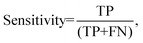A high-throughput serum Raman spectroscopy platform and methodology for colorectal cancer diagnostics
Cerys A.
Jenkins†
 *a,
Rhys A.
Jenkins
b,
Meleri M.
Pryse
b,
Kathryn A.
Welsby
b,
Maki
Jitsumura
c,
Catherine A.
Thornton
a,
Peter R.
Dunstan
*a,
Rhys A.
Jenkins
b,
Meleri M.
Pryse
b,
Kathryn A.
Welsby
b,
Maki
Jitsumura
c,
Catherine A.
Thornton
a,
Peter R.
Dunstan
 b and
Dean A.
Harris
c
b and
Dean A.
Harris
c
aSwansea University Medical School, Institute of Life Science 1, Swansea University, Swansea, UK. E-mail: cerys.jenkins@swansea.ac.uk; Tel: +01792 604843
bDepartment of Physics, Centre for Nanohealth, Swansea University, Swansea, UK
cDept of Colorectal Surgery, Morriston Hospital, Swansea, UK
First published on 1st November 2018
Abstract
Vibrational spectroscopic techniques such as Raman spectroscopy and Fourier transform infrared spectroscopy (FTIR) have huge potential for the analysis of biological specimens. The techniques allow the user to gain label-free, non-destructive biochemical information about a given sample. Previous studies using vibrational spectroscopy with the specific application of diagnosing colorectal diseases such as cancer have mainly focused on in vivo or in vitro studies of tissue specimens using microscopy or probe based techniques. There have been few studies of vibrational spectroscopic techniques based on the analysis of blood serum for the advancement of colorectal cancer diagnostics. With growing interest in the field of liquid biopsies, this study presents the development of a high-throughput (HT) serum Raman spectroscopy platform and methodology and compares dry and liquid data acquisition of serum samples. This work considers factors contributing to translatability of the methodologies such as HT design, inter-user variability and sample handling effects on diagnostic capability. The HT Raman methods were tested on a pilot dataset of serum from 30 cancer patients and 30 matched control patients using statistical analysis via cross-validated PLS-DA with a maximum achieved a sensitivity of 83% and specificity of 83% for detecting colorectal cancer.
1 Introduction
Colorectal cancer (CRC) is the fourth most common cancer in the UK and is the third most common cause of cancer death globally.1,2 There are difficulties associated with diagnosing CRC early as there are no specific ‘red flag’ symptoms.3 This results in large numbers of patients being referred on a 2 weeks wait ‘fast-track’ pathway to have cancer excluded in accordance with NICE guidance. The ‘high risk’ symptoms qualifying for the pathway have just a 3% positive predictive value which means that the majority of the large numbers of patients referred do not have cancer. Currently, the ‘gold standard’ diagnostic test for CRC is colonoscopy with sensitivity 95% and specificity 90%. As a specialised investigation requiring a skilled operator timely access to colonoscopy is restricted. The procedure is costly (£401, NHS England tariff4), carries a risk of complication, is often poorly tolerated by the patient and requires oral bowel cleansing preparation. There is therefore a pressing need for a less invasive yet highly accurate test for colorectal cancer to streamline the need for confirmatory testing and assist with earlier detection of cancer.Raman spectroscopy (RS) is a vibrational spectroscopy technique providing unique spectral characteristics from the scattering of incident light interacting with the sample in question. When used appropriately it is a non-destructive technique that has been previously reported in pilot studies for the detection of cancer in both in vitro and in vivo studies.5–9 Previously, emphasis has been made on the use of Raman spectroscopy for histopathology applications and it has been shown that Raman spectroscopy can be used to accurately differentiate between diseased and healthy tissue in gastrointestinal, oral, breast and brain cancers.10,11 More recently, there have been studies exploring the potential of Raman spectroscopy to analyse biofluids for disease detection using plasma, serum and urine.12–14 These are more accessible than tissue samples traditionally used to perform Raman analysis. However, despite protocols being published to try and standardise RS analysis of biological samples, there is a lack of large biofluid serum RS studies towards clinical translation compared to similar FTIR applications.15,16 This includes studies into inter-operator usability and the effect of different sampling modalities and pre-analytical considerations for biofluid RS where these have been reported for FTIR.17,18 Another potential hurdle of biofluid RS are the lack of high throughput (HT) systems for different sampling modes, such as liquid and dry samples.
This study presents the development of high throughput (HT) platforms for liquid and dry serum RS for colorectal cancer detection. Currently the referral of symptomatic patients into secondary care is very high with only 3–5% of referrals ultimately being diagnosed with CRC. This provides the rationale for a blood test approach using Raman Spectroscopy. The HT platform has been developed for use as a potential triage tool in primary care for symptomatic patients with suspected colorectal cancer. The method has been refined using cell free serum in line with traditional pathology laboratory techniques. Serum has better long term stability than whole blood without the spectral contamination from cellular or coagulation factors. The principles of the HT platforms developed within this study are not limited to serum however and would be applicable to any biofluid sample. The HT platforms are tested in a pilot study and compared in a cohort of 60 patients (30 cancer and 30 healthy) using partial least squares discriminant analysis (PLS-DA). This is in line with previous vibrational spectroscopic pilot studies for cancer detection.14,19–21 The work will investigate spectral reproducibility of serum Raman spectra in the liquid HT platform considering effects of sampling modality and freeze–thaw cycles on the diagnostic capability of the method. Clinical discrimination will be investigated and compared between methods using cross-validated PLS-DA. The results of this study show great potential for a HT Raman platform as a novel diagnostic tool for CRC detection.
2 Materials and methods
2.1 Ethical approval and patient recruitment
Informed consents were obtained from all participants of this study. Blood samples from all patients were collected after informed consent. Full ethical approval for all aspects of this study was granted by the Wales Research and Ethics Committee (REC reference 14/WA/0028). Serum samples from all patients were collected by trained phlebotomists through a collaboration with Abertawe Bro Morgannwg University Local Health Board (ABMU). Samples were collected and analysed in accordance with the UK policy framework for health and social care research. All samples in the work presented were collected from a secondary care patient population. Samples from patients with colorectal cancer were confirmed to be adenocarcinoma by a pathologist. Healthy control patient samples were collected after confirmation of a negative colonoscopy.2.2 Patient demographics
A total of 60 patients were recruited for this study into 2 sample groups. The study groups included early and late stage colorectal cancer patients (TNM stage T1–4) and control patients who were confirmed not to have colorectal cancer by a normal colonoscopy. Both cohorts were age and sex matched as seen in Table 1. All patients were fasted for at least 8 h prior to blood collection to eliminate the influence of the fed state.| Study group | Number patients | Mean age (years) | Number smokers | Number males | Number females |
|---|---|---|---|---|---|
| Cancer | 30 | 67.7 ± 9.7 | 8 | 15 | 15 |
| Control | 30 | 65.0 ± 12.8 | 3 | 13 | 17 |
| Total | 60 | 66.4 ± 11.3 | 11 | 28 | 32 |
2.3 Serum separation
Blood was collected via venous collection into Vacutainer SST collection tubes (BD, USA). Samples were spun at 1300 rcf for 10 min within 2 hours of collection. Fresh samples (250 μl aliquots) were then kept on ice until data collection within 24 h of blood collection as to prevent sample degradation. The remainder of the samples were aliquoted again at 250 μl and stored at −80 °C. Frozen samples were passively thawed on ice to room temperature and data was collected within 24 h of thawing.2.4 Spectroscopic data collection
The spectral acquisition utilises rapid but multiple exposures to collect individual spectra which are then accumulated into one representative spectrum. This allows for assessment of sample degradation. Analysis of the spectra as a function of exposure confirms that the powers detailed below were suitable to preserve the integrity of the serum samples. Use of this acquisition methodology allows self-consistent checks of sample integrity in relation to the diagnostic model development.
2.4.1.1 Substrate optimisation studies. Substrates were interrogated with the 785 nm laser line and with the 532 nm laser line with 165–170 mW and 45–55 mW of power, respectively. 5 spectra were collected and averaged for each substrate to demonstrate Raman spectral response allowing variances to be checked and effective cosmic ray identification/removal.
2.4.1.2 Optimisation of position for dry serum data collection. Serum samples (3 μl) were pipetted onto an aluminium foil substrate in duplicate and left to dry for 1 hour at room temperature. A 20× N Plan EPI objective (Leica, USA) was used to take point spectra across the droplet in a ‘snake’ configuration to create a Raman map. Samples were interrogated with 785 nm and 532 nm laser lines with 65–85 mW and 25–45 mW of power at the sample respectively. The mapping data were then used to optimise position across dried droplets for HT dry serum data collection.
2.4.1.3 Dry serum Raman data collection. Serum samples (3 μl) were pipetted onto an aluminium foil multi-well substrate in duplicate and left to dry for 1 hour at room temperature. Point spectra were collected across each dried droplet with 5 spectra being taken across the two droplets. Samples were interrogated with 785 nm and 532 nm laser light with 65–85 mW and 25–45 mW of power using a 20× N Plan EPI objective (Leica, USA). Total acquisition time for 532 nm and 785 nm was approximately 10 minutes.
2.4.1.4 Liquid serum Raman data collection. Liquid serum samples were excited with both the 785 nm and 532 nm sources with a 10× dry objective. Spectra were recorded from 200 μl liquid serum samples in the sample substrate and 5 repeat spectra were collected from a single sample. Samples were interrogated with approx. 65–85 mW of power from the 532 nm laser and 40 mW from the 785 nm laser. The total time for data collection was approximately 6 min and 12.5 min for the 532 nm and 785 nm laser sources including repeat spectra.
2.4.1.5 Temperature effect on spectral reproducibility. A liquid measurement platform has been developed for a high throughput environment, incorporating a temperature stabilised plate that houses a 40-well plate machined from 316L stainless steel. The stainless steel substrate was chosen for its good thermal conductivity, non-corrosive properties and minimal Raman response. The temperature stabilised liquid platform is used to control the temperature of the serum within the well plate with an accuracy better than ±0.25 °C, as measured via thermocouples within the platform.
The effect of temperature stabilisation on spectral reproducibility was evaluated in this work. Serum spectra were collected without the use of the temperature stabilised platform and compared to serum spectra taken using the platform. The temperature of the well plate is kept above the dew point to prevent condensation from forming when in use.
2.5 Data preprocessing
The raw Raman spectral data were subject to a spectral preprocessing routine consisting of wavenumber standardisation, background subtraction using a rolling circle filter algorithm and normalisation to the peak at 1004 cm−1 developed in house.222.6 Data analysis
Preprocessed data were used for spectral comparisons and for computing the mean and difference spectra to demonstrate spectral differences and make spectral comparisons.To highlight spectral variation due to inter-operator variability and the effects of temperature on the spectra, preprocessed spectra were subject to PCA analysis. PCA is an unsupervised analysis technique. PCA transforms spectral datasets under a matrix transformation such that the spectral variances are maximised. The transformed data are set up such that the principle components (PCs) of the transformation are in rank order of spectral variance. It is then possible to project the component scores and plot to investigate spectral differences. Loading plots on the PCs were also plotted to investigate the underlying spectral causes of the variation described by the PCs.
Processed spectra were also subject to partial least squares discriminant analysis (PLS-DA). Briefly, PLS-DA is a multivariate analysis technique that can be used to investigate causes of differences and variances within datasets.26 It is based on partial least squares regression (PLS) and can be used on datasets that have binary groups (e.g. cancer vs. control). PLS regression can be used to form a linear multivariate model between two matrices (X and Y), where in our case X is the spectral dataset and Y is a set of observable variables. The discriminant analysis or (PLS-DA) is used where Y is known and a PLS regression model is built between a dataset matrix (X) and a ‘label’ matrix (Y) where the ‘label’ matrix contains numbers that correspond to groups within the dataset e.g. (1 = Cancer, −1 = Control). By cross validating PLS-DA models classification performance can be measured for a given dataset in terms of a confusion matrix. Therefore, this technique lends itself well to investigating the effect of different pre-analytical techniques by allowing the user to both investigate the causes of variance within a dataset via loadings and PLS-DA scores but also to quantify the result via giving a numerical performance to measure the magnitude of the changes. All spectra were analysed on a spectrum-wise basis as the dataset is relatively small.27 The number of latent variables chosen in the classification model for each was different. The number selected minimised the cross validation (CV) error. Post CV, sensitivity and specificity for detecting colorectal cancer were calculated from CV confusion matrices for each technique as follows:
where TN is true negatives, TP is true positives, FP is false positives and FN is false negative results. The calculated sensitivities and specificities were then used as a measure to demonstrate model performance between methodologies.
3 Results and discussion
When developing HT methodologies for diagnostic serum Raman spectroscopic methods the substrates, measurement positions, treatment of the samples and treatment of the data must be simple and consistent. To develop the methodologies for dry and liquid HT Raman for CRC detection, optimisation was carried out on the measurement substrates, conditions and the way serum samples were treated before and during measurements as seen in Table 2. The effect of the different optimised methodologies on diagnostic capability and practicality were then compared against each other considering the optimal laboratory setting and also in terms of translating the techniques into a clinical setting.| Optimisation studies | |
| Laser source | 785 nm and 532 nm |
| Substrates | Glass, polypropylene, aluminium foil, calcium flouride |
| Measurement position (dry) | Position across droplet |
| Measurement environment (liquid) | Temperature stable vs. Non-temperature stabilised |
| Inter-user variation (dry and liquid) | User 1 vs. user 2 |
| Effects on diagnostic capability | |
| HT methods | Dry vs. liquid |
| Freeze–thaw cycles | Fresh vs. freeze–thawed |
| Dry spectroscopic comparison | HT liquid Raman |
| Liquid spectroscopic comparison | HT dry Raman |
3.1 Development of high-throughput dry serum Raman spectroscopy
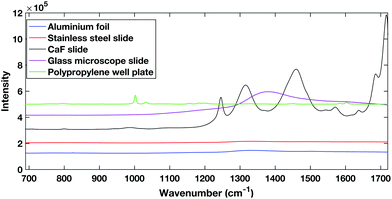 | ||
| Fig. 1 Comparison of different potential substrates for dry HT serum Raman spectroscopy. Spectra are offset for clarity. | ||
The polypropylene slides and the calcium fluoride slide both showed large spectral contributions. The calcium fluoride disk had been used previously and cleaned showing that despite the low expected spectral contribution, calcium fluoride disks can undergo degradation. The glass slides tested are cheap and are already used commonly in a clinical setting, however, they have a large spectral response when excited within the NIR region. The aluminium foil and stainless steel slides showed the lowest spectral contribution when excited with a 785 nm laser.
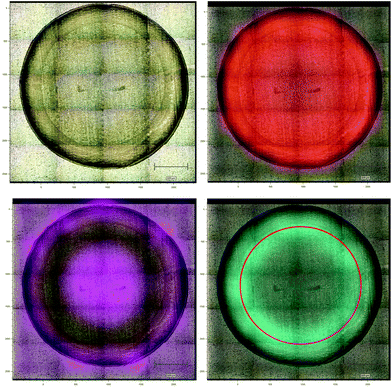
Aluminium foil previously has been reported as a potential substrate for biofluid analysis with Raman spectroscopy.16,28 However, it can be subject to warping or crumpling effects while a droplet dries onto it. To combat this the aluminium foil substrate in this work was pressed and dimpled into a multi-well design minimising the warping and crumpling effects. This reduced dry droplet cracking and allowed HT dry droplet data acquisition with excellent SNR and good spectral reproducibility as seen in Fig. 2. One drawback of using a dry serum RS process is that serum components appear to segregate during the drying process leaving an inhomogeneous film with inherent spectral variability. This process can vary across different substrates. Therefore, the region in which spectral measurements were to be taken was optimised using PCA mapping. Dried droplets were mapped over the fingerprint region (610 cm−1–1720 cm−1). PCA maps were then generated and superimposed onto white light images of the droplets using a non-mean centred PCA algorithm with the most variable areas having brighter mapped colours as seen in Fig. 3. As expected with a non-centred PCA the first PCA map shows homogeneity across the sample representing the mean dry spectrum. When investigating the map across PC2 it is clear that there is a darker and therefore less variable region across the droplet just inside the outer ring. This least variable region was therefore selected as the optimal region in which to take spectral measurements to reduce spectral variability due to the sample drying effects.
 | ||
| Fig. 2 Representative example of raw spectra data from the aluminium foil multi-well substrate (inset). | ||
3.2 Dry platform inter-user variability
For a HT technique to be considered translatable as a clinical diagnostic platform, it must be repeatable for multiple trained users. The inter user variability of the dry HT technique was tested by having two experienced users take measurements from the same dried droplet in the same laboratory within the same hour (to minimise the effects of external laboratory conditions). This was performed on the same instrument with the same instrument calibrations. Measurements were taken in immediate succession i.e. user 1 collected 5 spectra then user 2 collected 5 spectra. Therefore, the only experimental variability that should be introduced is the difference between the user measurements. Fig. 4 shows the PCA score plot for two users taking 5 individual spectra across the same dried droplet following the protocol for the positioning as set out in Fig. 3.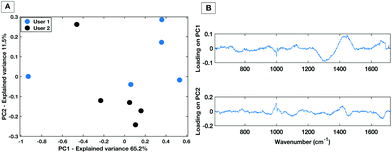 | ||
| Fig. 4 PC score plot showing PC1 vs. PC2 (A) and loading plot (B) for inter-user variability study for the dry HT platform. | ||
The PC1 vs. PC2 plot shows that between the two users there is a general separation across PC2 with the spectra from each user grouping together. This implies that the largest spectral variances are due to inter-user variability. The associated loading plot for PC1 and PC2 shows that there are spectral variances mostly caused by the 1300 cm−1–1400 cm−1 spectral region. This is associated with a spectral contribution from the aluminium substrate in this region (Fig. 1). Therefore, despite optimisation of the region across a dried droplet in which users take measurements there are still differences in the substrate spectral contribution. Repeated tests by the users confirmed that the dried droplet approach leads to variances caused by which operator was taking the measurements. Therefore, when taking spectral measurements by the dry platform the user would need to be kept consistent for a given dataset to ensure spectral comparability.
3.3 Development of high-throughput liquid serum Raman spectroscopy
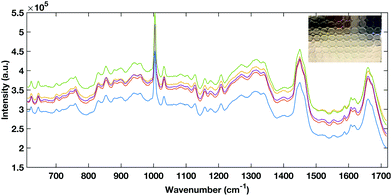
Multi-well HT measurement platforms are used across biological research to allow the analysis of multiple samples or replicates. Typically for biological applications polypropylene well plates are used. However, plastic multi well plates tend to have a large spectral contribution due to their structure which can ‘flood’ the biological spectral information as seen in Fig. 5. As aluminium had a low spectral contribution when testing dry substrates, an aluminium well plate was tested but tarnished and was subject to surface oxidisation.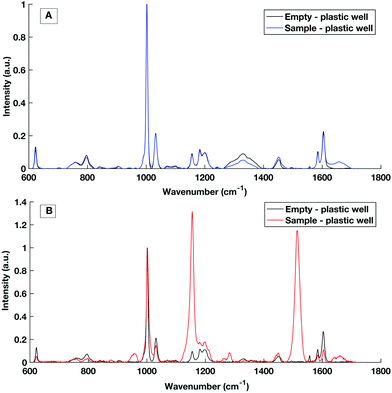 | ||
| Fig. 5 Raman spectral contribution of empty plastic multiwell plate with and without a serum sample interrogated with (A) 785 nm laser excitation and (B) 532 nm laser excitation. | ||
To avoid the spectral contribution from a plastic system a 40-well stainless steel substrate was developed for HT serum RS. The stainless steel well plate allows rapid spectral collection from multiple liquid samples, offers a low Raman background contribution and, via employing the same protocols for the cleaning of surgical instruments, offers a re-usable and contaminant-free platform for sample handling. Fig. 6 shows a representative example of raw liquid spectral data collected using the developed HT platform from 785 nm (a), 532 nm excited serum (b) and the well plate design (inset).
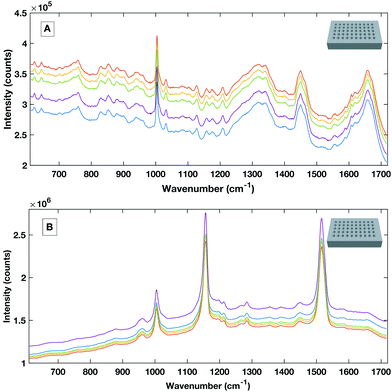 | ||
| Fig. 6 Example raw spectral data taken from the stainless steel HT platform showing good SNR rand spectral repeatability with the 785 nm excitation (A) and the 532 nm excitation (B). | ||
Liquid serum spectra collected using the platform with both 785 nm and 532 nm laser excitation show good SNR, minimal substrate contribution and also good spectral variability within repeat measurements. The stainless steel design was therefore used a simple, cost effective HT substrate.
3.4 Temperature and sample degradation study
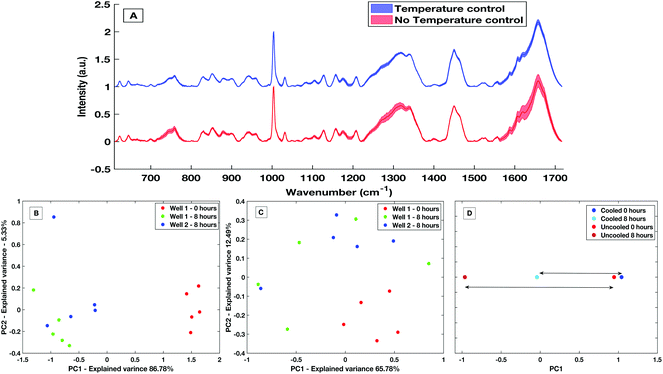
The development of a high throughput liquid platform allows up to 40 liquid samples to be loaded into the stainless-steel well plate at once and analysed via automated hardware and software protocols with no user interaction necessary between sample runs. Based on collecting five acquisitions per well, 8 hours is required to analyse the entire well plate. Efforts must be made to keep the liquid samples stable throughout this 8 hours period in which evaporation effects can lead to spectra deemed as unreliable from a diagnostic perspective, as is displayed in Fig. 7A. Sample temperature stabilisation is introduced to minimise evaporation and maintain sample volume and aid compositional integrity.To demonstrate the effectiveness within the HT liquid serum platform, a comparative study of spectra from un-stabilised (variable with room temperature and sampling conditions) and actively cooled samples taken over an 8 hours period was carried out. With no stabilization in place, a large decrease in the sample volume was observed with an evaporation rate of approximately 10 μl h−1. Evaporation is greatly reduced with an actively stabilised and cooled sample. Spectra variability is largely reduced by the presence of cooling over the course of 8 hours with a maximum and mean spectral standard deviation a factor of 2.9× and 1.9× smaller than when no cooling present, respectively. This variability is shown by the comparative standard deviation from the mean processed spectra in Fig. 7A. The mean spectral standard deviation with no temperature control was found to be 0.0265, compared to the cooled sample which had a mean spectral standard deviation of 0.0138. This reduction in spectral variance is further demonstrated via PCA analysis. Fig. 7B is a PC1 vs. PC2 score plot of the same sample taken 8 hours apart in a non-temperature stabilised setting. The PC1 vs. PC2 score plot shows that spectra taken at time 0 were significantly separated along PC1 compared to spectra taken after 8 hours, with PC1 accounting for 86.78% of the overall spectral variance for the non-stabilised spectra. When compared to PCA analysis of spectra taken on the temperature stabilized platform (Fig. 7C) the overall variance of the temperature stabilised spectra is lower and the spectra show less overall variance. This is further shown when directly comparing the overall variance along PC1 for the non-temperature stabilised and the cooled spectra (Fig. 7D), where the overall spectral variance in the spectra taken in the cooled platform is almost half that of the temperature non-stabilised spectra. Although there is still separation in the PC1 scores of the sample when cooling is present, this is an amount of separation also commonly seen within spectra-spectra variability.
3.5 Inter-operator variability – liquid
To demonstrate the minimisation in the inter-operator variability when using the liquid HT platform, spectral acquisition from a serum sample within the same well on the same day with equal volume of sample was repeated by two trained users. Fig. 8 shows PC1 vs. PC2 score plot and associated loadings on PC1 and PC2 for 5 repeat measurements from each user. The PC scatter plots show no evident spectral grouping between the users contributing to the spectra. The PC loading plots show that the main spectral variance (46.52%) is attributed to noise within the spectra as no distinguishable spectral features can be identified. Furthermore, compared to the inter-user variability of the dry platform (Fig. 4) the liquid platform shows an overall reduction in spectral variance. It can therefore be concluded that using the HT liquid platform the spectra are not affected by the trained system user so the liquid platform would lend itself well to translating into a clinical setting with more than one trained user.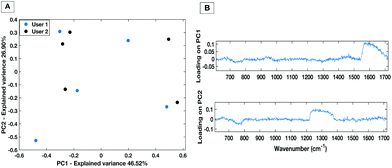 | ||
| Fig. 8 PC score plot (A) and loading plot (B) for inter-user variability study for the liquid HT platform. | ||
3.6 Direct comparison of the diagnostic capability of the HT Raman serum platforms
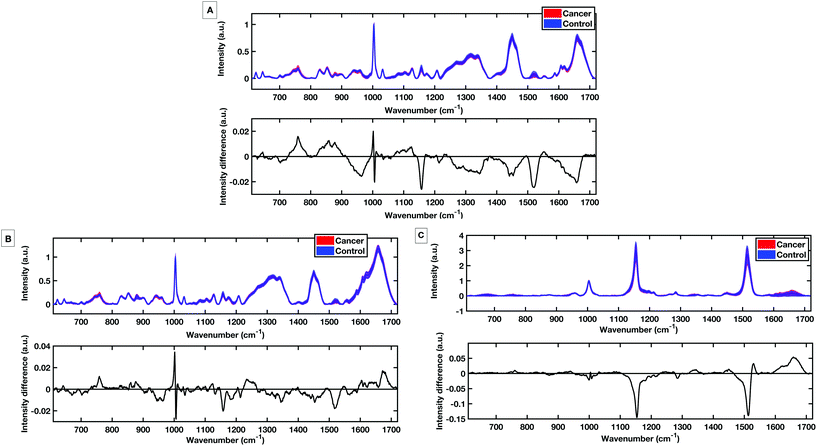
To evaluate the potential of the HT platforms developed in this work a pilot dataset of 60 patients was used to evaluate the diagnostic capability of the methods. Spectra were acquired from fresh (same day of blood draw) serum samples with; 30 patients with colorectal cancer determined by positive colonoscopies, and 30 patients with negative colonoscopies for colorectal cancer. Fig. 9 shows the mean, standard deviation and difference spectra produced for the pilot patient dataset from the HT dry serum platform and the HT liquid temperature stabilised platform. The mean difference spectra from all methods show differences in peaks at 1147 cm−1 and 1518 cm−1 tentatively assigned to carotenoids with control patients showing higher levels.12 Within the dry and liquid 785 nm spectra control intensities are higher in the C/C stretching bands (1447 cm−1), the shoulder of the phenylalanine band and higher peaks attributed to the Amide I region 1650–1660 cm−1 and the Amide III region between 1208 cm−1 and 1342 cm−1.18,29,30 The cancer spectra show higher levels in bands at 759 cm−1 and 853 cm−1, attributed to tyrosine as well differences at 1127 cm−1 attributed to phospholipids and lipoproteins.PLS-DA discriminatory models were produced for data from the dry and the liquid HT serum Raman platforms. PLS-DA model parameters were optimised according to minimising the cross-validation error within the models. All PLS-DA models were cross validated using k-fold cross validation with 5 folds. Receiver operating curves (ROC) were also generated from the predicted values from the 5-fold cross validation and the area under the curve (AUC) plotted for both the training set and the cross validated models.
Fig. 10A shows the cross-validated prediction scores against sample number for the 785 nm dry platform. The prediction shows a good separation between the PLS-DA scores for the cancer and the control patients. The loading plot for the 785 nm dry data shows that the LV1 shows spectral differences that match the difference plots generated in Fig. 9A and B. Fig. 10D shows that the cross-validated prediction scores for the liquid model is still well separated between the groups, but not as well as the dry data. The general trend and peak positions of the differences shown in the liquid loading on LV1 and the difference plot is also comparable. The HT liquid method with 532 nm excitation also shows good cross-validated separation but not quite as high as the 785 nm excitation data. However, the loading plot for LV1 for the 532 nm data matches almost exactly the difference plot shown in Fig. 9C. The area under the curve (AUC) for all of the cross validated PLS-DA models was higher than 0.8 indicating that all three techniques have potential to be considered ‘good’ learners.31 However, as reflected in the predictions vs. samples plots the 785 nm dry data had the highest AUC at 0.8834. Following the calculation of the cross validated PLS-DA models, sensitivities and specificities for the techniques to identify colorectal cancer within the serum samples was calculated for each HT Raman platform. Table 3 shows a comparison of the HT platforms in terms of the calculated CV sensitivities, specificities, analysis times and the effects of inter-operator variability.
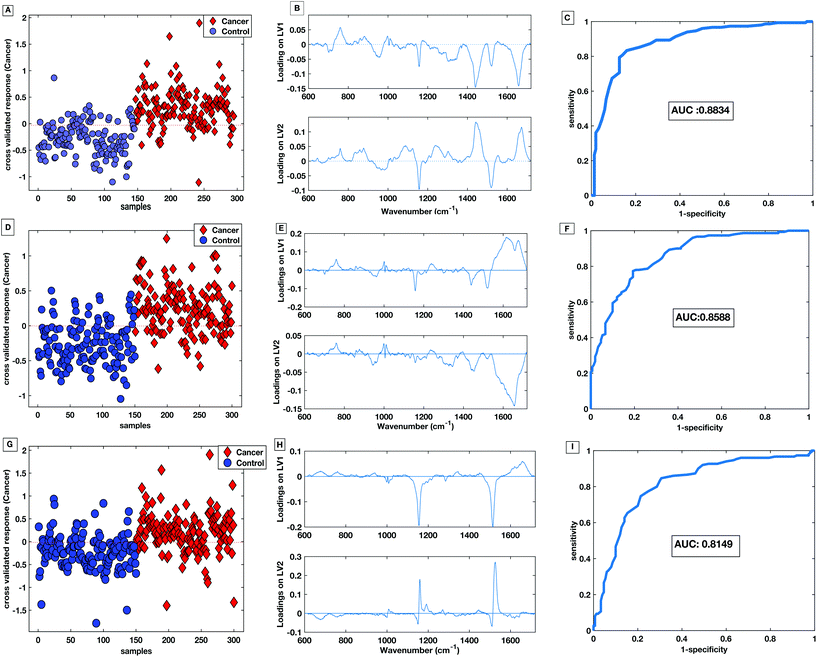 | ||
| Fig. 10 PLS-DA score plot with associated loadings on LV1 and LV2 and the CV ROC curve for 785 nm dry platform (A–C), 785 nm liquid platform (D–F) and 532 nm platform (G–I). | ||
| Sensitivity (%) | Specificity (%) | User variable? | Total time (min) | |
|---|---|---|---|---|
| 785 nm dry | 83 | 83 | Yes | 80 |
| 785 nm liquid | 77 | 81 | No | 22.5 |
| 532 nm liquid | 77 | 78 | No | 16 |
With these patient samples, the dry 785 nm methodology yields the most effective diagnostic results with the highest sensitivity, specificity and AUC. Therefore, within a research laboratory with one user, this method may be considered optimal. However, when extended to considering aspects of translation the dry methodology exhibited inter-user spectral variability which would potentially cause a large variation in diagnostic results.
The liquid serum platform showed higher specificity with 785 nm excitation than with 532 nm excitation. The sensitivities were equivalent for 785 nm and 532 nm excitation. This is likely to be due to the spectra sharing some common spectral bands such as the phenylalanine and the carotenoid associated bands as seen in Fig. 6. Fig. 10 parts B, E and H show the loadings associated to the scores for each PLS-DA model. The bands shared within the spectra are also common in the PLS-DA loadings associated to the spectral discrimination. The PLS-DA models constructed within this work highlight the effective reproducible processes in the measurement platform and analysis routines. This enables high levels of discrimination at both wavelengths, even though the spectra themselves are different.
Finally, despite the liquid methodologies having a slightly lower sensitivity and specificity than the dry methods they are not affected by inter-user variability. Moreover the overall analysis time for the liquid methods is also quicker as there is no need to wait for the samples to dry.
3.7 The effect of freeze-thawing samples on diagnostic capability of HT-Raman methods
Previous work has shown that in vibrational spectroscopic studies the preparation of a sample can affect spectra. For example, Lovergne et al. showed that freeze–thaw cycles affect spectral variability in plasma samples within FTIR studies.17 When using clinical samples, some sample sources may sometimes be available fresh and sometimes available after storage (freezing). To investigate the effect that freezing samples has on diagnostic capability serum samples were compared for both the dry and liquid HT measurement platforms for fresh samples and samples that had undergone a freeze–thaw cycle. One aliquot of each patient sample was used on day of collection for immediate Raman analysis (results presented above) and another was frozen at −80 °C. After one month of storage, frozen samples were thawed at room temperature and analysed. Data from the samples that had undergone a freeze thaw cycle were subject to PLS-DA analysis. Diagnostic models were then calculated, cross validated and compared to the models calculated for fresh serum samples. Table 4 demonstrates that the dry samples are affected more significantly by the freeze–thaw process and the liquid samples maintain a similarly high sensitivity, specificity and AUC as the fresh samples in Table 3. These results demonstrate a strong motivation for application of the method to a liquid sample on the basis of both reproducibility and sample handling flexibility.| Sensitivity (%) | Specificity (%) | Sensitivity change vs. fresh (%) | Specificity change vs. fresh (%) | |
|---|---|---|---|---|
| 785 nm dry | 72 | 78 | −11 | −5 |
| 785 nm liquid | 79 | 77 | +2 | −4 |
| 532 nm liquid | 77 | 77 | 0 | +1 |
4 Conclusions
HT Raman spectroscopy of serum to triage colorectal cancer referrals to colonoscopy offers a potentially cost saving, low risk clinical tool that could aid more rapid diagnosis of colorectal cancer. In this study the development of HT platforms for serum RS for the triage of colorectal referrals has been presented which has been previously presented as a critical research gap in colorectal cancer.32 Aluminium foil proved to be a cheap and effective substrate for drying serum samples and collecting spectra, when coupled to fresh serum samples the aluminium substrate proved the most diagnostically effective with a sensitivity and specificity for colorectal cancer of 83% and 83%. Furthermore, in terms of analysis time the HT platform takes just 10 min per sample excluding drying time. However, when considering translatability the dry platform did not perform as well when samples were analysed via different users or when samples had been frozen prior to analysis.To combat limitations with the dry HT platform, a stainless steel well plate with a temperature stabilisation stage has also been developed and optimised for liquid serum spectral acquisition. The stainless-steel well plate design shows minimal spectral contributions and allows automated sample collection for up to 40 patients per sample run. Furthermore, the platform is re-usable when subject to cleaning protocols consistent with surgical tools.
Stabilising the temperature of the liquid platform was shown to reduce the spectral variance and minimised evaporation effects allowing more reproducible data acquisition. The liquid HT platform was also compatible with more than one wavelength and showedless susceptibility to inter-user variation between spectra and was not as affected as the dry protocol by the freezing of samples. The maximum sensitivity and specificity achieved with the HT liquid platform was with the 785 nm laser using fresh samples at 77% and 81% respectively. The 532 nm excitation had lower discriminatory values than the 785 nm models but it had the quickest overall sampling time.
It is appreciated that one limitation of the work presented here is the lack of testing of diagnostic models with an independent testing set. However, the diagnostic values presented demonstrate the efficacy of the HT platforms developed and are encouraging for future studies. If HT serum RS is to be translated as a triage tool for colorectal referrals, diagnostic capability of the techniques, further studies with larger clinically relevant cohorts using more robust diagnostic algorithms and independent testing of patient cohorts will need to be carried out. Future work will include the evaluation of the HT diagnostic platforms in conjunction with machine learning based techniques in a large patient cohort. Furthermore, the effects of cancer stage, patient co-morbidities, medications and the limit of detection for serum Raman spectroscopy will be considered as well as the health economic considerations of HT serum RS as a triage tool for colorectal cancer referrals.
The new gains we have demonstrated in this study will continue to be advanced to establish early and effective diagnosis of colorectal cancer for patient benefit. The methodology is also being applied to validate Raman spectroscopy as key clinical tool for liquid serum biopsies.
Conflicts of interest
PRD, DAH and CAJ declare their involvement in CanSense Ltd, a recently incorporated cancer diagnosis spin-out company from Swansea University (company no: 11367637).Acknowledgements
This work has been funded through a PhD studentship funded by Cancer Research Wales (Registered Charitable Incorporated Organisation Number: 1167290). The authors are grateful for the technical support originating from the College of Science and Centre for Nanohealth at Swansea University.References
- https://www.cancerresearchuk.org/health-professional/cancer-statistics/incidence/common-cancers-compared .
- http://www.who.int/news-room/fact-sheets/detail/cancer .
- https://www.nice.org.uk/guidance/NG12 .
- https://improvement.nhs.uk/resources/proposed-national-tariff-prices-1718-1819/ .
- D. W. Shipp, F. Sinjab and I. Notingher, Adv. Opt. Photonics, 2017, 9, 315 CrossRef.
- M. Sattlecker, C. Bessant, J. Smith and N. Stone, Analyst, 2010, 135, 895–901 RSC.
- J. R. Hands, G. Clemens, R. Stables, K. Ashton, A. Brodbelt, C. Davis, T. P. Dawson, M. D. Jenkinson, R. W. Lea, C. Walker and M. J. Baker, J. Neuro-Oncol., 2016, 127, 463–472 CrossRef PubMed.
- P. D. Lewis, K. E. Lewis, R. Ghosal, S. Bayliss, A. J. Lloyd, J. Wills, R. Godfrey, P. Kloer and L. A. Mur, BMC Cancer, 2010, 10, 640 CrossRef CAS PubMed.
- G. Theophilou, K. M. Lima, P. L. Martin-Hirsch, H. F. Stringfellow and F. L. Martin, Analyst, 2016, 141, 585–594 RSC.
- K. Lau, M. Isabelle, G. R. Lloyd, O. Old, N. Shepherd, I. M. Bell, J. Dorney, A. Lewis, R. Gaifulina, M. Rodriguez-Justo, C. Kendall, N. Stone, G. Thomas and D. Reece, Int. Soc. Opt. Photonics, 2016, 9704, 97040B Search PubMed.
- C. A. Jenkins, P. D. Lewis, P. R. Dunstan and D. A. Harris, World J. Gastrointest. Oncol., 2016, 8, 427 CrossRef PubMed.
- D. K. R. Medipally, A. Maguire, J. Bryant, J. Armstrong, M. Dunne, M. Finn, F. M. Lyng and A. D. Meade, Analyst, 2017, 142, 1216–1226 RSC.
- K. W. C. Poon, F. M. Lyng, P. Knief, O. Howe, A. D. Meade, J. F. Curtin, H. J. Byrne and J. Vaughan, Analyst, 2012, 137, 1807 RSC.
- A. Sahu, S. Sawant, H. Mamgain and C. M. Krishna, Analyst, 2013, 138, 4161–4174 RSC.
- H. J. Butler, L. Ashton, B. Bird, G. Cinque, K. Curtis, J. Dorney, K. Esmonde-White, N. J. Fullwood, B. Gardner, P. L. Martin-Hirsch, M. J. Walsh, M. R. McAinsh, N. Stone and F. L. Martin, Nat. Protoc., 2016, 11, 664–687 CrossRef CAS PubMed.
- M. Paraskevaidi, C. L. M. Morais, K. M. Ashton, H. F. Stringfellow, P. L. Martin-hirsch and F. L. Martin, Analyst, 2018, 44, 1–4 Search PubMed.
- L. Lovergne, P. Bouzy, V. Untereiner, R. Garnotel, M. J. Baker, G. Thiéfin and G. D. Sockalingum, Faraday Discuss., 2016, 187, 521–537 RSC.
- M. J. Baker, J. Trevisan, P. Bassan, R. Bhargava, H. J. Butler, K. M. Dorling, P. R. Fielden, S. W. Fogarty, N. J. Fullwood, K. A. Heys, C. Hughes, P. Lasch, P. L. Martin-Hirsch, B. Obinaju, G. D. Sockalingum, J. Sulé-Suso, R. J. Strong, M. J. Walsh, B. R. Wood, P. Gardner and F. L. Martin, Nat. Protoc., 2014, 9, 1771–1791 CrossRef CAS PubMed.
- S. Feng, R. Chen, J. Lin, J. Pan, G. Chen, Y. Li, M. Cheng, Z. Huang, J. Chen and H. Zeng, Biosens. Bioelectron., 2010, 25, 2414–2419 CrossRef CAS PubMed.
- J. Pichardo-Molina, C. Frausto-Reyes, O. Barbosa-García, R. Huerta-Franco, J. González-Trujillo, C. Ramírez-Alvarado, G. Gutiérrez-Juárez and C. Medina-Gutiérrez, Lasers Med. Sci., 2007, 22, 229–236 CrossRef CAS PubMed.
- J. L. González-Solís, J. C. Martínez-Espinosa, L. A. Torres-González, A. Aguilar-Lemarroy, L. F. Jave-Suárez and P. Palomares-Anda, Lasers Med. Sci., 2014, 29, 979–985 CrossRef PubMed.
- T. M. James, M. Schlösser, R. J. Lewis, S. Fischer, B. Bornschein and H. H. Telle, Appl. Spectrosc., 2013, 67, 949–959 CrossRef CAS PubMed.
- A. Baer, S. Schmidt, S. Haensch, M. Eder, G. Mayer and M. J. Harrington, Nat. Commun., 2017, 8, 974 CrossRef PubMed.
- T. S. Németh, Biopolymer Research Trends, Nova Publishers, 2007 Search PubMed.
- H. J. Butler, L. Ashton, B. Bird, G. Cinque, K. Curtis, J. Dorney, K. Esmonde-White, N. J. Fullwood, B. Gardner and P. L. Martin-Hirsch, et al. , Nat. Protoc., 2016, 11, 664 CrossRef CAS PubMed.
- R. G. Brereton and G. R. Lloyd, J. Chemom., 2014, 28, 213–225 CrossRef CAS.
- C. Beleites, U. Neugebauer, T. Bocklitz, C. Krafft and J. Popp, Anal. Chim. Acta, 2013, 760, 25–33 CrossRef CAS PubMed.
- L. Cui, H. J. Butler, P. L. Martin-Hirsch and F. L. Martin, Anal. Methods, 2016, 8, 481–487 RSC.
- L. Silveira, R. d. C. F. Borges, R. S. Navarro, H. E. Giana, R. A. Zângaro, M. T. T. Pacheco and A. B. Fernandes, Lasers Med. Sci., 2017, 32, 787–795 CrossRef PubMed.
- D. Rohleder, G. Kocherscheidt, K. Gerber, W. Kiefer, W. Köhler, J. Möcks and W. Petrich, J. Biomed. Opt., 2005, 10, 031108 CrossRef CAS PubMed.
- A. Hanley and J. McNeil, Radiology, 1982, 143, 29–36 CrossRef PubMed.
- M. Lawler, D. Alsina, R. A. Adams, A. S. Anderson, G. Brown, N. S. Fearnhead, S. W. Fenwick, S. P. Halloran, D. Hochhauser, M. A. Hull, V. H. Koelzer, A. G. McNair, K. J. Monahan, I. Näthke, C. Norton, M. R. Novelli, R. J. Steele, A. L. Thomas, L. M. Wilde, R. H. Wilson and I. Tomlinson, Gut, 2018, 67, 179–193 CrossRef PubMed.
Footnote |
| † Present address: WestCHEM, Department of Pure and Applied Chemistry, Technology & Innovation Centre, University of Strathclyde, Glasgow, UK |
| This journal is © The Royal Society of Chemistry 2018 |