6 glioma cells.
Materials and methods
Materials
Zein protein, curcumin (purity >98%), dopamine hydrochloride (purity >98%), coumarin-6, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), (4-Amidinophenyl)-6-indolecar-bamidine dihydrochloride (DAPI) and CM-H2DCFDA (5-(and-6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate) were purchased from Sigma-Aldrich. G23 peptide with a C-terminal cysteine was synthesized by GL Biochem Ltd (Shanghai, China) with a purity of 93.6% as analyzed by HPLC and mass spectrometry. MAL-PEG5000-NH2TFA was purchased from JenKem technology Co., Ltd, (Beijing, China).Preparation of ZpD NPs, CUR-ZpD NPs and CUR-ZpD-G23 NPs
The preparation of curcumin (CUR)-loaded pD-coated zein nanoparticles (CUR-ZpD NPs) was based on a modified method of phase separation.35 Shortly, zein and CUR were dissolved in ethanol (80% v/v) at different weight ratios. Subsequently, 1 ml of the mixed solution was rapidly dispersed into 19 mL dopamine hydrochloride solution in Tris-HCl buffer (10 mM, pH 8.5) and stirred for 12 h at room temperature. Dopamine concentration was fixed at 0.5 mg mL−1 unless specified otherwise. The CUR-ZpD NPs were collected by centrifugation (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 30 min) and washed with deionized water for three times. For the preparation of Z-pD NPs, the same procedure was followed without CUR.
000g for 30 min) and washed with deionized water for three times. For the preparation of Z-pD NPs, the same procedure was followed without CUR.
For G23 functionalization of CUR-ZpD NPs, firstly, G23-cys was conjugated to MAL-PEG5K-NH2 in Tris-HCl buffer (10 mM, pH 7.4) for 3 h via a maleimide–thiol reaction. Subsequently, 2.5 mg ml−1 CUR-ZpD NPs were resuspended in Tris-HCl buffer (10 mM, pH 8), which contained 5 mg mL−1 PEG-G23. After 12 h stirring at room temperature, particles were collected by centrifugation and washed with deionized water.
Characterization of the NPs
Dynamic laser scattering (DLS) and zeta potential measurements of blank and CUR-loaded NPs were performed on a commercial laser light scattering instrument (Malvern ZEN3690, Malvern Instruments) at 25 °C and 90° scattering angle. The surface morphology of the nanoparticles was observed using a transmission electron microscope (Philips CM120) equipped with a 4k CCD camera at. Carbon film coated 200 mesh copper grids (Electron Microscopy Sciences) were glow-discharged in air. Fourier Transform Infrared Spectroscopy (FTIR) was carried out on a spectrometer (Tensor 27, Bruker, Germany), where the scanning range was 400–4000 cm−1, and the resolution was 4 cm−1.Drug loading and release
The drug loading (DL) and entrapment efficiency (EE) of curcumin were determined by high-performance liquid chromatography (HPLC). The HPLC system (Shimadzu, Kyoto, Japan) was equipped with a SIL-20AC autosampler, a LC-20AT pump and a SPD-20A absorbance detector. The 50 μL samples were injected and separated on a Vydac RP-C18 column (250 mm × 4.6 mm i.d., 5 μm particles, 300 Å pore size, Grace Vydac, Lokeren, Belgium) with a 20 or 30 min gradient of 2–90% acetonitrile in water/0.1% formic acid at a flow rate of 1 mL min−1. The levels of curcumin were quantified by calibration graph and dilution factor. The encapsulation efficiency (EE, %) and drug loading (DL, %) were calculated based on the following eqn (1) and (2), respectively.| EE (%) = Weight of CUR in NPs/Total weight of CUR × 100% | (1) |
| DL (%) = Weight of CUR in NPs/Total weight of the NPs × 100% | (2) |
To study the drug release, free curcumin and CUR-loaded NPs with an equivalent amount of CUR (500 μg) were placed in a dialysis tube (cut off 3.5 kDa) in 30 ml PBS containing 0.5% (w/v) Tween 80 at different pH values (7.4 or 5.0) in a 37 °C water bath due to the poor water solubility of free curcumin.36 At different intervals, 1 mL of external solution was withdrawn and replaced by 1 mL of fresh PBS solution with the same pH value. The amount of CUR in the samples was determined using HPLC. The cumulative release was calculated using the following eqn (3).
 | (3) |
Cell culture
Human cerebral microvascular endothelial (hCMEC/D3) cells were cultured in EBM-2 supplemented with 5% FBS as previously described.34 Rat C6 glioma cells were cultured in DMEM supplemented with 10% FBS and 1% penicillin–streptomycin at 37 °C in a humidified environment with 5% CO2.Cytotoxicity assay
Cell viability was measured using the MTT colorimetric assay. hCMEC/D3 cells (passage 30–38) were seeded at a density of 1 × 105 cells per cm2 in a 96-well plate precoated with 150 μg ml−1 rat tail collagen type-I, and grown for five days in 100 μl of culture medium. The C6 glioma cells were seeded in 96-well plates at a density of 5 × 103 cells per well and incubated for 24 h. Subsequently, the medium was replaced with fresh medium containing various NPs (either blank or CUR-loaded NPs) or CUR at different concentrations with further incubation for 24 h. 50 μL of a 0.5 mg ml−1 solution of MTT reagent dissolved in PBS was added to each well and the plates were incubated for an additional 3 h at 37 °C. After 3 h incubation, the medium containing MTT was removed and 200 μL of dimethyl sulfoxide (DMSO) was added to each well to dissolve the MTT formazan crystals formed. The optical density at 570 nm and 630 nm was measured in a microtiter plate reader SpectraMax M3 (Molecular Devices) after shaking for 15 min. Untreated cells were used as control. Cell viability was calculated as described in the following equation:where A570 is the absorbance at 570 nm and A630 is the absorbance at 630 nm.
Transcytosis assay
In order to measure the transendothelial transport of NPs a filter-free in vitro BBB model was used, as previously described.34 In brief, hCMEC/D3 cells (1 × 105 cells per cm2) were seeded onto collagen gels in a 24-well plate, and grown for five days in 1 ml of culture medium. The cell monolayers were washed once with prewarmed HBSS. Subsequently, 500 μl of coumarin-6 labelled ZpD NPs or ZpD-G23 NPs (1 μg ml−1) diluted in EBM-2 was added apically to the cells and incubated at 37 °C for 2, 4 and 8 h. Then, the apical medium was collected and the cells were washed with 500 μl prewarmed HBSS to collect residual NPs (apical fraction). The collagen gels were digested in 200 μl 0.25% (w/v) collagenase A in HBSS for 90 min at 37 °C followed by centrifugation at 200g for 5 min to pellet cells. The supernatant was collected and mixed with 400 μl EBM-2 (basolateral fraction). The cell pellet was soaked in 500 μl of ultrapure water for 10 min, and subsequently mixed with 500 μl of EBM-2 (cellular fraction). The three fractions were lyophilized and redissolved in 1 ml 50% DMSO and measured in triplicate using Fluostar-Optima microplate reader (BMG Labtech) with Ex/Em at 485 nm/520 nm. The fluorescence in the distinct apical, cellular, and basolateral fractions of cells without treatment was subtracted from the measured intensity values. The percentage of NPs fluorescence associated with the apical, cellular and basolateral fraction was expressed relative to the total fluorescent content present in all three fractions collectively.NP uptake in 2D C6 glioma cell cultures and drug penetration into 3D C6 glioma tumor spheroids
To trace the cellular uptake of the NPs, a fluorescent marker (coumarin-6) was encapsulated into NPs, using the same method as for NP loading with CUR. C6 glioma cells were seeded in a 12-well plate at a density of 5 × 104 cells per well and incubated for 24 h. The medium was then replaced with fresh medium containing coumarin-6 and C6-loaded ZpD NPs with an equal amount of coumarin-6 (0.1 μg ml−1) for 4 h. Cells were fixed using 3.7% paraformaldehyde and the cell nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI). Finally, the cells were washed with PBS (pH 7.4, three times), and directly subjected to fluorescence imaging using confocal laser scanning microscopy (CLSM) (Leica TCS SP8, Germany). The average fluorescence intensity of cells was measured using ImageJ software, and 100–200 cells (9 images in total) were selected from three independent experiments to obtain a mean value.C6 glioma cells (2 × 105 cells per well) were seeded in 6-well plates, which were coated with 1.5 ml of a sterile 2% agar, and cultured for approximately 4 days in a humidified incubator. When the diameter of tumor spheroids reached approximately 200 μm, tumor spheroids were treated with coumarin-6, C6-ZpD NPs and C6-ZpD-G23 NPs (0.5 μg mL−1 coumarin-6) for 4 h. Then, the culture medium was taken out and the tumor spheroids were suspended in 100 μl of PBS, after being washed with PBS twice, and transferred to a glass slide. Optical tumor spheroid sections were imaged by CLSM from top to bottom with 5 μm per section. 12 spheroids were analyzed per condition from 3 independent experiments.
Detection of reactive oxygen species (ROS)
Intracellular accumulation of ROS was detected using the ROS-sensitive probe CM-H2DCFDA. Briefly, C6 glioma cells were seeded at a density of 1 × 105 cells per well in a 12-well cell culture plate and let to adhere for 24 h. Then the C6 glioma cells were stimulated with CUR or CUR loaded NPs (CUR = 10 μg ml−1) for 4 h at 37 °C and then washed with PBS. After that, PBS containing 2.5 μM of CM-H2DCFDA was added at 37 °C for another 30 min and then washed with PBS twice. The cells were observed under the fluorescence microscope (Leica, Germany), using appropriate filters (Ex/Em: 485/520). The average fluorescence intensity of cells was measured using ImageJ software, and 1000–2000 cells (9 images in total) were selected from three independent experiments to obtain a mean value.Cell apoptosis assay
Apoptosis in C6 glioma cells was detected by annexin V and PI staining and flow cytometry (FCM). The cells (2 × 105 cells per well) were seeded into 6-well plates and incubated at 37 °C for 24 h. Then, the cells were treated with free curcumin, ZpD NPs, CUR-ZpD NPs and CUR-ZpD-G23 NPs for another 24 h, at a curcumin concentration of 1 μg ml−1. After washing three times with PBS, cells were stained with annexin V-fluorescein isothiocyanate (FITC-V) and propidium iodide (PI) and fluorescence was assessed by FCM instrument (BD FACSVerse) using 488 nm laser, Blue-D detector (PI: 586/42 filter) and Blue-E detector (FITC: 527/32 filter).Cell migration assay
Monolayers of confluent C6 glioma cells in 6-well plates were scratched with a 200 μl sterile pipette tip to create a wound. Cells were washed with PBS and then incubated in DMEM with 2% FBS plus free curcumin, ZpD NPs, CUR-ZpD NPs and CUR-ZpD-G23 NPs (curcumin was 1 μg ml−1) for 24 h. Images were obtained by light microscopy. The scratch area was calculated through ImageJ software, after which the migration of treated cells was compared to that of untreated cells and expressed as percentage.Soft agar colony assay
C6 glioma cells (2 × 103) were resuspended in 2 mL of DMEM supplemented with 10% FBS and 0.5% agarose at 37 °C. Cell suspensions were plated on top of a base layer comprised of 2 ml of DMEM+ 10% FBS and 0.8% agar in each well of a 6-well plate. Cells were then covered in 2 ml of complete medium and incubated for 2 days. Fresh medium containing curcumin, CUR-ZpD NPs or CUR-ZpD-G23 NPs (curcumin 5 μg ml−1) was added and incubated with the cells for another 7 days. Medium was replaced every 72 h. Following treatment, MTT (1 mg ml−1) was added and the number of C6 glioma cell colonies was counted using a light microscope.Colony forming assay
C6 glioma cells (1 × 105) were plated in 6-well plates and incubated overnight, then treated for 24 h with curcumin, CUR-ZpD NPs or CUR-ZpD-G23 NPs (curcumin 5 μg ml−1). Cells were then gently washed, trypsinized, and replated in 35 mm dishes at 500 cells per dish and maintained for an additional 7 days. Cell growth was estimated by colony counting followed by MTT assay.37,38 The percentage of colony forming efficiency was calculated from MTT absorbance values (at 570 nm and 630 nm), with viability of untreated cells set at 100%.Intravascular quantification of ZpD and G23-ZpD nanoparticles in zebrafish larvae
For in vivo visualization of ZpD and G23-ZpD nanoparticles, the NPs were covalently labeled with Cy3 (Lumiprobe GmbH (Germany). To this end, Cy3 amine (1 mg ml−1; 100 μl tris-HCl buffer) was added to ZpD NPs in the absence or presence of PEG-G23 (see ‘preparation of ZpD NPs’ above), respectively and kept stirring overnight. Particles were collected by centrifugation and washed with deionized water. The labeling efficiency was determined by Fluostar-Optima microplate reader (BMG Labtech) with a calibration curve of Cy3 amine in DMSO at the concentration of 0.05, 0.1, 0.2, 0.4, 0.8 and 1.6 μg mL−1. Fluorescence emission spectra of Cy3 from 550 to 700 nm were recorded at room temperature with an excitation wavelength at 540 nm. The excitation and emission band slits were 2 nm.Zebrafish (Danio rerio, Tg(mpeg:GFP)gl22 (ref. 39) or Tg(kdrl:GFP)s843 (ref. 40)), in compliance with the directives of the local animal welfare committee of Leiden University, were maintained and handled according to the guidelines from the Zebrafish Model Organism Database (http://zfin.org). Fertilization was performed by natural spawning at the beginning of the light period, and eggs were raised at 28.5 °C in egg water (60 g mL−1 Instant Ocean sea salts). Prior to injection, zebrafish embryos were embedded and anesthetized in 0.4% agarose containing 0.01% tricaine. Free Cy3-NH2 in MilliQ H2O (300 μM); 3 mg ml−1 Cy3-ZpD and 2 mg ml−1 Cy3-ZpD-G23 (both 300 μM Cy3) were injected with 1 nL volume in the duct of Cuvier at 3.5 days post fertilization (dpf) as described previously.41 Embryos were randomly picked from a dish of 10–20 successfully injected embryos (exclusion criteria were: no backward translocation of erythrocytes after injection and/or damage to the yolk ball). Fish were imaged two hours post injection. Confocal z-stacks were captured on a Leica TCS SP8 confocal microscope, using a 10× air objective (HCX PL FLUOTAR) or a 40× water-immersion objective (HCX APO L), using Ex/Em 488/500–520 nm for GFP and 532/550–570 nm for Cy3 fluorescence detection. For lateral view of whole embryo, four overlapping z-stacks were captured to cover the complete embryo. Laser intensity, gain, and offset settings were identical between stacks and sessions. Images were processed using the Fiji distribution of ImageJ (ImageJ 2.0.0-rc-69/1.52p; Java 1.8.0_172 [64-bit]). To enhance visualization of the biodistribution in lateral 10× images, Cy3 channel ‘Diplay Range Max’ was set from 255 to 126, equally for each fish. Quantification of Cy3 in the dorsal aorta (n = 3 larva per injection group) was performed by fluorescence intensity measurement of sum of four slices (axial resolution respectively 2.4 μm, z-step 1.2 μm) of three regions of interest (ROIs) (81.2 μm2) per larva (Fig. S5 and Table S1†). Averages of three ROIs per fish were calculated.
Statistical analysis
Experiments were repeated at least 3 times. Statistical analyses were performed using the GraphPad Prism Software 8.0 (La Jolla, CA, USA) and data are expressed as mean values ± standard deviation (SD). After having verified normal distribution of data (one-way ANOVA test), the differences between the median values were evaluated using a pairwise comparison through the Student's t-test where the minimal p value was set at 0.05.Results and discussion
Optimization of curcumin-loaded polydopamine (pD)-coated zein nanoparticles (CUR-ZpD NPs)
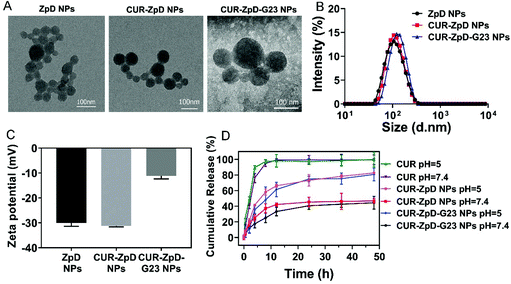
Polymerized dopamine (pD) is known to deposit on a nanoparticle surface and form a durable layer under an oxidative condition (pH8.5). The catechol and anthracene fractions on the surface of pD allow amine-terminated functional ligand incorporation through Schiff base reaction under alkaline condition.21 Fig. S1† shows the influence of dopamine concentration on the size and polydispersity (PDI) of zein NPs. Compared to pure zein particles, the dispersibility of particles increased after coating with polydopamine as was revealed by a lower PDI. The size of pD-coated NPs remained constant up to a concentration of 0.5 mg ml−1 dopamine, but increased significantly when the dopamine concentration increased to 1 mg ml−1. Therefore, 0.25 mg ml−1 and 0.5 mg ml−1 dopamine were used for the further optimization of curcumin-loaded pD-coated zein NPs. As shown in Table 1, the zein concentration was varied from 5 to 40 mg ml−1 to prepare nanoparticles. The resulting pD-coated zein NPs all showed a good dispersibility with low PDI (between 0.10 and 0.24) and a strong negative surface charge (between −15 mV and −36 mV), which was due to the deprotonation of the phenolic hydroxyl groups. The size of NPs increased from 53 nm to 140 nm and from 56 to 150 nm with an increasing concentration of zein when the concentration of dopamine was fixed at 0.25 mg ml−1 and 0.5 mg ml−1, respectively. The sample yield increased with an increasing zein concentration. In addition, the yields of NPs prepared with a dopamine concentration of 0.5 mg ml−1 was higher than that prepared with 0.25 mg ml−1 dopamine, reaching almost 78% when the concentration of zein was 20 mg ml−1. In the following drug encapsulation study, NPs were prepared with 20 mg ml−1 zein and coated using a 0.5 mg ml−1 dopamine solution. The weight ratio of zein to the hydrophobic drug molecule curcumin (CUR) was varied from 1 to 100 (see Table 2). Even though the loading efficiency (DL%) increased with an increasing CUR concentration, a corresponding decrease in the encapsulation efficiency (EE%) was also observed. In addition, the size and PDI of CUR-loaded NPs remained constant when NPs were prepared with zein![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CUR ratios from 5
CUR ratios from 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 100
1 to 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Therefore, to maximize EE% and DL% at a low dose of CUR, a zein
1. Therefore, to maximize EE% and DL% at a low dose of CUR, a zein![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CUR weight ratio of 5 was used to prepare CUR-loaded pD-coated zein (CUR-ZpD) NPs for our experiments.
CUR weight ratio of 5 was used to prepare CUR-loaded pD-coated zein (CUR-ZpD) NPs for our experiments.
| Dopamine (mg ml−1) | Zein (mg ml−1) | Size (nm) | PDI | Zeta potential | Yield (%) |
|---|---|---|---|---|---|
| 0.5 | 40 | 150.5 ± 1.6 | 0.10 ± 0.02 | −15.2 ± 0.2 | 79 ± 4 |
| 0.5 | 20 | 106.2 ± 0.4 | 0.13 ± 0.02 | −30.5 ± 1.3 | 78 ± 4 |
| 0.5 | 10 | 56.7 ± 2.3 | 0.17 ± 0.04 | −32.9 ± 1.2 | 46 ± 4 |
| 0.5 | 5 | 63.1 ± 0.6 | 0.24 ± 0.01 | −33.2 ± 0.8 | 44 ± 6 |
| 0.25 | 40 | 140.8 ± 0.8 | 0.11 ± 0.01 | −24.4 ± 0.3 | 56 ± 3 |
| 0.25 | 20 | 89.6 ± 0.3 | 0.15 ± 0.003 | −35.9 ± 0.4 | 51 ± 4 |
| 0.25 | 10 | 53.2 ± 0.4 | 0.14 ± 0.004 | −32.7 ± 0.2 | 36 ± 5 |
| 0.25 | 5 | 53.2 ± 0.5 | 0.24 ± 0.01 | −32.6 ± 0.8 | 36 ± 3 |
Zein![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Cur (weight ratio) Cur (weight ratio) |
Size (nm) | PDI | Zeta potential | EE (%) | DL (%) |
|---|---|---|---|---|---|
100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
102.3 ± 1.5 | 0.13 ± 0.02 | −39.3 ± 1.8 | 94.4 | 0.56 |
50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
117.5 ± 0.4 | 0.10 ± 0.01 | −44.8 ± 1.6 | 92.1 | 1.29 |
25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
117.0 ± 1.0 | 0.12 ± 0.01 | −38.7 ± 0.8 | 92.8 | 2.54 |
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
122.5 ± 0.4 | 0.11 ± 0.001 | −41.2 ± 2.6 | 86.8 | 5.02 |
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
106.3 ± 1.1 | 0.18 ± 0.01 | −37.2 ± 1.5 | 81.7 | 8.11 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
227.3 ± 6.2 | 0.34 + 0.01 | −39.9 ± 2.0 | 71.5 | 8.96 |
Characterization and in vitro drug release behavior of CUR-ZpD NPs
In order to overcome the blood brain barrier (BBB), nanoparticles were decorated with G23 peptide, which was reported in our previous work to mediate the transport of polymersomes over the BBB in vitro and in vivo.33,34 Electron microscopic investigation revealed that neither loading of ZpD NPs with curcumin nor functionalization of CUR-ZpD NPs with G23 peptide instigated morphological changes in the NPs (Fig. 1A). The observed adhesion between particles can be explained by aggregation of soft protein particles during the air drying process when preparing TEM samples. As determined with DLS, the particle size of ZpD NPs with or without CUR was about 100 nm, while that of G23-functionalized nanoparticles (CUR-ZpD-G23 NPs) increased to around 120 nm. The Zeta potential value increased from around −30 mV to −13 mV after G23 modification, indicating the incorporation of PEG-G23 (Fig. 1C).Next, FTIR was carried out to verify the chemical composition of the nanoparticle preparations. As shown in Fig. S2,† CUR-ZpD NPs and CUR-ZpD-G23 NPs exhibited a broad peak from 1631–1660 cm−1, which was associated with the amide bond I stretching of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of the zein protein. In addition, CUR-ZpD NPs and CUR-ZpD-G23 NPs showed a characteristic peak at 1280 cm−1, which corresponded with the aromatic C–O stretching vibrations of curcumin, indicating the encapsulation of curcumin in the nanoparticles. The intense peaks at 1036 cm−1 and 3228 cm−1 were assigned to the stretching vibrations of C–O–C of PEG and N–H of G23, which indicated that PEG-G23 was successfully conjugated to the surface of CUR-ZpD NPs, via Michael addition reaction between the catechol in pD and amine in NH2-PEG-G23.18,42
O of the zein protein. In addition, CUR-ZpD NPs and CUR-ZpD-G23 NPs showed a characteristic peak at 1280 cm−1, which corresponded with the aromatic C–O stretching vibrations of curcumin, indicating the encapsulation of curcumin in the nanoparticles. The intense peaks at 1036 cm−1 and 3228 cm−1 were assigned to the stretching vibrations of C–O–C of PEG and N–H of G23, which indicated that PEG-G23 was successfully conjugated to the surface of CUR-ZpD NPs, via Michael addition reaction between the catechol in pD and amine in NH2-PEG-G23.18,42
The curcumin release behavior of CUR-ZpD NPs and CUR-ZpD-G23 NPs was investigated at different pH (pH = 7.4 and pH = 5.0) at 37 °C for 48 h, which mimicked the physiological pH in normal tissues and intracellular endosomes, respectively (Fig. 1D). Free curcumin exhibited a burst release profile and reached 100% release within 8 h incubation irrespective of pH. In contrast to free curcumin, the CUR-loaded nanoparticles released curcumin in a sustained manner, which was attributed to the adherent pD polymer coating at the surface of the zein NPs, forming a diffusion barrier for the curcumin in the nanoparticles.43 The release profiles of CUR-ZpD NPs and CUR-ZpD-G23 NPs were similar. At pH 7.4, approximately 45% curcumin was released from CUR-ZpD NPs and CUR-ZpD-G23 NPs after 48 h. At pH 5, curcumin release from the nanoparticles reached 83% and 80%, respectively. This pH-sensitive curcumin release from the nanoparticles is most likely due to the degradation of the pD polymer coating in acidic media, which would facilitate drug release.20
Quantitative measurement of the transport of C6-ZpD-G23 NPs across a filter-free BBB model
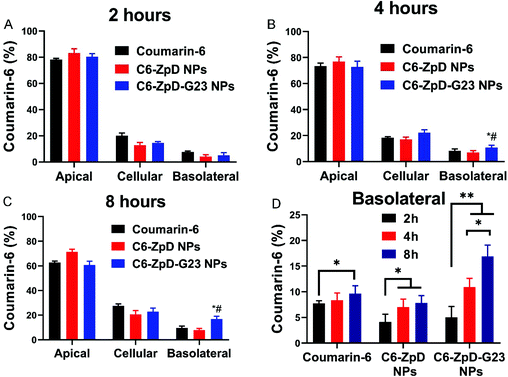
Next, the influence of NP functionalization with G23 peptide on the transcytosis capacity of ZpD NPs was investigated using a filter-free BBB model.34 As shown in Fig. 2A, the in vitro filter-free BBB model consists of a collagen gel covered with a polarized monolayer of brain microvascular endothelial (hCMEC/D3) cells. In this model the absence of a porous filter, which is commonly used in other (e.g. Transwell and microfluidic) BBB models, circumvents adherence of NPs to the filter and consequent inability of transcellular NP transport quantification. The NPs were labeled with coumarin-6 for fluorescence-based detection. Coumarin-6 (C6) and C6-labeled NPs were added apically (at an equal concentration of 1 μg mL−1 coumarin-6) to a polarized cell monolayer of hCMEC/D3 cells and incubated for 2, 4 and 8 h at 37 °C. At this concentration of C6, the cell viability of the hCMEC/D3 cells was unaffected (Fig. S3†). Incubation with C6-ZpD NPs resulted in an increase in basolateral fluorescence signal with an increase in incubation time, which was similar to the basolateral accumulation obtained with free C6 (Fig. 2A–C). In contrast, the basolateral accumulation of C6-ZpD-G23 NPs was significantly higher than that of free C6 and C6-ZpD after 4 h and 8 h of incubation (Fig. 2A–C). Specifically, after 8 h of incubation, 16.9 ± 2.2% C6-ZpD-G23 NPs accumulated at the basolateral side of the BBB, while the basolateral accumulation of free C6 and C6-ZpD NPs was 9.6 ± 1.5% and 7.8 ± 1.8%, respectively. This indicates that G23 functionalization of C6-ZpD NPs significantly enhanced their transendothelial transport, i.e., by 2-fold. However, since we can't exclude the possibility that the basolateral fluorescence signal that is measured following incubation of the BBB model with NPs is partly derived from ‘free’ C6 which has been released from the NPs and becomes basolaterally secreted, the absolute transcytosis percentages may be overestimated.Cellular uptake of CUR-ZpD and CUR-ZpD-G23 NPs in C6 glioma cells, and NPs penetration in 3D tumor spheroids
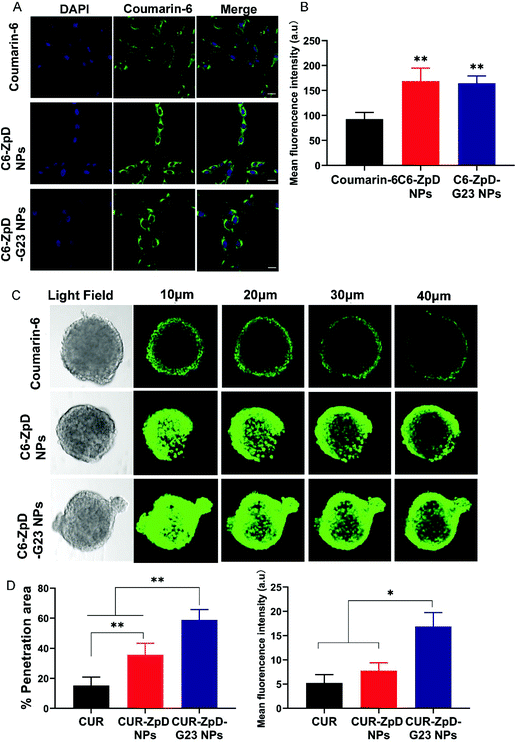
To investigate the cellular distribution of C6-labeled ZpD and ZpD-G23 NPs, glioma cells were treated with C6 or C6-labeled NPs for 4 h, and then stained with DAPI for the identification of the nucleus. As shown in Fig. 3A, the green fluorescence from NPs was localized in the cytoplasm and excluded from the nucleus. In addition, the coumarin-6 loaded NPs showed around 2-fold increase in green fluorescence intensity compared to free coumarin-6, indicating that the NPs were efficiently taken up by C6 glioma cells (Fig. 3A; quantification in B).To investigate whether the ZpD NPs may resolve the difficulties of poor drug distribution in tumors, we evaluated the drug penetration capacity of the NPs using 3D tumor spheroids, which better simulate the in vivo tumor environment than 2D cell cultures. After incubation of C6 glioma spheroids with NPs for 4 h, the penetration ability was assessed using confocal laser scanning microscopy (CLSM) (Fig. 3C). Low fluorescence signals were detected in the tumor spheroids, while a significant decrease in fluorescence intensity from the rim to the center was found in coumarin-6-treated spheroids. Excitingly, a more intense fluorescence signal was observed throughout the tumor spheroids after incubation with NPs, indicating that the ZpD nanocarriers promoted drug penetration in tumor tissue (Fig. 3C). Moreover, the C6-ZpD-G23 NPs were detected deeper into the tumor spheroid (58.8 ± 6.8% positive area) with a higher fluorescence intensity (MFI 16.9 ± 2.8%) than C6-ZpD NPs (35.8 ± 7.6% positive area, MFI 7.7 ± 1.6%) at 40 μm from the top of the spheroids (Fig. 3D). This indicates that G23 functionalization of the NPs resulted in more effective penetration of NPs in the tumor spheroid. Thus, deep tumor penetration was ascribed to the presence of the G23 peptide.
Cytotoxicity and clonogenic potential of glioma cells in presence of CUR-ZpD and CUR-ZpD-G23 NPs
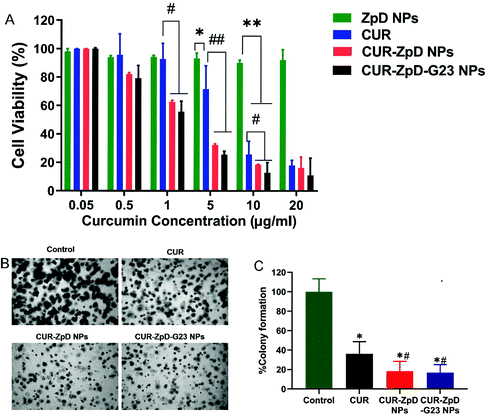
Since the ZdP NPs showed successful transport into C6 glioma cells and tumor spheroids, further experiments were done to analyze the in vitro dose-dependent anticancer efficacy of CUR-ZpD NPs on C6 glioma cells.First, an MTT assay was performed to assess the overall cell viability following treatment of C6 glioma cells with free CUR, empty ZpD NPs, CUR-ZpD NPs and CUR-ZpD-G23 NPs. Empty ZpD NPs showed no effect on C6 glioma cell viability (Fig. 4A), indicating that ZpD NPs themselves are non-toxic to C6 glioma cells. At low concentrations of CUR (≤1 μg mL−1) no significant decrease in cell viability was observed, which was consistent with previous research showing that curcumin has a non-cytotoxic effect at low concentrations.37,44 At concentrations >1 μg mL−1 CUR induced a dose-dependent reduction in cell viability. When CUR was formulated into ZpD NPs the reduction in cell viability was enhanced compared to treatment with the same dose of free CUR, and already present at a CUR concentration of 1 μg mL−1. This indicates that the CUR-loaded NPs showed a stronger inhibition effect on C6 glioma cells than free CUR, which may relate to the higher cellular uptake of NPs compared to free CUR (Fig. 3A). The antitumor activity of CUR-ZpD NPs and CUR-ZpD-G23 NPs was concentration dependent. The cell viability decreased to 32% and 25%, respectively, after incubation with CUR-ZpD NPs and CUR-ZpD-G23 NPs (CUR 5 μg ml−1) for 24 h when compared to untreated cells. As shown in Fig. S4,† C6 glioma cells showed reduced adherence when treated with 2.5 μg ml−1 and 5 μg mL−1 curcumin for 24 h, which was more pronounced, i.e., leading to cell rounding and detachment, when the curcumin was administered as a nanoparticle formulation. In our next experiments, we used a curcumin concentration of 5 μg ml−1.
To further demonstrate the anticancer activity of the curcumin-loaded ZpD NPs, we performed the soft agar colony-formation assay, which serves as a useful tool to test whether a given treatment can reduce the clonogenic survival of cancer cells. CUR treatment of C6 glioma cells followed by a 7-day incubation period significantly decreased the clonogenic proliferation of C6 glioma cells (to 36 ± 7% cf. to control), confirming the therapeutic effect of curcumin (Fig. 4B; quantification in C). Cell treatment with CUR-ZpD NPs and CUR-ZpD-G23 NPs presented a considerably further decline in the colony formation (to 18 ± 5% and 17 ± 6% cf. to control, respectively). Overall, our data suggest that the observed anticancer activity of CUR-loaded ZpD-NPs against C6 glioma cells resulted from the efficient uptake of the ZpD NPs by the cancer cells, followed by intracellular release of curcumin from the ZpD NPs.
C6 glioma cell migration
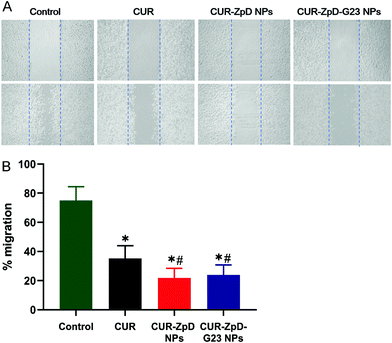
Next, a scratch assay was performed to evaluate if CUR-ZpD NPs have the potential to inhibit C6 glioma cell migration. Following treatment of C6 glioma cells with curcumin, cell migration was significantly lower compared to control (untreated) cells, i.e. 35.6 ± 8.7% and 75.0 ± 9.4%, respectively (Fig. 5A; quantified in B). C6 glioma cell migration following treatment with CUR-ZpD NPs and CUR-ZpD-G23 NPs groups were 21.8 ± 6.6% and 23.9 ± 6.9%, respectively (Fig. 5B). The results indicate that CUR-ZpD and CUR-ZpD-G23 NPs inhibited the migration of C6 glioma cells, thereby substantiating their tumor-suppressive effect.ROS activity and induction of apoptosis in C6 glioma cells in presence of CUR-ZpD and CUR-ZpD-G23 NPs
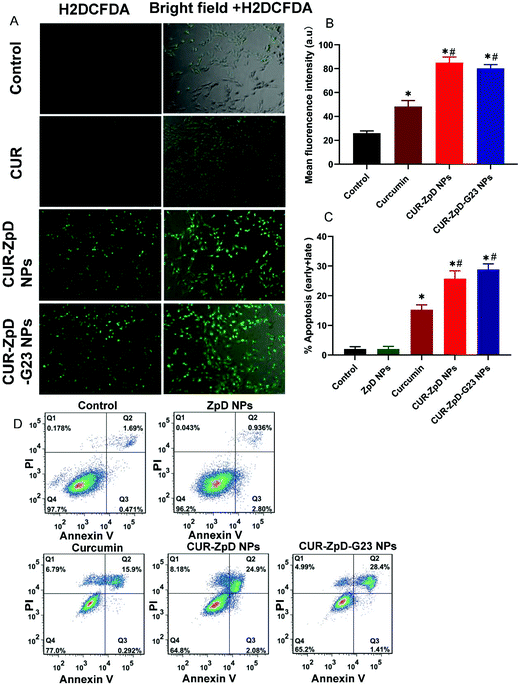
Because curcumin has been shown to induce apoptosis in cancer cells via the generation of reactive oxygen species (ROS),45,46 we next measured ROS activity and the extent of apoptosis in C6 glioma cells upon treatment with CUR, CUR-ZpD NPs and CUR-ZpD-G23 NPs.The ROS activity in C6 glioma cells was measured using H2DCFDA to (fluorescent) DCF conversion. As shown in Fig. 6A and B, 4 h incubation of C6 glioma cells with curcumin resulted in a 2-fold increase in cellular fluorescence intensity, i.e., ROS level. Incubation of cells with CUR-ZpD NPs and CUR-ZpD-G23 NPs showed a 4-fold increase in cellular fluorescence intensity compared to control cells, i.e., a 2-fold increase in intensity compared to CUR-treated cells, indicating higher ROS activity in C6 glioma cells after treatment with CUR-loaded NPs than with free curcumin (Fig. 6A; quantified in B).
Following CUR treatment, 15.2 ± 1.7% of the cell population was apoptotic, while treatment with CUR-ZpD and CUR-ZpD-G23 NPs resulted in 25.8 ± 2.6% and 28.9 ± 1.9% apoptotic cells, respectively (Fig. 6C). These data suggest that the CUR-ZpD and CUR-ZpD-G23 NPs more efficiently induce apoptosis than free curcumin, which is consistent with the results from the MTT cell viability assay (Fig. 4A).
Circulation of ZpD-G23 NPs in zebrafish larvae
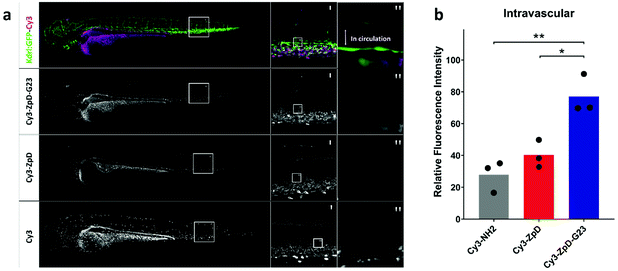
First attempts in vivo were made to assess whether ZpD(-G23) particles remain present in circulation after injection and don't form aggregates (>1 μm). Therefore, transgenic zebrafish larva (Danio Rerio, kdrl:GFP, 80 hours post fertilization) were used as in vivo model. The fish are transparent and have a developed green fluorescent vasculature, allowing high resolution localization of fluorescently-labeled nanoparticle formulations after injection (Fig. 7a, upper row). Fish were injected into the duct of Cuvier with 1 nL of Cy3-NH2, Cy3-ZpD, or Cy3-ZpD-G23 and imaged two hours post injection (Fig. 7a, left column). For all injections, fluorescence was observed in egg yolk, choroid plexus, ionocytes, and kidneys, which can be explained by the presence of free Cy3-NH2 (Fig. S5†). The tail, in which the dorsal aorta (Fig. 7a, middle column) is laterally orientated, allows for confocal stacks of intravascular presence of Cy3 (Fig. 7a, right column). Confocal microscopic investigation revealed that no circulating aggregates were present. Especially in the larvae injected with Cy3-ZpD-G23 increased intravascular fluorescence was observed compared to in larvae injected with free Cy3-NH2. This observation was quantified in multiple larvae (Fig. S6 and Table S1†), revealing a 49.1 ± 9.2% and 36.7 ± 8.7% increase in intravascular fluorescence in Cy3-ZpD-G23 injected larvae compared to larvae injected with Cy3-NH2 and Cy3-ZpD, respectively (Fig. 7b). From this in vivo study it can be concluded that Cy3-ZpD-G23 NPs circulate, because of the higher intravascular fluorescence intensity in comparison to free Cy3-NH2, without visible aggregation, and therefore demonstrates that Cy3-ZpD-G23 NPs have potential to reach its target when injected intravenously.Conclusions
A facile and robust approach to formulate G23-functionalized polydopamine-coated zein-curcumin (CUR-ZpD-G23) NPs is reported. The CUR-ZpD-G23 NPs were approximately 120 nm in diameter with a spherical shape. CLSM studies showed that the CUR-ZpD-G23 NPs exhibited excellent cellular uptake ability by C6 glioma cells and penetration ability into 3D tumor spheroids. G23-functionalized NPs demonstrated an increase in transcytosis across an in vitro BBB model compared to non-functionalized NPs, while CUR-loaded NPs demonstrated a concentration-dependent cytotoxicity in C6 glioma cells, and inhibited cell migration and colony formation. The CUR-loaded NPs increased cellular ROS production in C6 glioma cells and induced apoptosis. Following in vivo intravenous injection in zebrafish, ZpD-G23 NPs demonstrated the ability to circulate, which is a first prerequisite for their use in targeted drug delivery. We conclude that ZpD-G23 NPs seem promising for the delivery of hydrophobic drugs, including curcumin, to glioblastoma cells, which warrants further experimentation in in vivo glioblastoma models.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
H. Z. gratefully acknowledges financial support from the China Scholarship Council (No. 201706240273) and Stichting De Cock-Hadders (ID. 119800154).References
- S. M. Chang, J. G. Kuhn, H. I. Robins, S. C. Schold, A. M. Spence, M. S. Berger, M. P. Mehta, I. F. Pollack, C. Rankin and M. D. Prados, Cancer, 2001, 91, 417–422 CrossRef CAS.
- M. J. B. Taphoorn, R. Henriksson, A. Bottomley, T. Cloughesy, W. Wick, W. P. Mason, F. Saran, R. Nishikawa, M. Hilton, C. Theodore-Oklota, A. Ravelo and O. L. Chinot, J. Clin. Oncol., 2015, 33, 2166–2175 CrossRef CAS.
- W. A. Banks, Nat. Rev. Drug Discovery, 2016, 15, 275–292 CrossRef CAS.
- E. A. Neuwelt, B. Bauer, C. Fahlke, G. Fricker, C. Iadecola, D. Janigro, L. Leybaert, Z. Molnár, M. E. O'Donnell, J. T. Povlishock, N. R. Saunders, F. Sharp, D. Stanimirovic, R. J. Watts and L. R. Drewes, Nat. Rev. Neurosci., 2011, 12, 169–182 CrossRef CAS.
- T. D. Azad, J. Pan, I. D. Connolly, A. Remington, C. M. Wilson and G. A. Grant, Neurosurg. Focus, 2015, 38, 1–10 Search PubMed.
- H. L. Wong, X. Y. Wu and R. Bendayan, Adv. Drug Delivery Rev., 2012, 64, 686–700 CrossRef CAS.
- A. S. Wadajkar, J. G. Dancy, D. S. Hersh, P. Anastasiadis, N. L. Tran, G. F. Woodworth, J. A. Winkles and A. J. Kim, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2017, 9, e1439 Search PubMed.
- J. Xie, Z. Shen, Y. Anraku, K. Kataoka and X. Chen, Biomaterials, 2019, 224, 119491 CrossRef CAS.
- C. Saraiva, C. Praça, R. Ferreira, T. Santos, L. Ferreira and L. Bernardino, J. Controlled Release, 2016, 235, 34–47 CrossRef CAS.
- J. S. Michael, B.-S. Lee, M. Zhang and J. S. Yu, J. Transl. Int. Med., 2018, 6, 128–133 Search PubMed.
- S. Fu, M. Liang, Y. Wang, L. Cui, C. Gao, X. Chu, Q. Liu, Y. Feng, W. Gong, M. Yang, Z. Li, C. Yang, X. Xie, Y. Yang and C. Gao, ACS Appl. Mater. Interfaces, 2019, 11, 1841–1854 CrossRef CAS.
- Y. Luo, Z. Teng and Q. Wang, J. Agric. Food Chem., 2012, 60, 836–843 CrossRef CAS.
- S. Chen, Y. Han, L. Jian, W. Liao, Y. Zhang and Y. Gao, Carbohydr. Polym., 2020, 236, 116090 CrossRef CAS.
- C. C. Sun, H. Su, G. D. Zheng, W. J. Wang, E. Yuan and Q. F. Zhang, Food Chem., 2020, 330, 127245 CrossRef CAS.
- R. Penalva, C. J. González-Navarro, C. Gamazo, I. Esparza and J. M. Irache, Nanomedicine, 2017, 13, 103–110 CrossRef CAS.
- Q. Zhong, M. Jin, P. M. Davidson and S. Zivanovic, Food Chem., 2009, 115, 697–700 CrossRef CAS.
- J. Xue, Y. Zhang, G. Huang, J. Liu, M. Slavin and L. (Lucy) Yu, Food Hydrocolloids, 2018, 83, 25–35 CrossRef CAS.
- J. Park, T. F. Brust, H. J. Lee, S. C. Lee, V. J. Watts and Y. Yeo, ACS Nano, 2014, 8, 3347–3356 CrossRef CAS.
- F. Ji, H. Sun, Z. Qin, E. Zhang, J. Cui, J. Wang, S. Li and F. Yao, Polymers, 2018, 10, 1–17 Search PubMed.
- Q. Zheng, T. Lin, H. Wu, L. Guo, P. Ye, Y. Hao, Q. Guo, J. Jiang, F. Fu and G. Chen, Int. J. Pharm., 2014, 463, 22–26 CrossRef CAS.
- Z. Wang, Y. Duan and Y. Duan, J. Controlled Release, 2018, 290, 56–74 CrossRef CAS.
- B. B. Aggarwal and K. B. Harikumar, Int. J. Biochem. Cell Biol., 2009, 41, 40–59 CrossRef CAS.
- A. B. Kunnumakkara, et al. , J. Pharmacol., 2017, 174, 1325–1348 CAS.
- Z. Shabaninejad, M. H. Pourhanifeh, A. Movahedpour, R. Mottaghi, A. Nickdasti, E. Mortezapour, A. Shafiee, S. Hajighadimi, S. Moradizarmehri, M. Sadeghian, S. M. Mousavi and H. Mirzaei, Eur. J. Med. Chem., 2020, 188, 112040 CrossRef CAS.
- D. Lelli, C. Pedone and A. Sahebkar, Biomed. Pharmacother., 2017, 88, 832–834 CrossRef CAS.
- T. S. Ramasamy, A. Z. Ayob, H. H. L. Myint, S. Thiagarajah and F. Amini, Cancer Cell Int., 2015, 15, 1–15 CrossRef.
- P. Maiti, A. Al-Gharaibeh, N. Kolli and G. L. Dunbar, Oxid. Med. Cell. Longevity, 2017, 2017, 1–17 CrossRef.
- J. Hong, Y. Liu, Y. Xiao, X. Yang, W. Su, M. Zhang, Y. Liao, H. Kuang and X. Wang, Drug Delivery, 2017, 24, 109–120 CrossRef CAS.
- K. Stojanov, J. V. Georgieva, R. P. Brinkhuis, J. C. Van Hest, F. P. Rutjes, R. A. J. O. Dierckx, E. F. J. De Vries and I. S. Zuhorn, Mol. Pharm., 2012, 9, 1620–1627 CrossRef CAS.
- M. M. Haroon, G. H. Dar, D. Jeyalakshmi, U. Venkatraman, K. Saba, N. Rangaraj, A. B. Patel and V. Gopal, J. Controlled Release, 2016, 228, 120–131 CrossRef CAS.
- M. M. Haroon, K. Saba, V. H. Boddedda, J. M. Kumar, A. B. Patel and V. Gopal, J. Biosci., 2019, 44, 1 CrossRef CAS.
- C.-H. Su, C.-Y. Tsai, B. Tomanek, W.-Y. Chen and F.-Y. Cheng, Nanoscale, 2016, 8, 7866–7870 RSC.
- J. V. Georgieva, R. P. Brinkhuis, K. Stojanov, C. A. G. M. Weijers, H. Zuilhof, F. P. J. T. Rutjes, D. Hoekstra, J. C. M. van Hest and I. S. Zuhorn, Angew. Chem., Int. Ed., 2012, 51, 8339–8342 CrossRef CAS.
- E. De Jong, D. S. Williams, L. K. E. A. Abdelmohsen, J. C. M. Van Hest and I. S. Zuhorn, J. Controlled Release, 2018, 289, 14–22 CrossRef CAS.
- S. Li, Z. Li, J. Pang, J. Chen, H. Wang, Q. Xie and Y. Jiang, Ind. Eng. Chem. Res., 2018, 57, 590–599 CrossRef CAS.
- Y. Huang, L. Hu, S. Huang, W. Xu, J. Wan, D. Wang, G. Zheng and Z. Xia, Int. J. Nanomed., 2018, 13, 8309–8323 CrossRef CAS.
- A. Zanotto-Filho, E. Braganhol, M. I. Edelweiss, G. A. Behr, R. Zanin, R. Schröder, A. Simões-Pires, A. M. O. Battastini and J. C. F. Moreira, J. Nutr. Biochem., 2012, 23, 591–601 CrossRef CAS.
- A. Singh, W. Kim, Y. Kim, K. Jeong, C. S. Kang, Y. S. Kim, J. Koh, S. D. Mahajan, P. N. Prasad and S. Kim, Adv. Funct. Mater., 2016, 26, 7057–7066 CrossRef CAS.
- F. Ellett, L. Pase, J. W. Hayman, A. Andrianopoulos and G. J. Lieschke, Blood, 2011, 117(4), e49–e56 CrossRef CAS.
- S. W. Jin, D. Beis, T. Mitchell, J. N. Chen and D. Y. Stainier, Development, 2005, 132(23), 5199–5209 CrossRef CAS.
- F. Campbell, F. L. Bos, S. Sieber, G. Arias-Alpizar, B. E. Koch, J. Huwyler, A. Kros and J. Bussmann, ACS Nano, 2018, 12(3), 2138–2150 CrossRef CAS.
- Z. Cui, K. W. K. Yeung, S. Wu, X. Yang, P. K. Chu, X. Liu, J. Li and L. Tan, ACS Nano, 2017, 11, 11250–11263 CrossRef.
- P. B. Lee, H. Dellatore, S. M. Miller and W. M. Messersmith, Science, 2007, 318, 426–430 CrossRef.
- M. H. Seyithanoğlu, A. Abdallah, S. Kitiş, E. M. Güler, A. Koçyiğit, T. T. Dündar and M. Gündağ Papaker, Cell. Mol. Biol., 2019, 65, 101–108 CrossRef.
- B. Kim, H. S. Kim, E.-J. Jung, J. Y. Lee, B. K. Tsang, J. M. Lim and Y. S. Song, Mol. Carcinog., 2016, 55, 918–928 CrossRef CAS.
- T. Yu, J. Dohl, F. Elenberg, Y. Chen and P. Deuster, J. Cell. Physiol., 2019, 234, 6371–6381 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0bm01536a |
| This journal is © The Royal Society of Chemistry 2021 |