Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Hydrogen bonding networks of nalidixic acid–copper(II) complexes†
Catarina
Bravo
 ab,
Filipa
Galego
a and
Vânia
André
ab,
Filipa
Galego
a and
Vânia
André
 *ab
*ab
aCentro de Química Estrutural, Instituto Superior Técnico, Universidade de Lisboa, Av. Rovisco Pais, 1, 1049-001 Lisboa, Portugal. E-mail: vaniandre@tecnico.ulisboa.pt
bAssociação do Instituto Superior Técnico para a Investigação e Desenvolvimento (IST-ID), Av. Rovisco Pais, 1, 1049-003 Lisboa, Portugal
First published on 9th November 2019
Abstract
The usage of quinolone antibiotics has been threatened by antimicrobial resistance, and therefore it is of utmost importance to revitalize these drugs with approaches that may lead to improved properties and increased efficiency. Herein, the formation of hydrogen bonding networks of nalidixic acid–Cu(II) complexes is presented as a possible pathway to achieve that goal.
The increasing interest in copper complexes has been motivated by their potential applications for medical/pharmaceutical purposes,1 as they have been revealing promising activity as antimicrobial,2 antiviral,3 anti-inflammatory4 and even anticancer agents.5 Copper can readily exist in Cu(I) and Cu(II) oxidation states, and this property endows this trace metal with the ability to participate as a catalytic cofactor in several vital biological processes, from bacteria to humans.6,7
Modifying biologically active structures used in medical therapeutics to improve their efficiency and bioavailability is becoming an increasingly recurring approach. Metallopharmaceuticals have been evidenced as an outstanding alternative, increasing considerably the activity of the drugs, and often displaying different mechanisms of action and improving their bioavailability.8 Moulton and Ma, for instance, have already proved that the use of discrete coordination complexes in drug delivery is a good approach to alter the lipophilicity and solubility of pharmaceutically active species.9,10 Lately, the antimicrobial approach of these drugs has attracted special interest considering that intracellular metals strongly influence the activity of antibiotics.8
Quinolones are a family of widely prescribed antibiotics with a broad spectrum of activity and act by inhibiting supercoiling of bacterial DNA by binding to bacterial topoisomerase II (DNA gyrase) and topoisomerase IV in Gram-positive species.11 This effect seems to be induced by a metal-dependent mechanism, mediated by the increase of enzyme–DNA cleavage complexes, stressing the importance of the interaction of this drug with a transition metal ion present in the cytoplasm, such as copper(II), to induce DNA cleavage.12,13 In fact, studies of quinolone–copper complexes have shown changes in the physicochemical properties of pharmaceuticals, such as the decrease of antibiotics' hardness14 and improved lipophilicity, allowing modification in the potency and even in the specificity of the drug.2,10
Nalidixic acid (NALD) was the first quinolone antibiotic used to treat urinary tract infections caused mostly by Gram-negative bacteria. This quinolone was taken off the pharmaceutical market due to its low solubility and was substituted with improved similar active pharmaceutical ingredients.15,16 However, the increasing occurrence of acquired bacterial resistance, observed in different bacterial species treated with quinolones, has been threatening the usage of this drug class,17 and therefore, improving this drug's activity by changing its mechanism of action and physicochemical properties can be a good alternative to work around the problem.
The chelation of nalidixic acid to copper has shown to be a promising alternative to improve the efficiency of the drug.16,18 For instance, biological studies on a reported copper complex with nalidixic acid and 1,10-phenanthroline (Phen), [Cu(Phen)(NALD)(H2O)][NO3]·3H2O, have shown that it interacts with DNA and other nucleic acids such as rRNA and mRNA,19 without the need to bind to enzymes.20,21 This interaction is influenced by different DNA–drug interactions including the formation of hydrogen bonds.12,16,18 Hydrogen bonding has manifold possibilities of donor and acceptor group-interactions between the drug and the DNA helix, and depending on the drug's characteristics, the interaction might be completely different.22,23 In metallopharmaceuticals, for instance, the presence of different counterions in the drug24 or even the presence of solvent molecules on its structure might have an important role in its biological activity. One interesting undervalued characteristic in drugs is perhaps a drug structure with water bridges; although they are not a common feature in the structures of many drugs,23 they have proved to be quite relevant to the activity of several compounds and to the way they interact with DNA through hydrogen-bonding.25,26
Hence, it is important to fully understand the crystal structures of compounds, investigate the effects of intramolecular hydrogen donor functional groups and study how the presence of different counterions and water molecules can influence intermolecular interactions and even the structure of the compound. In other words, a better understanding of the packing of structures mostly dependent on intermolecular hydrogen bonding effects in these cases could be a possible route to have insight into different properties of the drug.
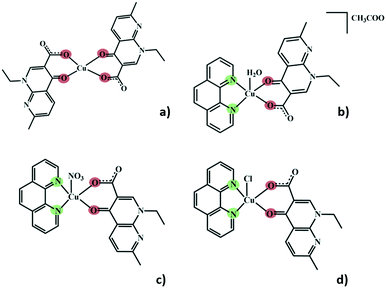
Herein, we present a crystallography focused study of five different structures of Cu(II) complexes comprising nalidixic acid, four of which also include 1,10-phenanthroline as a secondary ligand. The different Cu(II) salts and 1,10-phenanthroline have been shown to influence the global packing of these compounds, and the importance of the water molecules in the formation of hydrogen bonding frameworks became evident. In all these five structures, nalidixic acid is deprotonated and coordinates to the metal via the carbonyl group and one oxygen atom of the carboxylate moiety. 1,10-Phenanthroline coordinates via both N-sites (Fig. 1).
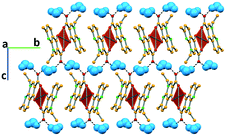
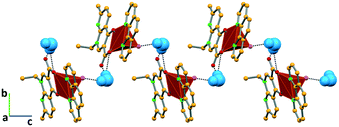
In complex I ([Cu(NALD)2]·H2O), Cu(II) lies on an inversion centre and it assumes a square planar geometry and a water molecule establishes hydrogen bonds between two carboxylate groups (O–Hw⋯OCOO−, 3.189(5) and 2.861(5) Å) connecting consecutive complexes and giving rise to 2D hydrogen bonding frameworks (Fig. 2).
The use of 1,10-phenanthroline induces the change into a square pyramidal geometry (τ ≈ 0) of the metal centre, in which the apical position is differently occupied, depending on the metal salt used during the synthesis. In complex II ([Cu(NALD)(Phen)(H2O)]+·[CH3COO]−·3H2O), copper acetate was used and the apical position is occupied by a water molecule, with the acetate anion cocrystallizing as a counterion along with two water molecules. In complex III ([Cu(NALD)(Phen)(NO3)]·H2O), the nitrate from the salt occupies the apical position and a non-coordinated water molecule is also present in the structure. Complexes IV ([Cu(NALD)(Phen)(Cl)]·H2O) and V ([Cu(NALD)(Phen)(Cl)]·4H2O) resulted from experiments with copper chloride, and, in both cases, the chloride occupies the apical position in the Cu(II) coordination geometry; the difference in the chemical composition of both of these complexes is that IV is monohydrated and V is tetrahydrated.
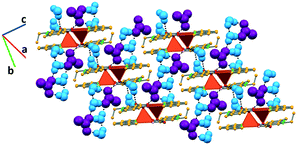
In complex II, the coordinated water molecule establishes hydrogen bonds with an acetate anion (O–Hw⋯OAc, 2.672(6) Å) and another complex (O–Hw⋯O, 2.796(6) Å). The acetate anion further interacts with three hydration water molecules (O–Hw⋯OAc, 2.770(8), 2.790(8) and 2.785(8) Å). The third crystallographically independent hydration water molecule bridges together two complexes by interacting with the coordinated water of one complex (O–Hw⋯Ow, 2.886(8) Å) and the carboxylate moiety of nalidixic acid of the other complex (O–Hw⋯OCOO−, 2.994(7) Å). Furthermore, two of the hydration water molecules interact among them (O–Hw⋯Ow, 2.931(8) Å), forming a D2 discrete chain, according to the Infantes and Motherwell27 notation for water clusters. All these interactions give rise to a 2D hydrogen bonding framework, in which pairs of complexes orient themselves in an antiparallel fashion due to direct hydrogen bonds among them and are connected to other pairs by an extended hydrogen bonded system (Fig. 3).
In complex III, the water molecule bridges consecutive complex units by establishing hydrogen bonds with the nitrate (O–Hw⋯ONO3−, 2.907(8) Å) and the carboxylate (O–Hw⋯OCOO−, 2.877(7) Å) moieties of the consecutive complexes, giving rise to 1D frameworks (Fig. 4).
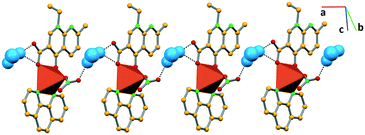
Similarly, in complex IV, the water molecule connects the succeeding complexes via hydrogen bonding with the carboxylate moiety of nalidixic acid (O–Hw⋯OCOO− 2.985(9) Å) of one complex and the coordinated chloride (O–Hw⋯Cl 3.297(7) Å) of another complex, forming a 1D network that grows along the c axis (Fig. 5).
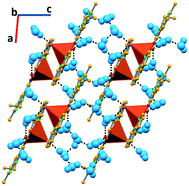
In complex V, due to the higher number of water molecules, the supramolecular arrangement is again based on a higher number of hydrogen bonds. One water molecule directly links two complexes via O–Hw⋯Cl and O–Hw⋯OCOO− hydrogen bonds (3.173(8) and 2.728(10) Å, respectively). The chloride is further connected to a second water molecule (O–Hw⋯Cl, 3.307(11) Å), as well as the carboxylate moiety of nalidixic acid (O–Hw⋯OCOO−, 2.762(12) Å). Water molecules further interact among them (O–Hw⋯Ow, 2.03(10), 1.86(12), 1.93, and 2.00 Å), developing a 2D hydrogen bonding framework (Fig. 6). Using the Infantes and Motherwell27 notation for water clusters, it is possible to say that water clusters are based on R6 rings and D2 discrete chains.
A more detailed study of the interactions between atoms can be done, referring to the analysis of the Hirshfeld surfaces28–30 and the 2D fingerprint plots.31 Using this approach, it is possible to say that the hydrogen bond interactions (O–H interactions) discussed before represent a higher percentage of the total interactions in complex III (29%) and a lower one in complex IV (12.6%). It is worth noting that in complex IV, only one O–H⋯O hydrogen bond is reported, as Cl competes with O to establish intermolecular contacts, justifying the lower percentage in complex IV. It is also interesting that the structures with the higher percentage of O–H interactions correspond to the ones with the higher percentage of filled space (72.9% and 71.4% for complexes I and III, respectively, versus 70.8%, 70.2% and 69% for complexes II, IV and V, respectively).
The H–H interactions range from 39.1% in complex III to 49.1% in complex II of the total Hirshfeld surface, with the presence of the acetate being responsible for the increased percentage in complex II.
The C–H interactions are also prominent, especially in complex II (19.2%), assuming a lower percentage in complex I (6.8%). Apart from the above-mentioned interactions, the π⋯π (C–C) contacts represent approximately 10% (9.1–11.1%) of the total interactions, except for complex II in which they are quite less significant (0.6%). The presence of the acetate (in complex II) is responsible for the non-alignment of the aromatic moieties precluding the π⋯π (C–C) interactions.
Lone-pair⋯π (O–C) interactions are also observed (1.2–3.3%). The C–N interactions are considerable for complexes I, II, IV and V (2.1–3.3%), but negligible for complex III (0.4%). N–O contacts represent 1.6 and 1.5% of the total interactions in complexes I and III, and are negligible for the other structures. For complexes IV and V, we must further consider the Cl–H interactions (11.5 and 9.8%, respectively).
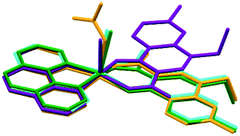
Regarding the conformation of complexes II to V, it is interesting to note that the main difference in the position of nalidixic acid is detected in II. Complex II is the only complex in which the counterion does not coordinate to Cu(II) and this seems to influence the orientation in which our active pharmaceutical ingredient coordinates to the metal centre (Fig. 7).
Infrared spectroscopy data corroborate the crystal structure determined from single crystal X-ray diffraction data, proving the presence of the expected functional groups in the complexes. The broad bands observed in the 3200–3501 cm−1 region are attributed to the hydrogen bonds found in all the structures as well as the stretching vibration of the hydration water molecules.
Complexes I and II are obtained as pure phases, and therefore further characterization is possible. Complex III is obtained with NH4NO3. Complex IV is also obtained concomitantly with complex V.
Compounds I and II are stable until temperatures close to 100 °C, as proven by differential scanning calorimetry (DSC), thermogravimetric analysis (TGA) and/or hot-stage microscopy (HSM). The calculations of the mass losses detected in the TGA are in agreement with the expected values for the water loss, at around 100 °C.
Conclusions
Within this work, five new nalidixic acid–Cu(II) complexes were disclosed, with 1,10-phenanthroline as a second ligand in four of them. Mechanochemistry has been proven once again13 to be a great technique for the synthesis of these compounds.‡ All of the complexes form 1D or 2D hydrogen bonding frameworks and the interactions with water are crucial for their stability. This type of framework may change the type of biological interaction with DNA and should be further studied.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Authors acknowledge Fundação para a Ciência e a Tecnologia (FCT, Portugal) (projects UID/QUI/00100/2019 and PTDC/QUI-OUT/30988/2017 and contract under DL No. 57/2016 regulation) and FEDER, Portugal2020 and Lisboa2020 for funding (project LISBOA-01-0145-FEDER-030988). Dr Auguste Fernandes is acknowledged for the FTIR, DSC and TGA data.Notes and references
- C. Duncan and A. R. White, Metallomics, 2012, 4, 127–138 RSC.
- S. Chandraleka, K. Ramya, G. Chandramohan, D. Dhanasekaran, A. Priyadharshini and A. Panneerselvam, J. Saudi Chem. Soc., 2014, 18, 953–962 CrossRef.
- N. Shahabadi and P. Fatahi, DNA Cell Biol., 2012, 31, 1328–1334 CrossRef CAS PubMed.
- J. E. Weder, C. T. Dillon, T. W. Hambley, B. J. Kennedy, P. A. Lay, J. R. Biffin, H. L. Regtop and N. M. Davies, Coord. Chem. Rev., 2002, 232, 95–126 CrossRef CAS.
- C. Marzano, M. Pellei, F. Tisato and C. Santini, Anti-Cancer Agents Med. Chem., 2009, 9, 185–211 CrossRef CAS PubMed.
- I. Iakovidis, I. Delimaris and S. M. Piperakis, Mol. Biol. Int., 2011, 2011, 1–13 CrossRef PubMed.
- T. J. Meyer, J. Ramlall, P. Thu and N. Gadura, International Journal of Biological, Biomolecular, Agricultural, Food and Biotechnological Engineering, 2015, 9, 273–278 Search PubMed.
- K. Poole, Curr. Trends Microbiol., 2017, 25, 820–832 CrossRef CAS PubMed.
- Z. Ma and B. Moulton, Coord. Chem. Rev., 2011, 255, 1623–1641 CrossRef CAS.
- Z. B. Ma and B. Moulton, Mol. Pharmaceutics, 2007, 4, 373–385 CrossRef CAS PubMed.
- P. Ball, in The Quinolones, ed. V. T. Andriole, Academic Press, San Diego, 3rd edn, 2000, pp. 1–31 Search PubMed.
- E. Y. Bivián-Castro, M. G. López, M. Pedraza-Reyes, S. Bernès and G. M. Díaz, Bioinorg. Chem. Appl., 2009, 2009, 1–8 CrossRef PubMed.
- V. Uivarosi, Molecules, 2013, 18, 11153–11197 CrossRef CAS PubMed.
- J. Robles, J. Martín-Polo, L. Alvarez-Valtierra, L. Hinojosa and G. Mendoza-Díaz, Met.-Based Drugs, 2000, 7, 301–311 CrossRef CAS PubMed.
- V. Andre, F. Galego and M. Martins, Cryst. Growth Des., 2018, 18, 2067–2081 CrossRef CAS.
- I. Sousa, V. Claro, J. L. Pereira, A. L. Amaral, L. Cunha-Silva, B. de Castro, M. J. Feio, E. Pereira and P. Gameiro, J. Inorg. Biochem., 2012, 110, 64–71 CrossRef CAS PubMed.
- K. J. Aldred, R. J. Kerns and N. Osheroff, Biochemistry, 2014, 53, 1565–1574 CrossRef CAS PubMed.
- H. Farrokhpour, H. Hadadzadeh, F. Darabi, F. Abyar, H. A. Rudbari and T. Ahmadi-Bagheri, RSC Adv., 2014, 4, 35390–35404 RSC.
- N. Ramírez-Ramírez, G. Mendoza-Díaz and M. Pedraza-Reyes, Bioinorg. Chem. Appl., 2003, 1, 25–34 CrossRef PubMed.
- N. Ramirez-Ramirez, G. Mendoza-Díaz, F. Gutiérrez-Corona and M. Pedraza-Reyes, J. Biol. Inorg. Chem., 1998, 3, 188–194 CrossRef CAS.
- G. Mendoza-Díaz, L. M. R. Martinez-Aguilera, R. Perez-Alonso, X. Solans and R. Moreno-Esparza, Inorg. Chim. Acta, 1987, 138, 41–47 CrossRef.
- L. Tabernero, J. Bella and C. Aleman, Nucleic Acids Res., 1996, 24, 3458–3466 CrossRef CAS PubMed.
- S. K. Panigrahi and G. R. Desiraju, J. Biosci., 2007, 32, 677–691 CrossRef CAS PubMed.
- P. Díaz, J. Benet-Buchholz, R. Vilar and A. J. P. White, Inorg. Chem., 2006, 45, 1617–1626 CrossRef PubMed.
- D. G. Brown, M. R. Sanderson, J. V. Skelly, T. C. Jenkins, T. Brown, E. Garman, D. I. Stuart and S. Neidle, EMBO J., 1990, 9, 1329–1334 CrossRef CAS PubMed.
- A. Canals, M. Purciolas, J. Aymani and M. Coll, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2005, 61, 1009–1012 CrossRef PubMed.
- L. Infantes and S. Motherwell, CrystEngComm, 2002, 4, 454–461 RSC.
- J. McKinnon, M. Spackman and A. Mitchell, Acta Crystallogr., Sect. B: Struct. Sci., 2005, 60, 627–668 CrossRef PubMed.
- M. Servati-Gargari, S. K. Seth, R. L. LaDuca, O. Z. Yesilel, A. Pochodylo, A. Bauzá, B. C. Jana, T. Arslan, A. Frontera and G. Mahmoudi, Inorg. Chim. Acta, 2015, 438, 220–231 CrossRef CAS.
- J. Dalal, N. Sinha, H. Yadav and B. Kumar, RSC Adv., 2015, 5, 57735–57748 RSC.
- R. Soman, S. Sujatha, S. De, V. C. Rojisha, P. Parameswaran, B. Varghese and C. Arunkumar, Eur. J. Inorg. Chem., 2014, 2014, 2653–2662 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. CCDC 1938699–1938703 for complexes I to V. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c9ce01057b |
| ‡ Mechanochemistry (liquid-assisted grinding) and solution synthesis have been used in the synthesis of these compounds. In both cases ammonia was used to promote the deprotonation of nalidixic acid and facilitate the coordination to Cu(II). |
| This journal is © The Royal Society of Chemistry 2019 |