Open Access Article
Open Access ArticleBiological evaluation of native streptococcal competence stimulating peptides reveals potential crosstalk between Streptococcus mitis and Streptococcus pneumoniae and a new scaffold for the development of S. pneumoniae quorum sensing modulators†
Tahmina Ahmed
Milly
and
Yftah
Tal-Gan
 *
*
Department of Chemistry, University of Nevada, Reno, 1664 N. Virginia Street, Reno, Nevada 89557, USA. E-mail: ytalgan@unr.edu
First published on 29th May 2020
Abstract
Streptococcus pneumoniae, an opportunistic human pathogen, acquires genes from its neighboring species of the mitis group of streptococci, which confer antibiotic resistances and allow it to produce diverse virulence factors. Most species of the mitis group are naturally competent, and they utilize the competence stimulating peptide (CSP) and the CSP-dependent competence regulon, a conserved quorum sensing (QS) circuit, to regulate their competence behavior. The dependence of the mitis group on this communication pathway makes QS a potential target to control their behavior. In this work, we sought to evaluate the impact of the native pheromones of the adjacent species of S. pneumoniae to modulate the activity of the S. pneumoniae competence regulon. Our results revealed the potential role of S. mitis as a modulator of QS in S. pneumoniae. Most importantly, our analysis also revealed that by using the native pheromone of S. mitis as a template, highly potent pan-group agonists and antagonists of the pneumococcal competence regulon could be developed. The newly developed QS modulators may have therapeutic utility in treating pneumococcal infections.
Streptococcus pneumoniae (pneumococcus), a member of the mitis group of streptococci, is an opportunistic human pathogen that is a major cause of a variety of diseases, including the deadly pneumonia, bacteremia, meningitis, sepsis and otitis media.1 It has been estimated that more than 22
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths and 445
000 deaths and 445![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 hospitalizations, with a $3.5 billion direct medical cost, occur annually in the United States as a result of pneumococcal infections.2 Furthermore, the increasing rate of resistance development against several antibiotics, including vancomycin, quinolones, macrolides and beta lactams by the recombinogenic pneumococcal strains necessitates the development of alternative strategies to attenuate pneumococcal infections.3,4 Pneumococcus utilizes the competence regulon, a conserved quorum sensing (QS) circuit (Fig. 1), to acquire genetic material from the neighboring environment, including genes that confer antibiotic resistance, as well as to regulate a wide range of functions related to pathogenicity such as virulence factor production and biofilm formation.5–9 Thus, intercepting this QS circuitry could be utilized as a potential non-antibiotic therapeutic strategy to control pneumococcal infections without affecting the survival of the bacteria, thus minimizing the selective pressure for resistance development.10–16
000 hospitalizations, with a $3.5 billion direct medical cost, occur annually in the United States as a result of pneumococcal infections.2 Furthermore, the increasing rate of resistance development against several antibiotics, including vancomycin, quinolones, macrolides and beta lactams by the recombinogenic pneumococcal strains necessitates the development of alternative strategies to attenuate pneumococcal infections.3,4 Pneumococcus utilizes the competence regulon, a conserved quorum sensing (QS) circuit (Fig. 1), to acquire genetic material from the neighboring environment, including genes that confer antibiotic resistance, as well as to regulate a wide range of functions related to pathogenicity such as virulence factor production and biofilm formation.5–9 Thus, intercepting this QS circuitry could be utilized as a potential non-antibiotic therapeutic strategy to control pneumococcal infections without affecting the survival of the bacteria, thus minimizing the selective pressure for resistance development.10–16
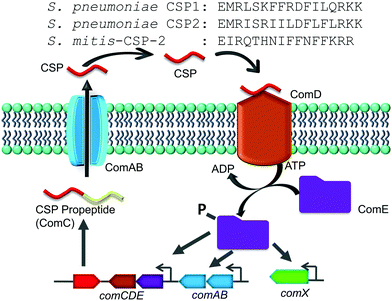
The competence regulon in pneumococcus relies on the production of, secretion of and response to a 17-amino-acid competence stimulating peptide (CSP, Fig. 1) signal.5 Different strains of pneumococcus can produce different CSP signals and the two major forms of CSP (CSP1 and CSP2) share 50% homology with each other.17 Each CSP signal is associated with a cognate transmembrane histidine kinase receptor, termed ComD (ComD1 and ComD2, respectively).18 The competence regulon QS circuitry (Fig. 1) involves the secretion of mature CSP signal into the extracellular environment with the help of a dedicated ABC transporter, ComAB.5 As the bacteria grow, the concentration of CSP increases, and once a threshold concentration is attained, the CSP pheromone can bind to and activate the ComD receptor.8 ComD activation prompts the phosphorylation of the cytoplasmic response regulator, ComE, which then acts as a transcription factor to upregulate the transcription of the QS circuitry genes (comABCDE) as well as the comX gene, the master regulator of the QS circuitry that controls the different QS-regulated phenotypes.6–8
Similar to S. pneumoniae, other species of the mitis and the anginosus streptococcal groups are naturally competent. Regardless of the specific CSP pheromone involved, these species utilize a similar pheromone-dependent competence regulon to upregulate the expression of competence genes required for DNA uptake and recombination.19 In the anginosus group, different species utilize the same CSP signaling molecule, while a large variation of CSP pheromones have been reported within the same species in the mitis group, specifically among Streptococcus mitis strains.20–22 The mitis group of streptococci is composed of 13 species; most of them are commensal bacteria, with S. pneumoniae also being a notorious opportunistic pathogen.20,21 The recombination events among the mitis group of streptococci have been shown to be instrumental in the development of antibiotic resistance. Specifically, interspecies gene transfer of both antimicrobial resistance and virulence genes was observed between S. pneumoniae and its neighboring species, S. mitis, which shares more than 80% of its genes with S. pneumoniae.21,23 In this study we aimed to evaluate the impact that native CSP signals of streptococci of the mitis and anginosus groups have on the activity of the S. pneumoniae competence regulon. To this end, we chemically synthesized different reported native streptococcal CSP pheromones of the mitis and anginosus groups and tested their ability to activate/inhibit the S. pneumoniae ComD1 and ComD2 receptors. Our initial analysis revealed that one of the CSP pheromones of S. mitis, S. mitis-CSP-2 (Fig. 1), can activate both S. pneumoniae ComD receptors. We therefore chose S. mitis-CSP-2 as the lead scaffold for the development of pan-group S. pneumoniae QS modulators. Indeed, using S. mitis-CSP-2 as a template, we were able to develop potent pan group QS inhibitors of S. pneumoniae QS. Moreover, our results revealed potential crosstalk between S. mitis and S. pneumoniae.
Results and discussion
Synthesis and initial screening of native streptococcal competence pheromones (CSPs)
In this work, we first set out to evaluate potential crosstalk between species of the mitis group by testing the ability of native streptococcal competence pheromones (CSPs) to modulate the activity of the S. pneumoniae competence regulon QS circuitry. To this end, we chemically synthesized different native CSPs of the most common commensal bacterial strains of the mitis group (see Table 1 for strain information and CSP sequences). To explore potential crosstalk between the different groups of streptococci, we decided to include CSP pheromones from the anginosus group. Since most of the species of the anginosus group, namely S. anginosus, S. constellatus and S. intermedius, produce the same CSP signal,21 we synthesized only one CSP signal (S. intermedius-CSP) representing the anginosus group of streptococci (Table 1). Lastly, in the initial screening assay, we included the two CSP forms of S. mutans, S. mutans-21-CSP (pre-mature extracellular signal) and S. mutans-18-CSP (mature extracellular signal),24 to test for potential crosstalk between opportunistic pathogens. All of the CSP pheromones (Table 1) were constructed using established solid-phase peptide synthesis (SPPS) protocols on 4-benzyloxybenzyl alcohol (Wang) resin.25 The peptides were then cleaved and purified to homogeneity using semipreparative RP-HPLC (>95% purity), and their identity was confirmed using mass spectrometry (see the Experimental section and ESI† for full experimental details).| Group | Name of CSP | Strain | CSP sequence |
|---|---|---|---|
| TType strain. GGenome sequence available. | |||
| Mitis | S. pneumoniae CSP1 | S. pneumoniae D39 | EMRLSKFFRDFILQRKK |
| S. pneumoniae CSP2 | S. pneumoniae TIGR4 | EMRISRIILDFLFLRKK | |
| S. mitis-CSP-1 | S. mitis NCTC 8033 (SK321) | ESRLPKIRFDFIFPRKK | |
| S. mitis-CSP-2 | S. mitis CCUG 31611TG (NCTC 12261) | EIRQTHNIFFNFFKRR | |
| S. cristatus-CSP | S. cristatus ACTC 51100G | DLRNIFLKIKFKKK | |
| S. oligofermentans-CSP | S. oligofermentans LMG 21535T | DSRNIFLKIKFKKK | |
| S. oralis-CSP | S. oralis SK255 | DWRISETIRNLIFPRRK | |
| S. gordonii-CSP-1 | S. gordonii NCTC 3165 | SQKGVYASQRSFVPSWFRKIFRN | |
| S. gordonii-CSP-2 | S. gordonii NCTC 7865T | DIRHRINNSIWRDIFLKRK | |
| S. gordonii challis-CSP | S. gordonii challis CHIG | DVRSNKIRLWWENIFFNKK | |
| S. sanguinis-CSP | S. sanguinis SK36G (S. sanguinis NCTC 7863) | DLRGVPNPWGWIFGR | |
| Anginosus | S. intermedius-CSP | S. intermedius NCTC 11 324T | DSRIRMGFDFSKLFGK |
| Mutans | S. mutans-18-CSP | S. mutans UA159G | SGSLSTFFRLFNRSFTQA |
| S. mutans-21-CSP | S. mutans UA159G | SGSLSTFFRLFNRSFTQALGK | |
Next, we tested the ability of the native streptococcal CSPs to modulate the activity of the S. pneumoniae ComD receptors (both ComD1 and ComD2) using a β-galactosidase cell-based bacterial reporter assay. Two β-gal reporter strains, D39pcomX::lacZ and TIGR4pcomX::lacZ, previously constructed by Lau and co-workers, were utilized to evaluate the QS modulation in both groups of S. pneumoniae.26 These reporter systems carry the lacZ gene under the control of the comX promoter; thus, upon QS activation, lacZ will be expressed, allowing the monitoring of the QS response by measuring the β-galactosidase activity (see the Experimental section for details). We then conducted an initial screening of all the native CSP pheromones at high peptide concentration (10 μM; see Fig. S-1, S-6, S-9, and S-14, ESI†). The initial screening results revealed that of the twelve CSP signals tested, only one native CSP pheromone, S. mitis-CSP-2, was able to fully activate both the S. pneumoniae ComD receptors (>75% activation compared to the native S. pneumoniae CSPs; Fig. S-1 and S-9, ESI†). The EC50 values of this peptide were determined through dose response curves and found to be 663 nM and 635 nM when activating the S. pneumoniae ComD1 and ComD2 receptors, respectively (Table 2). Although these EC50 values are 12- to 64-fold higher than the native pneumococcus CSPs, the values are still biologically relevant as the native CSP concentrations in streptococci supernatants were found to be in the micromolar range.15 Since none of the other native peptides were capable of activating/inhibiting the S. pneumoniae QS circuitry, we selected the S. mitis-CSP-2 pheromone as a template for the development of pan-group S. pneumoniae QS modulators that could be utilized to attenuate pneumococcal infections.
| Peptide name | Peptide sequence | S. pneumoniae ComD1 | S. pneumoniae ComD2 | ||
|---|---|---|---|---|---|
| EC50b (nM) | 95% CIc | EC50b (nM) | 95% CIc | ||
| a See the Experimental section for full experimental details and the (ESI) for plots of agonism dose response curves. b EC50 values were determined by testing peptides over a range of concentrations. c 95% confidence interval. d EC50 values of S. pneumoniae CSP1 and CSP2 from ref. 27. e EC50 not determined due to the analogue's low induction in the primary agonism screening assay. | |||||
| S. pneumoniae CSP1d | EMRLSKFFRDFILQRKK | 10.3 | 6.27–16.8 | 526 | 498–556 |
| S. pneumoniae CSP2d | EMRISRIILDFLFLRKK | 1650 | 1190–2300 | 50.7 | 40.6–63.2 |
| S. mitis-CSP-2 | EIRQTHNIFFNFFKRR | 663 | 608–722 | 635 | 426–947 |
| S. mitis-CSP-2-I2M | EMRQTHNIFFNFFKRR | 87.7 | 79.1–97.3 | 136 | 89.0–208 |
| S. mitis-CSP-2-F10D | EIRQTHNIFDNFFKRR | —e | — | >1000 | — |
| S. mitis-CSP-2-Q4L | EIRLTHNIFFNFFKRR | 88.2 | 64.6–121 | 252 | 231–275 |
| S. mitis-CSP-2-N7F | EIRQTHFIFFNFFKRR | 140 | 113–172 | 471 | 459–482 |
| S. mitis-CSP-2-I8F | EIRQTHNFFFNFFKRR | 128 | 93.2–176 | 209 | 139–313 |
| S. mitis-CSP-2-N11F | EIRQTHNIFFFFFKRR | 4.63 | 2.43–8.84 | 220 | 194–248 |
| S. mitis-CSP-2-F12I | EIRQTHNIFFNIFKRR | —e | — | >1000 | — |
| S. mitis-CSP-2-Q4I | EIRITHNIFFNFFKRR | —e | — | — | — |
| S. mitis-CSP-2-N7I | EIRQTHIIFFNFFKRR | 101 | 79.5–129 | 211 | 147–303 |
| S. mitis-CSP-2-F12L | EIRQTHNIFFNLFKRR | 54.3 | 34.7–84.7 | 813 | 539–1230 |
Point modification S. mitis-CSP-2 analogues
Previously, the Tal-Gan Laboratory had conducted a detailed structure–activity relationship (SAR) analysis of the two S. pneumoniae signal variants, CSP1 and CSP2, against the two S. pneumoniae ComD receptors, ComD1 and ComD2, and identified the critical residues involved in ComD1 and ComD2 receptor binding and activation.27–29 Based on the SAR results of both the CSP1 and CSP2 scaffolds in S. pneumoniae, we designed ten S. mitis-CSP-2 analogues bearing a point modification in key positions to match the corresponding residue in S. pneumoniae CSP1 or CSP2 pheromones and examined their ability to modulate the competence regulon of S. pneumoniae (Table 2). Since S. mitis-CSP-2 and the two CSP signals of S. pneumoniae differ in many positions (Fig. 1 and Table 1), we chose the key positions contributing to the S. pneumoniae receptor binding and activation, all of which are in the N-terminus and the central region of S. pneumoniae CSPs, and systematically replaced each residue in S. mitis-CSP-2 with the residues found in S. pneumoniae CSP1 or CSP2. We excluded the modifications related to the C-terminus of the pheromones as the C-terminal residues of S. pneumoniae CSP1 and CSP2 were found to be dispensable.27The initial screening of the S. mitis-CSP-2 point-modification analogues revealed that most of the mutated analogues exhibited higher potency against both the S. pneumoniae ComD1 and ComD2 receptors compared to the native S. mitis-CSP-2 (Fig. S-2 and S-10, (ESI†) and Table 2). Specifically, this analysis revealed that S. mitis-CSP-2-I2M, a point-mutated analogue that introduced the conserved residue, Met2, that is present in both the S. pneumoniae pheromones, has increased potency against both the S. pneumoniae ComD receptors compared to the native S. mitis-CSP-2. These results support previous observations that Met2 plays a critical role in binding and activation in both the S. pneumoniae receptors.27,30 On the contrary, the second S. mitis-CSP-2-based analogue bearing a conserved residue present in both S. pneumoniae CSPs, S. mitis-CSP-2-F10D, was found to activate only the S. pneumoniae ComD2 receptor, although only at a high concentration (>1000 nM, Table 2). Looking at the S. pneumoniae CSP1-based modifications, alterations of the hydrophobic residues in the central region of S. mitis at positions 4, 7, 8, 11 and 12 in S. mitis-CSP-2 with the corresponding residues in S. pneumoniae CSP1 revealed that, with the exception of S. mitis-CSP-2-F12I, all the resulting analogues exhibit enhanced activity against ComD1 compared to the native S. mitis-CSP-2. Importantly, the replacement of the hydrophilic residue Asn at position 11 in S. mitis-CSP-2 with the hydrophobic residue Phe resulted in an analogue (S. mitis-CSP-2-N11F) with >140-fold increased potency compared to the native S. mitis-CSP-2 signal against the S. pneumoniae ComD1 receptor, exhibiting comparable activity to the pneumococcal native peptide, S. pneumoniae CSP1. With the exception of S. mitis-CSP-2-F12I, all the resulting analogues also exhibited increased potency compared to the native S. mitis-CSP-2 signal against the S. pneumoniae ComD2 receptor (Table 2).
Moving to the S. pneumoniae CSP2-based modifications (Q4I, N7I, F12L), the initial screening revealed that only S. mitis-CSP-2-N7I exhibits enhanced potency compared to native S. mitis-CSP-2 against both pneumococcal ComD receptors, highlighting the important role the hydrophobic residues play in both pneumococcal receptor binding and activation. On the contrary, S. mitis-CSP-2-Q4I was found to be relatively inactive against both the S. pneumoniae ComD receptors (Table 2).
Multiple modifications S. mitis-CSP-2 analogues
The point modification study revealed several important S. mitis-CSP-2-based S. pneumoniae QS modulators. We therefore set out to utilize the recently acquired SAR insight to rationally design S. pneumoniae QS modulators with enhanced potencies. To this end, we synthesized a library of double and triple modification analogues where we incorporated multiple beneficial point modifications into a single analogue. Expectedly, the initial evaluation revealed that most of the doubly modified analogues are more active than the native S. mitis-CSP-2 signal against both S. pneumoniae ComD receptors. However, while some dual-modified analogues exhibited slightly increased or similar potency relative to the single replacement analogues, other double modification analogues resulted in reduction in potency compared to the single point modification analogues (Table 3). Specifically, the combination of the Q4L modification with other central region hydrophobic modifications (N7F, N7I, I8F, or N11F) resulted in doubly-modified analogues with reduced potency compared to the monosubstituted analogues against both pneumococcal ComD receptors. Most importantly, we identified one double modification analogue, S. mitis-CSP-2-N7II8F, that can bind and activate both pneumococcal ComD receptors with enhanced potencies compared to the single replacement analogues (compare the EC50 values of S. mitis-CSP-2-N7I and S. mitis-CSP-2-I8F against the pneumococcal ComD1 and ComD2 receptors in Table 2 with the EC50 values of S. mitis-CSP-2-N7II8F against the pneumococcal ComD1 and ComD2 receptors in Table 3), making it the most potent S. mitis-CSP-2-based pan-group pneumococcal QS activator identified in the double modification library. Interestingly, a similar double-modification where Asn7 was replaced with phenylalanine, the native residue in S. pneumoniae CSP1, resulted in a double-modified analogue, S. mitis-CSP-2-N7FI8F, that lost its activity against the pneumococcal ComD1 receptor, while maintaining its activity against the pneumococcal ComD2 receptor (Table 3).| Peptide name | Peptide sequence | S. pneumoniae ComD1 | S. pneumoniae ComD2 | ||
|---|---|---|---|---|---|
| EC50b (nM) | 95% CIc | EC50b (nM) | 95% CIc | ||
| a See the Experimental section for full experimental details and the ESI for plots of agonism dose response curves. b EC50 values were determined by testing peptides over a range of concentrations. c 95% confidence interval. d EC50 values of S. pneumoniae CSP1 and CSP2 from ref. 27. e EC50 not determined due to the analogue's low induction in the primary agonism screening assay.* Key analogues discussed in the text are italicized. | |||||
| S. pneumoniae CSP1d | EMRLSKFFRDFILQRKK | 10.3 | 6.27–16.8 | 526 | 498–556 |
| S. pneumoniae CSP2d | EMRISRIILDFLFLRKK | 1650 | 1190–2300 | 50.7 | 40.6–63.2 |
| S. mitis-CSP-2 | EIRQTHNIFFNFFKRR | 663 | 608–722 | 635 | 426–947 |
| S. mitis-CSP-2-I2MQ4L | EMRLTHNIFFNFFKRR | 49.9 | 42.4–58.9 | 141 | 76.8–257 |
| S. mitis-CSP-2-I2MN7F | EMRQTHFIFFNFFKRR | 131 | 76.5–225 | 123 | 91.7–166 |
| S. mitis-CSP-2-I2MN7I | EMRQTHIIFFNFFKRR | 448 | 345–582 | 54.8 | 51.7–58.0 |
| S. mitis-CSP-2-I2MI8F | EMRQTHNFFFNFFKRR | 151 | 87.1–262 | 83.2 | 65.9–105 |
| S. mitis-CSP-2-I2MN11F | EMRQTHNIFFFFFKRR | 17.7 | 11.5–27.4 | 753 | 573–990 |
| S. mitis-CSP-2-I2MF12L | EMRQTHNIFFNLFKRR | 25.8 | 13.1–50.8 | 146 | 81.3–261 |
| S. mitis-CSP-2-Q4LN7F | EIR L TH F IFFNFFKRR | 371 | 293–469 | 322 | 257–402 |
| S. mitis-CSP-2-Q4LN7I | EIR L TH I IFFNFFKRR | 348 | 278–436 | 315 | 229–433 |
| S. mitis-CSP-2-Q4LI8F | EIR L THN F FFNFFKRR | 280 | 153–513 | 370 | 275–499 |
| S. mitis-CSP-2-Q4LN11F | EIR L THNIFF F FFKRR | 9.38 | 8.44–10.4 | 434 | 308–609 |
| S. mitis-CSP-2-Q4LF12L | EIRLTHNIFFNLFKRR | 68.7 | 50.6–93.2 | 202 | 193–211 |
| S. mitis-CSP-2-N7FI8F | EIRQTH FF FFNFFKRR | — | — | 24.6 | 21.9–27.6 |
| S. mitis-CSP-2-N7FN11F | EIRQTHFIFFFFFKRR | —e | — | —e | — |
| S. mitis-CSP-2-N7FF12L | EIRQTHFIFFNLFKRR | 156 | 84.9–287 | 533 | 262–1083 |
| S. mitis-CSP-2-N7II8F | EIRQTH IF FFNFFKRR | 87.2 | 56.8–134 | 22.8 | 13.1–40.0 |
| S. mitis-CSP-2-N7IN11F | EIRQTHIIFFFFFKRR | —e | — | —e | — |
| S. mitis-CSP-2-N7IF12L | EIRQTHIIFFNLFKRR | 613 | 506–743 | 321 | 216–475 |
| S. mitis-CSP-2-I8FN11F | EIRQTHNFFFFFFKRR | 12.2 | 5.73–26.0 | 284 | 188–429 |
| S. mitis-CSP-2-I8FF12L | EIRQTHNFFFNLFKRR | 160 | 104–248 | 202 | 145–281 |
| S. mitis-CSP-2-N11FF12L | EIRQTHNIFFFLFKRR | 4.97 | 4.12–5.99 | 127 | 117–137 |
| S. mitis-CSP-2-I2MQ4LN7F | EMRLTHFIFFNFFKRR | 137 | 105–178 | 75.6 | 69.2–82.7 |
| S. mitis-CSP-2-I2MI8FN11F | E M RQTHN F FF F FFKRR | 6.95 | 4.69–10.3 | 26.2 | 14.1–49.0 |
| S. mitis-CSP-2-I2MN7FF12L | EMRQTHFIFFNLFKRR | 72.8 | 46.7–114 | 112 | 76.4–163 |
| S. mitis-CSP-2-I2MN7II8F | E M RQTH IF FFNFFKRR | 61.6 | 46.1–82.3 | 2.67 | 1.91–3.73 |
| S. mitis-CSP-2-I2MQ4LF12L | EMRLTHNIFFNLFKRR | 17.1 | 15.8–18.5 | 139 | 92.6–210 |
| S. mitis-CSP-2-I2MQ4LI8F | E M R L THN F FFNFFKRR | 42.0 | 25.7–68.5 | 30.0 | 15.1–59.3 |
| S. mitis-CSP-2-I2MQ4LN7I | EMRLTHIIFFNFFKRR | 155 | 141–170 | 187 | 150–233 |
| S. mitis-CSP-2-I2MQ4LN11F | EMRLTHNIFFFFFKRR | 14.8 | 11.3–19.4 | 188 | 126–281 |
| S. mitis-CSP-2-Q4LN7FI8F | EIRLTHFFFFNFFKRR | 427 | 402–454 | 87.2 | 69.1–110 |
| S. mitis-CSP-2-Q4LN7II8F | EIRLTHIFFFNFFKRR | 242 | 120–489 | —e | — |
| S. mitis-CSP-2-Q4LN7FN11F | EIRLTHFIFFFFFKRR | —e | — | —e | — |
| S. mitis-CSP-2-I2MN7FI8F | E M RQTH FF FFNFFKRR | 63.6 | 34.1–119 | 13.5 | 11.7–15.5 |
| S. mitis-CSP-2-Q4LN7FF12L | EIRLTHFIFFNLFKRR | 426 | 292–622 | —e | — |
| S. mitis-CSP-2-N7FI8FF12L | EIRQTHFFFFNLFKRR | 101 | 63.5–160 | 67.6 | 36.2–126 |
| S. mitis-CSP-2-N7II8FN11F | EIRQTHIFFFFFFKRR | —e | — | —e | — |
| S. mitis-CSP-2-N7II8FF12L | EIRQTHIFFFNLFKRR | 891 | 793–1001 | —e | — |
| S. mitis-CSP-2-I2MN7IN11F | EMRQTHIIFFFFFKRR | 568 | 429–750 | —e | — |
| S. mitis-CSP-2-N7FI8FN11F | EIRQTHFFFFFFFKRR | —e | — | —e | — |
Moving to the triple-modified analogues, it appears that the majority of combinations were not tolerated well, as most analogues exhibited similar or reduced activities compared to the single- or doubly-modified analogues against the pneumococcal ComD receptors (Table 3). However, the triple-modified library also revealed four S. mitis-CSP-2-based analogues, S. mitis-CSP-2-I2MI8FN11F, S. mitis-CSP-2-I2MN7II8F, S. mitis-CSP-2-I2MQ4LI8F, and S. mitis-CSP-2-I2MN7FI8F, with activities at the low nanomolar range (EC50 values <100 nM) against both pneumococcal ComD receptors (Table 3). All four analogues share the same two modifications, I2M and I8F, suggesting that these two modifications are critical for pan-group reactivity of the S. mitis-CSP-2 scaffold against the pneumococcus ComD receptors.
Development of S. mitis-CSP-2-based pneumococcal pan-group QS inhibitors
It has been shown that the interception of the pneumococcal competence regulon by either targeting CSP export or disrupting the native CSP:ComD interactions can be utilized to attenuate QS-dependent pneumococcus pathogenicity.26,27,31–33 All the previously developed CSP-based inhibitors share the same key modification, the replacement of glutamic acid at position 1 with alanine.26–29,32–35 We therefore reasoned that the S. mitis-CSP-2 template can also be converted into a pneumococcus ComD inhibitor by incorporating the same E1A modification. To this end, we first replaced Glu1 with alanine in the native S. mitis-CSP-2 to afford S. mitis-CSP-2-E1A and found that the resulting analogue can effectively inhibit the pneumococcus ComD1 receptor, albeit at a 6-fold higher concentration than S. pneumoniae CSP1-E1A (IC50 value of 497 nM), while losing all activity against the S. pneumoniae ComD2 receptor (Table 4).| Peptide name | Peptide sequence | S. pneumoniae ComD1 | S. pneumoniae ComD2 | ||
|---|---|---|---|---|---|
| IC50b (nM) | 95% CIc | IC50b (nM) | 95% CIc | ||
| a See the Experimental section for full experimental details and the ESI for plots of antagonism dose response curves. b IC50 values were determined by testing peptides over a range of concentrations. c 95% confidence interval. d IC50 not determined due to the analogue's low induction in the primary antagonism screening assay. e IC50 value of S. pneumoniae CSP1-E1A from ref. 27. | |||||
| S. pneumoniae CSP1-E1Ae | AMRLSKFFRDFILQRKK | 85.7 | 50.8–145 | —d | —d |
| S. mitis-CSP-2-E1A | AIRQTHNIFFNFFKRR | 497 | 422–585 | —d | — |
| S. mitis-CSP-2-E1AI2M | AMRQTHNIFFNFFKRR | 85.4 | 57.4–127 | —d | — |
| S. mitis-CSP-2-E1AN11F | AIRQTHNIFFFFFKRR | —d | — | —d | — |
| S. mitis-CSP-2-E1AI2MQ4L | AMRLTHNIFFNFFKRR | 54.2 | 40.6–72.2 | —d | — |
| S. mitis-CSP-2-E1AN11FF12L | AIRQTHNIFFFLFKRR | —d | — | —d | — |
| S. mitis-CSP-2-E1AN7II8F | AIRQTHIFFFNFFKRR | 204 | 133–311 | 135 | 82.5–222 |
| S. mitis-CSP-2-E1AI2MF12L | AMRQTHNIFFNLFKRR | —d | — | —d | — |
| S. mitis-CSP-2-E1AI2MI8F | AMRQTHNFFFNFFKRR | 143 | 88.9–229 | —d | — |
| S. mitis-CSP-2-E1AI2MN7FI8F | AMRQTHFFFFNFFKRR | 294 | 263–328 | 418 | 235–744 |
| S. mitis-CSP-2-E1AI2MN7II8F | AMRQTHIFFFNFFKRR | 141 | 81.8–243 | 32.9 | 16.4–66.0 |
| S. mitis-CSP-2-E1AI2MQ4LI8F | AMRLTHNFFFNFFKRR | 317 | 238–423 | —d | — |
| S. mitis-CSP-2-E1AI2MI8FN11F | AMRQTHNFFFFFFKRR | —d | — | —d | — |
Next, we selected the top S. mitis-CSP-2 modified analogues identified in this study and incorporated the E1A modification. We hypothesized that combining these modifications together with the E1A mutation would result in S. mitis-CSP-2-based analogues that maintain the binding affinity to the pneumococcal ComD receptors but may not be able to activate the receptors, leading to competitive pan-group antagonists of the S. pneumoniae competence regulon. To test our hypothesis, we synthesized eleven S. mitis-CSP-2 analogues bearing the E1A modification. These analogues included two S. mitis-CSP-2 single-modified analogues, five S. mitis-CSP-2 double-modified analogues and four S. mitis-CSP-2 triple-modified analogues, all bearing also the E1A modification, and tested their ability to modulate the QS circuitry in both pneumococcal specificity groups (Table 4). Our analysis revealed that all the S. mitis-CSP-2-E1A-based analogues that had the N11F modification were inactive against both pneumococcal ComD receptors (Table 4). Since the N11F modification resulted in highly potent QS activators, the lack of inhibitory activity for all the E1A-based analogues bearing this modification highlights the stringent and different requirements for ComD receptor inhibition compared to receptor activation. Most importantly, from the S. mitis-CSP-2-E1A-based library we identified three nanomolar range antagonists of both pneumococcus pherotypes, S. mitis-CSP-2-E1AN7II8F, S. mitis-CSP-2-E1AI2MN7II8F and S. mitis-CSP-2-E1AI2MN7FI8F (Table 4). These potent S. mitis-CSP-2-based pan-group inhibitors of the pneumococcus competence regulon are an important addition to the arsenal of chemical tools available to study pneumococcal behavior and attenuate pneumococcal infections.
Conclusions
In conclusion, we set out to investigate potential crosstalk between streptococci species and utilize the identified signaling molecules to develop novel pneumococcal QS modulators. First, our analysis revealed that the majority of CSP signals from closely related streptococci of the mitis group, as well as CSPs from other groups of streptococci, were unable to modulate QS in S. pneumoniae, highlighting the relatively high specificity of the pneumococcus ComD receptors to their cognate native CSP signals. Second, our analysis also revealed a potential role of S. mitis as a modulator of the competence regulon in S. pneumoniae. Our rationally-designed S. mitis-CSP-2-based analogues, generated using the SAR results of both the CSP1 and CSP2 scaffolds in S. pneumoniae,27 yielded several potent pan-group agonists of pneumococcal QS. Moreover, our analysis revealed that by using the S. mitis-CSP-2 pheromone as a template, it is possible to develop potent antagonists of the pneumococcal competence regulon. Most importantly, our work revealed the first pan-group S. mitis-CSP-2-based ComD inhibitors of S. pneumoniae with activities in the nanomolar range.Since S. mitis has the potential to influence the regulation of pneumococcus phenotypes associated with virulence and infectivity, developing highly potent QS modulators using the native CSP pheromone of S. mitis, S. mitis-CSP-2, with enhanced potency could lead to a complementary strategy to attenuate pneumococcal infections. Furthermore, these privileged scaffolds could influence the competence regulon in S. mitis, thus providing novel chemical tools capable of modulating QS in multiple species. Indeed, experiments aimed at identifying the effect of the S. mitis-CSP-2 analogues on the competence regulon in S. mitis are ongoing in our laboratory and will be reported in due course.
Experimental section
Chemical reagents and instrumentation
All the chemical reagents and solvents were purchased from Sigma-Aldrich or Chem-Impex and used without further purification. Water (18 MΩ) was purified using a Millipore Analyzer Feed System. Solid-phase resins were purchased from Advanced Chem Tech.Reversed-phase high-performance liquid chromatography (RP-HPLC) was performed using a Shimadzu system equipped with a CBM-20A communications bus module, two LC-20AT pumps, an SIL-20A auto sampler, an SPD-20A UV/vis detector, a CTO-20A column oven, and an FRC-10A fraction collector. The matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) data were obtained using a Bruker Microflex spectrometer equipped with a 60 Hz nitrogen laser and a reflectron. In positive ion mode, the acceleration voltage on Ion Source 1 was 19.01 kV. The exact mass (EM) data were obtained using an Agilent Technologies 6230 TOF LC/MS spectrometer. The samples were sprayed with a capillary voltage of 3500 V and the electrospray ionization (ESI) source parameters were as follows: gas temperature of 325 °C at a drying gas flow rate of 8 L min−1 at a pressure of 35 psi.
Peptide synthesis
All the CSP analogues were synthesized using the standard Fmoc-based solid-phase peptide synthesis (SPPS) procedures on 4-benzyloxybenzyl alcohol (Wang) resin. Pre-loaded Fmoc-L-Arg (Pbf) Wang resin (0.305 mmol g−1), Fmoc-L-Lys (Boc) Wang resin (0.59 mmol g−1), or Fmoc-Asn (Trt) Wang resin (0.332 mmol g−1) was used for peptides that required an arginine, lysine, or asparagine at the C-terminus, respectively. CSP and analogues were synthesized by using the Liberty1 automated peptide synthesizer (CEM Corporation) (for the full procedures, see the ESI†).Peptide purification by HPLC
Crude peptides were purified with RP-HPLC. The crude peptide was dissolved using ACN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O (1
H2O (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) (volume of ACN in water depends on the solubility of the peptide) and purified in 5 mL portions on a semipreparative Phenomenex Kinetex C18 column (5 μm, 10 mm × 250 mm, 110 Å) with a 5 mL min−1 flow rate; mobile phase A = 18 MΩ water + 0.1% TFA and mobile phase B = ACN + 0.1% TFA. The collected fraction was frozen using an acetone/dry ice bath, lyophilized overnight, and dissolved again in 5 mL ACN
3) (volume of ACN in water depends on the solubility of the peptide) and purified in 5 mL portions on a semipreparative Phenomenex Kinetex C18 column (5 μm, 10 mm × 250 mm, 110 Å) with a 5 mL min−1 flow rate; mobile phase A = 18 MΩ water + 0.1% TFA and mobile phase B = ACN + 0.1% TFA. The collected fraction was frozen using an acetone/dry ice bath, lyophilized overnight, and dissolved again in 5 mL ACN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O (1
H2O (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) for a secondary purification run. Preparative HPLC methods were used to separate the crude peptide mixture to different chemical components using the following linear gradient: (first prep 5% B → 45% B over 40 min and second prep 25% B → 38% B over 45 min). Fraction purity was determined through analysis with a Phenomenex Kinetex analytical C18 column (5 μm, 4.6 mm × 250 mm, 110 Å) using a linear gradient (5% B → 95% B over 27 min). Purities were determined by the integration of peaks with UV detection at 220 nm. Only peptide fractions that were purified to homogeneity (>95%) were used for the biological assays.
3) for a secondary purification run. Preparative HPLC methods were used to separate the crude peptide mixture to different chemical components using the following linear gradient: (first prep 5% B → 45% B over 40 min and second prep 25% B → 38% B over 45 min). Fraction purity was determined through analysis with a Phenomenex Kinetex analytical C18 column (5 μm, 4.6 mm × 250 mm, 110 Å) using a linear gradient (5% B → 95% B over 27 min). Purities were determined by the integration of peaks with UV detection at 220 nm. Only peptide fractions that were purified to homogeneity (>95%) were used for the biological assays.
Biological assays
Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This work was supported by a grant from the National Institutes of Health (R35GM128651). S. pneumoniae D39pcomX::lacZ and TIGR4pcomX::lacZ reporter strains were generous gifts from G. W. Lau (University of Illinois at Urbana-Champaign).References
- S. Mehr and N. Wood, Paediatr. Respir. Rev., 2012, 13, 258–264 Search PubMed.
- S. S. Huang, K. M. Johnson, G. T. Ray, P. Wroe, T. A. Lieu, M. R. Moore, E. R. Zell, J. A. Linder, C. G. Grijalva and J. P. Metlay, Vaccine, 2011, 29, 3398–3412 CrossRef PubMed.
- J. Cornick and S. Bentley, Microbes Infect., 2012, 14, 573–583 CrossRef CAS PubMed.
- N. J. Croucher, S. R. Harris, C. Fraser, M. A. Quail, J. Burton, M. van der Linden, L. McGee, A. von Gottberg, J. H. Song and K. S. Ko, Science, 2011, 331, 430–434 CrossRef CAS PubMed.
- L. Håvarstein, G. Coomaraswamy and D. A. Morrison, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 11140–11144 CrossRef PubMed.
- G. W. Lau, S. Haataja, M. Lonetto, S. E. Kensit, A. Marra, A. P. Bryant, D. McDevitt, D. A. Morrison and D. W. Holden, Mol. Microbiol., 2001, 40, 555–571 CrossRef CAS PubMed.
- L. Zhu, J. Lin, Z. Kuang, J. E. Vidal and G. W. Lau, Mol. Microbiol., 2015, 97, 151–165 CrossRef CAS PubMed.
- M. R. Oggioni, C. Trappetti, A. Kadioglu, M. Cassone, F. Iannelli, S. Ricci, P. W. Andrew and G. Pozzi, Mol. Microbiol., 2006, 61, 1196–1210 Search PubMed.
- D. L. Hava and A. Camilli, Mol. Microbiol., 2002, 45, 1389–1406 CAS.
- S. T. Rutherford and B. L. Bassler, Cold Spring Harbor Perspect. Med., 2012, 2, a012427 Search PubMed.
- D. N. McBrayer, B. K. Gantman, C. D. Cameron and Y. Tal-Gan, Org. Lett., 2017, 19, 3295–3298 CrossRef CAS PubMed.
- B. LaSarre and M. J. Federle, Microbiol. Mol. Biol. Rev., 2013, 77, 73–111 CrossRef CAS PubMed.
- Y. Tal-Gan, D. M. Stacy, M. K. Foegen, D. W. Koenig and H. E. Blackwell, J. Am. Chem. Soc., 2013, 135, 7869–7882 CrossRef CAS PubMed.
- Y. Tal-Gan, M. Ivancic, G. Cornilescu, T. Yang and H. E. Blackwell, Angew. Chem., Int. Ed., 2016, 55, 8913–8917 CrossRef CAS PubMed.
- A. Harrington and Y. Tal-Gan, J. Bacteriol., 2018, 200, e00709–e00717 CrossRef CAS PubMed.
- R. W. Mull, A. Harrington, L. A. Sanchez and Y. Tal-Gan, Curr. Top. Med. Chem., 2018, 18, 625–644 Search PubMed.
- O. Johnsborg, P. E. Kristiansen, T. Blomqvist and L. S. Håvarstein, J. Bacteriol., 2006, 188, 1744–1749 Search PubMed.
- G. Pozzi, L. Masala, F. Iannelli, R. Manganelli, L. Havarstein, L. Piccoli, D. Simon and D. Morrison, J. Bacteriol., 1996, 178, 6087–6090 CrossRef CAS PubMed.
- G. Salvadori, R. Junges, R. Khan, H. A. Åmdal, D. A. Morrison and F. C. Petersen, Methods Mol. Biol., 2017, 1537, 219–232 CrossRef CAS PubMed.
- M. Kilian, K. Poulsen, T. Blomqvist, L. S. Håvarstein, M. Bek-Thomsen, H. Tettelin and U. B. Sørensen, PLoS One, 2008, 3, e2683 CrossRef PubMed.
- L. S. Håvarstein, R. Hakenbeck and P. Gaustad, J. Bacteriol., 1997, 179, 6589–6594 CrossRef PubMed.
- G. Salvadori, R. Junges, D. A. Morrison and F. C. Petersen, Front. Microbiol., 2016, 7, 1009 Search PubMed.
- S. Shekhar, R. Khan, D. M. Ferreira, E. Mitsi, E. German, G. H. Rørvik, D. Berild, K. Schenck, K. Kwon and F. Petersen, Front. Immunol., 2018, 9, 747 CrossRef PubMed.
- C. R. Bikash, S. R. Hamry and Y. Tal-Gan, ACS Infect. Dis., 2018, 4, 1385–1394 Search PubMed.
- W. C. Chan and P. D. White, Fmoc solid phase peptide synthesis: a practical approach, Oxford University Press, Oxford, 2000 Search PubMed.
- L. Zhu and G. W. Lau, PLoS Pathog., 2011, 7, e1002241 CrossRef CAS PubMed.
- Y. Yang, B. Koirala, L. A. Sanchez, N. R. Phillips, S. R. Hamry and Y. Tal-Gan, ACS Chem. Biol., 2017, 12, 1141–1151 Search PubMed.
- B. Koirala, N. R. Phillips and Y. Tal-Gan, ACS Med. Chem. Lett., 2019, 10, 880–886 CrossRef CAS PubMed.
- Y. Yang and Y. Tal-Gan, Bioorg. Chem., 2019, 89, 102987 CrossRef CAS PubMed.
- Y. Yang, G. Cornilescu and Y. Tal-Gan, Biochemistry, 2018, 57, 5359–5369 CrossRef CAS PubMed.
- A. Domenech, A. R. Brochado, V. Sender, K. Hentrich, B. Henriques-Normark, A. Typas and J. W. Veening, Cell Host Microbe, 2020, 27, 544–555 CrossRef CAS PubMed e543.
- B. Koirala, J. Lin, G. W. Lau and Y. Tal-Gan, ChemBioChem, 2018, 19, 2380–2386 CrossRef CAS PubMed.
- Y. Yang, J. Lin, A. Harrington, G. Cornilescu, G. W. Lau and Y. Tal-Gan, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 1689–1699 Search PubMed.
- B. Koirala, R. A. Hillman, E. K. Tiwold, M. A. Bertucci and Y. Tal-Gan, Beilstein J. Org. Chem., 2018, 14, 1769–1777 CrossRef CAS PubMed.
- B. Koirala and Y. Tal-Gan, ChemBioChem, 2020, 21, 340–345 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Full details of experimental procedures, peptide characterization, initial screening results, and dose–response curves for all CSP analogues (PDF). See DOI: 10.1039/d0cb00012d |
| This journal is © The Royal Society of Chemistry 2020 |