Supramolecular conjugated polymer materials for organelle imaging in living cells†
Rong
Hu
,
Shengliang
Li
,
Huan
Lu
,
Libing
Liu
*,
Fengting
Lv
and
Shu
Wang
*
Beijing National Laboratory for Molecular Science, Key Laboratory of Organic Solids, Institute of Chemistry, Chinese Academy of Sciences, Beijing, 100190, P. R. China. E-mail: wangshu@iccas.ac.cn; Fax: +86-10-62636680; Tel: +86-10-62636680
First published on 17th April 2017
Abstract
A novel supramolecular system based on conjugated polymers was developed to realize specific targeting and imaging of organelles in living cells. The carboxyl group-modified polythiophene derivative (PT) forms a complex with six histidine equipped Tat peptide via Ni2+ ion chelation, hydrogen bonds and electrostatic interactions (PT/Ni2+/Tat-His6). The complex can reassemble with RFP-His10 due to the relatively high affinity of Ni2+ towards His10-tag compared to His6-tag. Thus, the PT/Ni2+/Tat-His6 complex could selectively label and image organelles expressing His10-tag in living cells with high stability.
The living eukaryotic cells are comprised of different organelles with specialized functions. These organelles are suspended in the cytoplasm and generate a specific micro-environment, in which are found various chemicals and chemical reactions fundamental to their functions.1,2 Thus specific labelling and imaging of organelles in living cells is of great significance for tracing their biological process and regulating their function. Small-molecule organic dyes and quantum dots (QDs) are the most widely used fluorescent probes, and multifarious commercial organelle stains have been developed to help identify the location and function of proteins and targets inside cells.3–6 However, the photobleaching of small-molecule organic dyes and cytotoxicity of QDs from heavy metal leaching cannot be neglected. Fluorescent proteins, possessing high brightness and specificity, are another kind of reporters, where complicated gene encoding and sequencing are needed.7,8
Conjugated polymers (CPs) are a kind of macromolecules with an extended conjugated system and exhibit outstanding optical properties.9–15 They are preferred over small-molecule organic dyes and QDs for in vitro and in vivo bioimaging applications due to their high fluorescent brightness, outstanding light harvesting ability, low cytotoxicity, good photostability, and diverse modification of side chains.16,17 CP-based non-specific cell imaging was firstly studied with no specific targeting elements linked to CPs but with CPs directly used for imaging through nonselective cellular uptake.18–20 Recently, CP-based targeted cell imaging was developed by modifying CPs with specific recognition groups (such as antibody, drugs, sugar and peptide) to realize selective binding and imaging.21–28 To the best of our knowledge, although CPs have been extensively used for cell imaging, specific cellular organelle imaging using the molecular design strategy is seldom reported. To address this challenge, here we have developed a novel supramolecular strategy to realize specific organelle imaging of living cells using a polythiophene (PT) derivative through specific targeting of intracellular histidine (His)-tagged proteins with controlled expression in organelles.
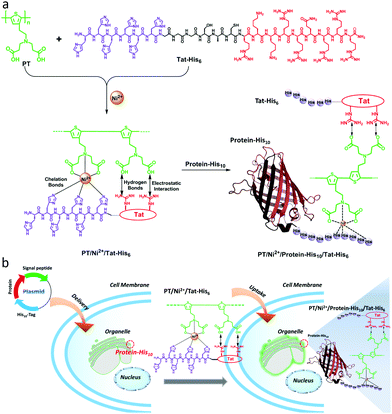
Carboxyl groups, especially N-nitrilotriacetic acid (NTA) derivatives, possess excellent affinity towards histidine (His)-tagged proteins through coordination with nickel ions (Ni2+). As a result, NTA/His-tag pairs have been widely used in biology and biotechnology for protein purification and intracellular protein labeling in living cells.29–34 As shown in Scheme 1a, in this work, a supramolecular complex (PT/Ni2+/Tat-His6) was prepared by the assembly of a carboxyl group modified polythiophene derivative17 with six histidine equipped transactivator of transcription (Tat) of HIV-1 (Tat-His6) via chelation with Ni2+ ions, hydrogen bonds and electrostatic interactions. The Tat unit as cell-penetrating peptide is responsible for cellular uptake with high efficiency and minimal cytotoxicity.35–37 Since the affinity of Ni2+ towards His10-tag is much higher (10-fold) than that towards His6-tag,35,38 the His10-tagged protein can reassemble with PT/Ni2+/Tat-His6 to form the PT/Ni2+/Protein-His10/Tat-His6 complex via chelation with Ni2+ ions. As shown in Scheme 1b, when the well designed plasmid including an organelle-specific gene is transfected into cells, the protein with His10-tag is expressed in the specific organelle. Upon uptake of the PT/Ni2+/Tat-His6 complex into cells, the PT/Ni2+/Protein-His10/Tat-His6 complex forms by a reassembly process, as a result of which targeted labelling and imaging of organelles with high specificity is realized.
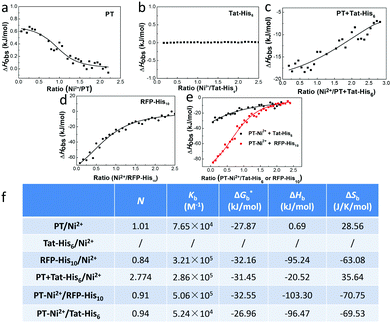
We first studied the mechanism of interactions among different components within the complex. Isothermal titration microcalorimetry (ITC) was employed to understand the thermodynamics of the supramolecular complex, and the measurements were operated in PBS buffer. Fig. 1a reveals that PT can chelate with Ni2+ (Kb = 7.65 × 104 M−1). While the observed enthalpy of Ni2+ with Tat-His6 shows no obvious change in the process under low concentration (Fig. 1b), a distinct enthalpy change can be observed with increasing concentration (Fig. S1, ESI†). The reason may be that Tat-His6 is not able to interact with Ni2+via chelation due to its high density of positive charge. As both Tat-His6 and Ni2+ are positively charged, they cannot coordinate successfully owing to electrostatic repulsion, even though His6-tag possesses high affinity to Ni2+. However, by titrating Ni2+ into PT/Tat-His6 solution, as shown in Fig. 1c, an intense binding affinity (Kb = 2.86 × 105 M−1) is obtained and is stronger than that of Ni2+ with PT, which proves the assistance of PT in the coordination process of Tat-His6 with Ni2+. Our assumption is that the binding of PT with Tat-His6 neutralized the positive charge on TAT-His6 and reduced the electrostatic repulsion between Ni2+ and Tat-His6, making the coordination possible. We also evaluated the binding ability of Ni2+ with RFP-His10 (Fig. 1d); the enthalpy of this system changes apparently, indicating that RFP-His10 can coordinate with Ni2+ effectively (Kb = 3.21 × 105 M−1). We further studied the binding affinity of RFP-His10 and Tat-His6 with the complex formed by PT and Ni2+ (PT–Ni2+), as shown in Fig. 1e. The fitting results reveal that RFP-His10 possesses a relatively high affinity towards PT–Ni2+ (Kb = 5.06 × 105 M−1) compared to Tat-His6 (Kb = 5.24 × 104 M−1). Besides, the ΔHobs values of these two systems are negative, demonstrating that the interaction of PT–Ni2+ with RFP-His10 or Tat-His6 is an exothermic process, which means coordination, hydrogen bonds and electrostatic interactions are the dominant force for the coordination process of Tat-His6 with Ni2+ and that of RFP-His10 with Ni2+. The calculated ITC results are shown in Fig. 1f, and the thermodynamic parameters also show that the coordination of Ni2+ with PT has no obvious influence on the chelation of Ni2+ with RFP-His10. These results reveal that with the help of PT, Tat-His6 can coordinate with Ni2+, and the affinity of RFP-His10 towards PT–Ni2+ is much higher than that of Tat-His6, making it feasible that the chelation of Tat-His6 with Ni2+ can be replaced by RFP-His10 to form a reassembly complex.
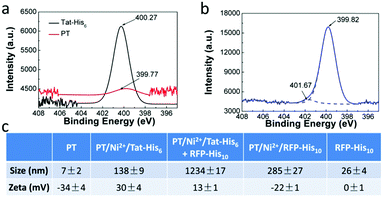
X-ray photoelectron spectroscopy (XPS) was employed to characterize the interactions in PT/Ni2+/Tat-His6. As shown in Fig. 2a and b, after the formation of the PT/Ni2+/Tat-His6 complex, the nitrogen binding energy peak of Tat-His6 (400.27 eV) shifts to a lower binding energy state (399.82 eV), as the content of the amine (–NH–) decreases and that of the imine (–N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ) increases. It was reported that the amine can convert to imine based on coordination, and the binding energy of amine nitrogen is higher than that of imine nitrogen.39,40 At the same time, the nitrogen binding energy of PT (399.77 eV) shifts to a higher state (401.67 eV) on account of the coordination with Ni2+. Combined with ITC results, we can figure out that Ni2+ can effectively chelate with His6-tag and PT in the PT/Ni2+/Tat-His6 complex. It is well established that Ni2+ exhibits a much higher affinity towards His10-tag than towards His6-tag, and we further evaluated the binding ability of RFP-His10 to the PT/Ni2+/Tat-His6 complex by using dynamic light scattering (DLS).
) increases. It was reported that the amine can convert to imine based on coordination, and the binding energy of amine nitrogen is higher than that of imine nitrogen.39,40 At the same time, the nitrogen binding energy of PT (399.77 eV) shifts to a higher state (401.67 eV) on account of the coordination with Ni2+. Combined with ITC results, we can figure out that Ni2+ can effectively chelate with His6-tag and PT in the PT/Ni2+/Tat-His6 complex. It is well established that Ni2+ exhibits a much higher affinity towards His10-tag than towards His6-tag, and we further evaluated the binding ability of RFP-His10 to the PT/Ni2+/Tat-His6 complex by using dynamic light scattering (DLS).
We measured the size and zeta potential of PT before and after the formation of the complex. As shown in Fig. 2c, the zeta potential of PT shifts from −34 mV to 30 mV, and the size increases obviously from 7 nm to 138 nm after the formation of the PT/Ni2+/Tat-His6 complex. We also studied the size and zeta potential of the PT/Ni2+/Tat-His6 complex in the absence and presence of RFP-His10. The size of the complex increases conspicuously with the diameter changing from 138 nm to 1234 nm after the addition of RFP-His10, while the size of RFP-His10 alone is only 26 nm in water. The size of PT/Ni2+/RFP-His10 is only 285 nm, which demonstrates that a new complex different from PT/Ni2+/RFP-His10 has been formed. In addition, the zeta potential of the new complex shifts from 33 mV to 13 mV because RFP (pI value: 5.1) possesses a net negative charge in neutral medium. Table S1 (ESI†) shows the size and zeta potential of PT/Ni2+ and PT/Tat-His6. Based on these results, we can prove that the PT/Ni2+/Tat-His6 complex is assembled based on chelation, hydrogen bonds and electrostatic interactions, and protein-His10 can bind with the PT/Ni2+/Tat-His6 complex to form a new complex, PT/Ni2+/Protein-His10/Tat-His6.
The aqueous solution of PT exhibits an absorption maximum at 410 nm with an extinction coefficient of 6.7 × 103 M−1 cm−1, and the emission maximum peak is 561 nm with a fluorescence quantum efficiency (QE) of 4.8%.17 As shown in Fig. 3, the fluorescence spectrum of the PT/Ni2+/Tat-His6 complex is quite similar to that of PT with a QE of 3%. After verifying the interactions within the complex, the penetration ability of the complex into HeLa cells was explored. Note that the Tat sequence is responsible for fast and efficient internalization. At the beginning of the imaging experiment, the cytotoxicity of the PT/Ni2+/Tat-His6 complex was evaluated against HeLa cells by using MTT cell-survival assay. Fig. S2 (ESI†) shows that the PT/Ni2+/Tat-His6 complex possesses high biocompatibility in the range of 0–50 μM in RUs. Before incubation with the complexes, HeLa cells were pretreated with chloroquine to effectively inhibit endosomal sequestration and enhance transduction. As shown in Fig. S3 (ESI†), the PT/Ni2+/Tat-His6 complex is taken up by HeLa cells efficiently. Control experiments show that PT alone, PT/Ni2+ or PT/Tat-His6 without loading with Ni2+ fails to cross the cell membrane, which demonstrates that Tat peptide and Ni2+ chelation play vital roles in cell penetration for the PT/Ni2+/Tat-His6 complex.
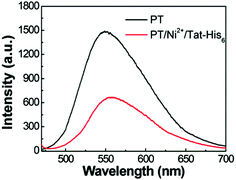 | ||
| Fig. 3 Fluorescence spectra of PT and the PT/Ni2+/Tat-His6 complex in aqueous solution. The excitation wavelength is 450 nm. | ||
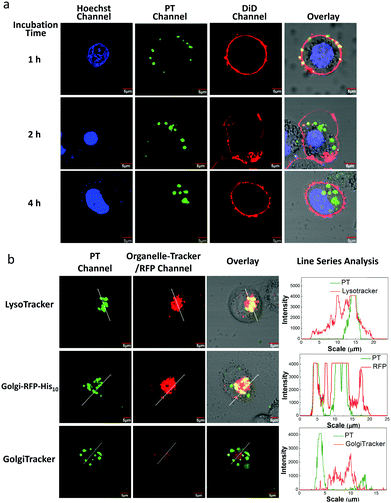
To examine the internalization speed of the PT/Ni2+/Tat-His6 complex, HeLa cells were incubated with the complex for 1, 2 and 4 h, and then the PT was colocalized with DiD (cell membrane dye) and Hoechst 33342 (nuclear dye). Confocal laser scanning microscope (CLSM) images show that the PT/Ni2+/Tat-His6 complex interacts with the cell membrane after incubation for one hour followed by internalization by cells within another one hour and finally localizes in the cytoplasm (Fig. 4a). We further investigated the potential of the PT/Ni2+/Tat-His6 complex for specific targeting and imaging of organelles in living cells. For this experiment, we selected a signal peptide that localizes at the Golgi apparatus and fused it with a fluorescent protein (RFP) that possessed a recombinant C-terminal His10-tag (Golgi-RFP-His10). HeLa cells without His10-tagged protein expression were incubated with the PT/Ni2+/Tat-His6 complex followed by further colocalization and line series analysis with LysoTracker under CLSM. As shown in Fig. 4b, perfect colocalization is detected between PT and LysoTracker. On the other hand, in the case of transfected HeLa cells expressing the RFP-His10 residue in the Golgi apparatus, the fluorescence of PT and Golgi-RFP-His10 accurately overlaps, and relatively few changes of emission intensity are observed. As a control, HeLa cells without the expression of His10-tagged protein were treated with the PT/Ni2+/Tat-His6 complex under the same condition. We found that PT exhibited poor colocalization with GolgiTracker, indicating that PT was not able to localize to the Golgi apparatus in cells without His10-tag upon uptake into cells. We studied the co-localization between LysoTracker and PT/Ni2+/Tat-His6 complex in HeLa cells expressing RFP-His10-tag in Golgi. As shown in Fig. S4 (ESI†), poor colocalization was obtained, which revealed that the complex could realize endosomal escape. Furthermore, HeLa cells were treated with chloroquine which can support endosomal escape effectively. TAT peptide possessing positive charge also enables endosomal escape through the “proton-sponge effect”. As a result, even though the PT/Ni2+/Tat-His6 complex was delivered into cells via an endocytosis process, intracellular escape would occur efficiently. Based on these observations, specific targeting and imaging of the Golgi apparatus was realized using the assembly–reassembly strategy.
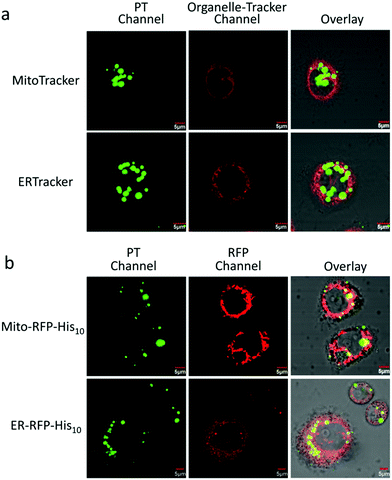
To evaluate the stability of the PT/Ni2+/Golgi-RFP-His10/Tat-His6 complex inside cells, cells were incubated for 24 hours and colocalization studies were performed as well. As shown in Fig. S5 (ESI†), compared to 4 hours incubation with the PT/Ni2+/Tat-His6 complex, the same high degree of colocalization of PT with Golgi-RFP-His10 is detected. In addition, similar brightness is also observed, which demonstrates that the PT/Ni2+/Golgi-RFP-His10/Tat-His6 assembly possesses excellent stabilization inside cells. After confirming the specific target of RFP-His10 fused in the Golgi apparatus, we also evaluated the specific binding ability towards RFP-His10 fused in other organelles. As shown in Fig. 5a, no colocalization of the PT with ERTracker or MitoTracker is detected in HeLa cells without the expression of His10-tagged protein. We genetically fused His10-tag to RFP and transiently expressed it in the endoplasmic reticulum (ER-RFP-His10) and mitochondria (Mito-RFP-His10). As shown in Fig. 5b, red fluorescence from RFP and green fluorescence from PT are observed with perfect colocalization in cells. Similar results are obtained in mitochondria expressing RFP-His10. These observations clearly demonstrate that the PT/Ni2+/Tat-His6 complex can label intracellular His10-tagged protein in various organelles with high specificity. Thus, targeted labelling and imaging of organelles with high specificity is realized using a supramolecular reassembly technique.
Conclusions
In summary, we have developed a supramolecular assembly based on conjugated polymers to realize specific organelle imaging. A complex (PT/Ni2+/Tat-His6) was prepared by the assembly of a carboxyl group modified polythiophene derivative with six histidine equipped Tat peptide via chelation with Ni2+ ions, hydrogen bonds and electrostatic interactions. The complex can form reassembly with RFP-His10 due to the relatively high affinity of Ni2+ towards His10-tag compared to His6-tag. Thus, the PT/Ni2+/Tat-His6 complex can selectively label and image organelles expressing His10-tag in living cells with high stability. This assembly–reassembly process holds a good ability for tracing protein traffic, regulating intracellular signal pathways and monitoring protein dynamics.The authors are grateful to the National Natural Science Foundation of China (No. 21373243, 21473220).
Notes and references
- D. L. Nelson and M. M. Cox, Lehninger Principles of Biochemistry, Worth publishers, New York, 2000 Search PubMed.
- S. D. Wiedner, L. N. Anderson, N. C. Sadler, W. B. Chrisler, V. K. Kodali, R. D. Smith and A. T. Wright, Angew. Chem., Int. Ed., 2014, 53, 2919–2922 CrossRef CAS PubMed.
- X. Michalet, F. F. Pinaud, L. A. Bentolila, J. M. Tsay, S. Doose, J. J. Li, G. Sundaresan, A. M. Wu, S. S. Gambhir and S. Weiss, Science, 2005, 307, 538–544 CrossRef CAS PubMed.
- I. L. Medintz, H. T. Uyeda, E. R. Goldman and H. Mattoussi, Nat. Mater., 2005, 4, 435–446 CrossRef CAS PubMed.
- B. N. G. Giepmans, S. R. Adams, M. H. Ellisman and R. Y. Tsien, Science, 2006, 312, 217–224 CrossRef CAS PubMed.
- M. Fernandez-Suarez and A. Y. Ting, Nat. Rev. Mol. Cell Biol., 2008, 9, 929–943 CrossRef CAS PubMed.
- M. Chalfie, Y. Tu, G. Euskirchen, W. W. Ward and D. C. Prasher, Science, 1994, 263, 802–805 CAS.
- L. He, C. P. Tan, R. R. Ye, Y. Z. Zhao, Y. H. Liu, Q. Zhao, L. N. Ji and Z. W. Mao, Angew. Chem., Int. Ed., 2014, 53, 12137–12141 CrossRef CAS PubMed.
- C. Zhu, L. Liu, Q. Yang, F. Lv and S. Wang, Chem. Rev., 2012, 112, 4687–4735 CrossRef CAS PubMed.
- L. H. Feng, C. L. Zhu, H. X. Yuan, L. B. Liu, F. T. Lv and S. Wang, Chem. Soc. Rev., 2013, 42, 6620–6633 RSC.
- S. W. Thomas, G. D. Joly and T. M. Swager, Chem. Rev., 2007, 107, 1339–1386 CrossRef CAS PubMed.
- J. Pecher and S. Mecking, Chem. Rev., 2010, 110, 6260–6279 CrossRef CAS PubMed.
- H. A. Ho, A. Najari and M. Leclerc, Acc. Chem. Res., 2008, 41, 168–178 CrossRef CAS PubMed.
- K. Y. Pu and B. Liu, Adv. Funct. Mater., 2011, 21, 3408–3423 CrossRef CAS.
- X. Q. Shen, L. Li, A. C. M. Chan, S. Q. Yao and Q.-H. Xu, Adv. Opt. Mater., 2013, 1, 92–96 CrossRef.
- C. F. Wu, S. J. Hansen, Q. O. Hou, J. B. Yu, M. Zeigler, Y. H. Jin, D. R. Burnham, J. McNeill, J. M. Olson and D. T. Chiu, Angew. Chem., Int. Ed., 2011, 50, 3430–3434 CrossRef CAS PubMed.
- C. Xing, Q. Xu, H. Tang, L. Liu and S. Wang, J. Am. Chem. Soc., 2009, 131, 13117–13124 CrossRef CAS PubMed.
- R. L. McRae, R. L. Phillips, I. B. Kim, U. H. F. Bunz and C. J. Fahrni, J. Am. Chem. Soc., 2008, 130, 7851–7853 CrossRef CAS PubMed.
- J. H. Moon, W. McDaniel, P. MacLean and L. E. Hancock, Angew. Chem., Int. Ed., 2007, 46, 8223–8225 CrossRef CAS PubMed.
- P. Bjork, K. P. R. Nilsson, L. Lenner, B. Kagedal, B. Persson, O. Inganas and J. Jonasson, Mol. Cell. Probes, 2007, 21, 329–337 CrossRef PubMed.
- K. lee, J. Lee, E. J. Jeong, A. Kronk, K. S. J. Elenitoba-Johnson, M. S. Lim and J. Kim, Adv. Mater., 2012, 24, 2479–2484 CrossRef CAS PubMed.
- C. F. Wu, S. J. Hansen, Q. O. Hou, J. B. Yu, M. Zeigler, Y. H. Jin, D. R. Burnham, J. D. McNeill, J. M. Olson and D. T. Chiu, Angew. Chem., Int. Ed., 2011, 50, 3430–3434 CrossRef CAS PubMed.
- K. Y. Pu, K. Li and B. Liu, Chem. Mater., 2010, 22, 6736–6741 CrossRef CAS.
- L. H. Feng, L. B. Liu, F. T. Lv and S. Wang, Adv. Mater., 2014, 26, 3926–3930 CrossRef CAS PubMed.
- B. Wang, C. L. Zhu, L. B. Liu and S. Wang, Polym. Chem., 2013, 4, 5212–5215 RSC.
- C. F. Xing, G. M. Yang, L. B. Liu, Q. Yang, F. T. Lv and S. Wang, Small, 2012, 8, 525–529 CrossRef CAS PubMed.
- S. Deshayes, H. Cabral, T. Ishii, Y. Miura, S. Kobayashi, T. Yamashita, A. Matsumoto, Y. J. Miyahara, N. Nishiyama and K. Kataoka, J. Am. Chem. Soc., 2013, 135, 15501–15507 CrossRef CAS PubMed.
- C. F. Wu, Y. H. Jin, T. Schneider, D. R. Burnham, P. B. Smith and D. T. Chiu, Angew. Chem., Int. Ed., 2010, 49, 9436–9440 CrossRef CAS PubMed.
- S. Lata and J. Piehler, Anal. Chem., 2005, 77, 1096–1105 CrossRef CAS PubMed.
- E. G. Guignet, R. Hovius and H. Vogel, Nat. Biotechnol., 2004, 22, 440–444 CrossRef CAS PubMed.
- C. T. Hauser and R. Y. Tsien, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 3693–3697 CrossRef CAS PubMed.
- C. R. Goldsmith, J. Jaworski, M. Sheng and S. J. Lippard, J. Am. Chem. Soc., 2006, 128, 418–419 CrossRef CAS PubMed.
- Y.-T. Lai, Y.-Y. Hu, L. Chang, Y. Yang, A. Chao, Z.-Y. Du, J. A. Tanner, M.-L. Chye, C. Qian, K.-M. Ng, H. Li and H. Sun, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 2948–2953 CrossRef CAS PubMed.
- R. K. June, K. Gogoi, A. Eguchi, X.-S. Cui and S. F. Dowdy, J. Am. Chem. Soc., 2010, 132, 10680–10682 CrossRef CAS PubMed.
- R. Wieneke, N. Laboria, M. Rajan, A. Kollmannsperger, F. Natale, M. C. Cardoso and R. Tampe, J. Am. Chem. Soc., 2014, 136, 13975–13978 CrossRef CAS PubMed.
- H. Yezid, K. Konate, S. Debaisieux, A. Bonhoure and B. Beaumelle, J. Biol. Chem., 2009, 284, 22736–22746 CrossRef CAS PubMed.
- H. D. Herce, A. E. Garcia and M. C. Cardoso, J. Am. Chem. Soc., 2014, 136, 17459–17467 CrossRef CAS PubMed.
- S. Lata, A. Reichel, R. Brock, R. Tampe and J. Piehler, J. Am. Chem. Soc., 2005, 127, 10205–10215 CrossRef CAS PubMed.
- Z. Wang, C. Sun, G. Vegesna, H. Liu, Y. Liu, J. Li and X. Zeng, Biosens. Bioelectron., 2013, 46, 183–189 CrossRef CAS PubMed.
- G. Yang, X. Zeng and X. Xia, J. Instrum. Anal., 2002, 21, 56–58 CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Detailed experimental procedures and additional Table S1 and Fig. S1–S4. See DOI: 10.1039/c7qm00105c |
| This journal is © the Partner Organisations 2017 |