Open Access Article
Open Access ArticleVersatile ruthenium complexes based on 2,2′-bipyridine modified peptoids†
Maria
Baskin
a,
Larisa
Panz
b and
Galia
Maayan
*a
aSchulich Faculty of Chemistry, Technion-Israel Institute of Technology, Haifa, 32000, Israel. E-mail: gm92@tx.technion.ac.il
bOrganic Mass Spectrometry Laboratory, Technion-Israel institute of technology, Haifa 32000, Israel
First published on 20th June 2016
Abstract
Helical peptoids bearing 2,2′-bipyridine form ruthenium complexes via intermolecular binding to linear peptoid strands or intramolecular binding to a cyclic scaffold. Ru(ii) binding promoted changes in the conformational order of the peptoids, and chiral induction from the peptoids to their metal center was observed.
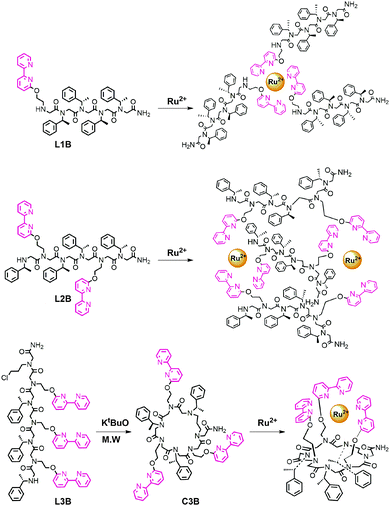
One of the most extensively used ligands in coordination chemistry is 2,2′-bipyridine (bipy),1 a bidentate chelator capable of forming complexes with various metal ions including Ru2+.2 In the last two decades, bipy derivatives were also incorporated within peptides3 and peptide mimics,4,5 and their metal-binding properties were described. The combination between bipy and peptidomimetic scaffolds allowed coordination of versatile metal cations to peptides3b and high binding affinities for metal ions6 targeting various biological and chemical applications.6–10 Thus, the introduction of bipy to peptidomimetics holds promise as a platform for the construction of unique metallofoldamers.11 Peptoids,12N-substituted glycine oligomers, are a class of foldamers13 that can adopt helical secondary structures with a helical pitch of three residues per turn via the incorporation of bulky chiral side chains within the oligomer sequence.14 Peptoids can be efficiently synthesized from primary amines on a solid support via the two-step “submonomer” approach,15 resulting in highly versatile scaffolds. This synthesis should enable not only the facile incorporation of metal-binding ligands but also a simple tuning of the ligand(s) position, quantity and identity, towards the creation of various metallopeptoids.16 Herein we explore the incorporation of bipy within the peptoid sequence and study three bipy modified helical peptoids – a linear pentamer bearing one bipy ligand (L1B), a linear hexamer with two bipy ligands (L2B) and a cyclic hexamer having three bipy ligands (C3B) – for the creation of versatile peptoid–ruthenium architectures as the intermolecular complexes (L1B)3Ru and (L2B)3Ru2, and the intramolecular complex (C3B)Ru (Fig. 1).
 | ||
| Fig. 1 Chemical structures of peptoid oligomers bearing 2,2′-bipyridine units and their anticipated ruthenium complexes. | ||
Initial attempts to incorporate 6-methylamine-2,2′-bipyridine17 within several peptoid sequences were not successful, probably due to extensive side reactions on the nitrogen atoms.18 We therefore decided to synthesize the bipy amine derivative 2-(2,2′-bipyridin-6-yloxy) ethylamine according to a previously published procedure for the synthesis of 2-(2,2′:6′,2′′-terpyridin-4′-yloxy) ethylamine.19 The compatibility of this bipy derivative (Nbp) with the “submonomer” method was tested by its incorporation at the third position of a peptoid tetramer containing methyl benzyl synthons at the other positions. HPLC analysis at 214 nm of the crude peptoid after each step – Nbp displacement, subsequent acylation and the successive methyl benzylamine displacement (Fig. S16, ESI†), as well as ESI-MS analysis of the crude peptoid tetramer, confirmed the successful incorporation of this bipy derivative within the sequence.
Peptoid oligomers L1B and L2B were designed such that each contains four bulky chiral side chains, which are sufficient for attaining helicity in oligomers of this length.14d In L2B, the ligands were placed at positions i and i + 3, which match the pitch of the helix, in order to orient these groups in proximity on the same face of the backbone and facilitated the creation of the intermolecular complex depicted in Fig. 1.20 Peptoid C3B was designed as a hexamer with three bipy ligands in alternating positions on the sequence such that they will all face the same side of the macrocycle plain and facilitate an intramolecular binding.21 The linear peptoids L1B, L2B and L3B, as well as the dimer Di-L1B (see the ESI†), incorporating Nbp, (S)-(−)-1-phenylethylamine (Nspe) and chloropropylamine (Npl, only L3B) as synthons were synthesized on the solid support employing the “submonomer” protocol. The peptoid C3B was prepared by a microwave-assisted cyclization of peptoid L3B following a method that we have recently published.22 All the peptoids were analysed and purified by HPLC, and their sequences were confirmed by ESI-MS. The Ru2+ complexes (L1B)3Ru, (L2B)3Ru2, (C3B)Ru and (Di-L1B)3Ru were synthesized using a modification of a previously reported protocol,23 by adding 1 equiv. of RuCl3 hydrate to 3 equiv. of L1B or Di-L1B, 1.5 equiv. of L2B and 1 equiv. of C3B, respectively in ethanol under reflux conditions. After a few hours of stirring, the pale yellow colour of the peptoid solution turned blue in the case of L1B and Di-L1B, and red-brown in the case of L2B and C3B. The final colour of all the complexes was red-orange. These complexes were purified and analysed by HPLC, and their identity was confirmed by detailed mass spectrometry studies including computed and experimental isotopic envelope analysis (see the ESI†). These studies show that all the Cl− ligands were replaced by the bipy-peptoids, supporting the suggested binding mode of the complexes presented in Fig. 1.
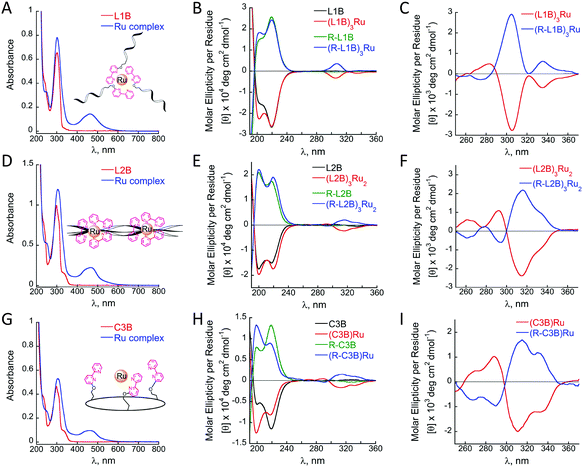
Metal free peptoids L1B, L2B and C3B exhibit absorption bands near λ = 302, 299 and 298 nm, respectively, in acetonitrile, corresponding to the π–π* transition of bipy. The complexes (L1B)3Ru, (L2B)3Ru2 and (C3B)Ru reveal shifts in these absorption bands with values of λ = 303, 305 and 305 nm, respectively, and additional bands near λ = 463, 461 and 464 nm, respectively, which correspond to the metal-to-ligand charge transfer (MLCT) bands of the ruthenium polypyridine complexes (Fig. 2A, D and G).
Circular dichroism (CD) measurements revealed some changes upon the formation of the complexes (L1B)3Ru and (L2B)3Ru2, and significant changes upon (C3B)Ru formation. Solutions of all three complexes showed an increase, relative to the metal-free peptoids, in the ellipticity near 200 nm while only the complex (C3B)Ru exhibited a major decrease, relative to the metal-free peptoid, in the magnitude of the CD signal near 218 nm (Fig. 2B, E and H, black and red lines).
It has been previously demonstrated that Nspe peptoids adopt right-handed helices,14b that have characteristic CD spectra with bands near 190 and 200 nm, the latter has been associated with the trans-amide bond conformation, and 218 nm, which is associated with the cis-amide bond conformation.14 Both peptoids L1B and C3B exhibit CD spectra with an intense band at 218 nm (Fig. 2B and H, red lines), indicating that helices with only cis-amide bonds are the major population in solution. Upon formation of the complexes (L1B)3Ru and (C3B)Ru an increase in the band near 200 nm was obtained (Fig. 2B and H, red lines), indicating that the helices with only cis-amide bonds are no longer the dominant population in solution. For (L1B)3Ru, this increase implies a decrease in the overall conformational order of the peptoid upon ruthenium binding.24 In the case of (C3B)Ru, there is also a decrease in the band near 218 nm, suggesting that the conformational order is almost diminished due to a significant alteration of the helical backbone after ruthenium complexation. The CD spectrum of L2B exhibits similar intensities of both bands near 200 and 218 nm (Fig. 2E, black line), reflecting the conformational heterogeneity of this peptoid. A solution of its ruthenium complex exhibited increases, relative to the metal-free peptoid, in the magnitude of both of these CD signals (Fig. 2E, red line), suggesting an increase in its overall conformational order as the magnitude of the signal in this case, reflecting the degree of helicity.8d,e,16b
Ruthenium binding to all three peptoids also produced new bands in the region between 260 and 360 nm corresponding to the π–π* transition of bipy. These bands, however, were very weak and did not show the expected Cotton effects. In order to probe this point we repeated the CD measurements using higher solution concentrations (200 μM instead of 30 or 100 μM) of (L1B)3Ru, (L2B)3Ru2, and (C3B)Ru and indeed obtained stronger CD signals that showed clear Cotton effects with minima at λmax = 305, 316 and 314 nm, respectively (Fig. 2C, F and I, red lines). These signals reflect the induction of chirality from the peptoid scaffold to the metal center,16b and imply at more preferable stereochemistry of the Δ isomer over the Λ isomer.25 The bipy π–π* CD signal of (C3B)Ru exhibits an exciton couplet,26 with a minimum at 314 nm and a maximum at 289 nm, crossing ε = 0 near 296 nm. In contrast, (L2B)3Ru2 exhibits a weaker exciton couplet, which is completely absent in the case of (L1B)3Ru. This can be attributed to reduced conformational constraints in the intermolecular complexes, which can diminish the effect of the dipole–dipole interactions responsible for an exciton couplet. In order to further explore the preference of stereochemistry, we prepared three more peptoids using Nrpe monomers instead of Nspe monomers, namely R-L1B, R-L2B and R-C3B, and their corresponding Ru2+ complexes (R-L1B)3Ru, (R-L2B)3Ru2 and (R-C3B)Ru. These complexes were purified and analysed by HPLC, characterized by UV and their identity was confirmed by detailed MS studies, exhibiting the same absorbance bands and masses as their corresponding Nspe containing Ru–peptoids (see the ESI†). CD measurements using 200 μM solutions of these complexes displayed the exact opposite spectra and Cotton effect relative to the Nspe containing peptoids–Ru complexes (Fig. 2C, F and I, blue lines). These results demonstrate that the stereochemistry of the Ru2+ center can indeed be controlled and dictated by the chirality of the peptoid scaffold.
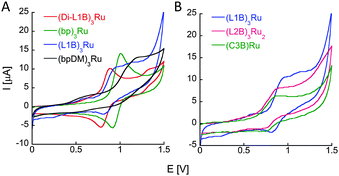
The redox properties of the new ruthenium complexes were realized from cyclic voltammetry. (L1B)3Ru, (L2B)3Ru2, (C3B)Ru and (Di-L1B)3Ru exhibit a reversible oxidation process of the metal centered Ru2+/3+ couple at Eoxp of 0.94 V, 0.88 V, 0.90 V and 0.88 V vs. Ag/AgNO3 respectively. These oxidation potentials imply that all the bipy ligands from the peptoids are coordinating Ru2+ and there are no Cl− ions bound as ligands.27 These oxidation potentials are negatively shifted compared to that of (bp)3Ru, which is centered around 1.02 V (Fig. 3A, green line), probably due to the electron withdrawing nature of the amides. To probe this point, we have synthesized 2-(2,2′-bipyridin-6-yloxy) ethyl(dimethyl)amine (bp-DM), which contains the methoxy group but lacks the amide. Its Ru2+ complex (bp-DM)3Ru was prepared, purified and characterized by UV and MS spectroscopies, and its CV showed an oxidation potential at Eoxp of 1.22 V, which expresses a positive shift compared to Ru(bipy)3, probably due to the donating character of the methoxy group. Notably, similar intensities of the current flows are obtained in identical concentrations of the complexes (L1B)3Ru and (C3B)Ru (0.5 mM) and in as twice as less concentrated solution of (L2B)3Ru2 (0.25 mM). We therefore propose that the two Ru centers in (L2B)3Ru2 are oxidized simultaneously, thus giving rise to only one oxidation peak.
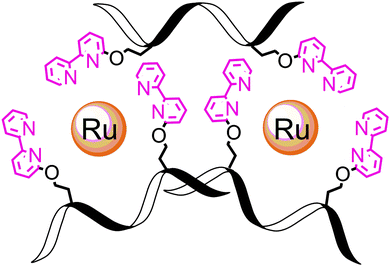
Finally, we note that L2B can bind ruthenium in an intramolecular fashion leading to a different (L2B)3Ru2 complex in which two intramolecular (L2B)2Ru complexes bind an additional L2B peptoid (Scheme 1). The weak exciton couplet exhibited by (L2B)3Ru2 and the similarity in the oxidation potentials of (L2B)3Ru2 and (C3B)Ru that demonstrate their similar central structure,8c support the existence of such a complex. We can therefore assume that both the intermolecular (L2B)3Ru2 and the intramolecular/intermolecular (L2B)3Ru2 are present in solution.
To conclude, this work describes the facile incorporation of bipy into the peptoid sequence for the design and synthesis of versatile bipy modified peptoid–ruthenium complexes. These include a helical triplex via intermolecular metal binding and a unique cyclometallopeptoid complex via intramolecular metal ligation of three pendant groups. Our results demonstrate that the dual effect of ruthenium binding; the chirality of the peptoid establishes an asymmetric environment about the metal center while metal complexation causes modifications in the conformational order of the peptoid backbone. Specifically, CD analysis indicates that the stereochemistry of the Ru2+ center is influenced by the chirality of the peptoid scaffold. We are currently studying other bipy-modified metallopeptoids and exploring the photophysical properties of peptoid–ruthenium complexes towards photocatalysis.
Notes and references
- C. Kaes, A. Katz and M. W. Hosseini, Chem. Rev., 2000, 100, 3553–3590 CrossRef CAS PubMed.
- K. Kalyanasundaram, Coord. Chem. Rev., 1982, 46, 159–244 CrossRef CAS.
- (a) B. Imperiali and S. L. Fisher, J. Org. Chem., 1992, 57, 757–759 CrossRef CAS; (b) B. Imperiali, T. J. Prins and S. L. Fisher, J. Org. Chem., 1993, 58, 1613–1616 CrossRef CAS; (c) B. Imperiali and S. L. Fisher, J. Am. Chem. Soc., 1991, 113, 8527–8528 CrossRef CAS.
- (a) D. L. Popescu, T. J. Parolin and C. Achim, J. Am. Chem. Soc., 2003, 125, 6354–6355 CrossRef CAS PubMed; (b) J. I. Yeh, E. Pohl, D. Truan, W. He, G. M. Sheldrick, S. Du and C. Achim, Chem. – Eur. J., 2010, 16, 11867–11875 CrossRef CAS PubMed; (c) R. M. Franzini, R. M. Watson, G. K. Patra, R. M. Breece, D. L. Tierney, M. P. Hendrich and C. Achim, Inorg. Chem., 2000, 45, 9798–9811 CrossRef PubMed.
- J. Lee, D. G. Udugamasooriya, H.-S. Lim and T. Kodadek, Nat. Chem. Biol., 2010, 6, 258–260 CrossRef CAS PubMed.
- R. P. Cheng, S. L. Fisher and B. Imperiali, J. Am. Chem. Soc., 1996, 118, 11349–11356 CrossRef CAS.
- A. Torrado and B. Imperiali, J. Org. Chem., 1996, 61, 8940–8948 CrossRef CAS PubMed.
- (a) C. P. Myers and M. E. Williams, Coord. Chem. Rev., 2010, 254, 2416–2428 CrossRef CAS; (b) C. P. Myers, B. P. Gilmartin and M. E. Williams, Inorg. Chem., 2008, 47, 6738–6747 CrossRef CAS PubMed; (c) M. A. Case, M. R. Ghadiri, M. W. Mutz and G. L. Mc Lendon, Chirality, 1998, 10, 35–40 CrossRef CAS PubMed; (d) M. R. Ghadiri, C. Soares and C. Choi, J. Am. Chem. Soc., 1992, 114, 825–831 CrossRef CAS; (e) M. Lieberman, M. Tabet and T. Sasaki, J. Am. Chem. Soc., 1994, 116, 5035–5044 CrossRef CAS.
- M. R. Ghadiri and A. K. Fernholz, J. Am. Chem. Soc., 1990, 112, 9633–9635 CrossRef CAS.
- (a) M. Gochin, V. Khorosheva and M. A. Case, J. Am. Chem. Soc., 2002, 124, 11018–11028 CrossRef CAS PubMed; (b) S. Tashiro and M. Shionoya, Chem. Lett., 2013, 42, 456–462 CrossRef CAS.
- (a) G. Maayan, Eur. J. Org. Chem., 2009, 5699–5710 CrossRef CAS; (b) Metallofoldamers. Supramolecular Architectures from Helicates to biomimetics, ed. G. Maayan and M. Albrecht, John Wiley & Sons, Ltd, 2013 Search PubMed; (c) J. P. Miller, M. S. Melicher and A. Schepartz, J. Am. Chem. Soc., 2014, 136, 14726–14729 CrossRef CAS PubMed.
- A. S. Knight, E. Y. Zhou, M. B. Francis and R. N. Zuckermann, Adv. Mater., 2015, 38, 5665–5691 CrossRef PubMed.
- (a) S. H. Gellman, Acc. Chem. Res., 1998, 31, 173–180 CrossRef CAS; (b) Foldamers: structure, properties, and applications, ed. S. Hecht, and I. Huc, Wiley-VCH, Weinheim, 2007 Search PubMed; (c) D. Seebach and J. Cardiner, Acc. Chem. Res., 2008, 41, 1366–1375 CrossRef CAS PubMed; (d) D. J. Hill, M. J. Mio, R. B. Prince, T. S. Hughes and J. S. Moore, Chem. Rev., 2001, 101, 3893–4012 CrossRef CAS PubMed; (e) S. A. Fowler and H. E. Blackwell, Org. Biomol. Chem., 2009, 7, 1508–1524 RSC; (f) C. M. Goodman, S. Choi, S. Shandler and W. F. DeGrado, Nat. Chem. Biol., 2007, 3, 252–262 CrossRef CAS PubMed.
- (a) C. W. Wu, et al. , J. Am. Chem. Soc., 2003, 125, 13525–13530 CrossRef CAS PubMed; (b) P. Armand, K. Kirshenbaum, A. Falicov, R. L. Jr. Dunbrack, K. A. Dill, R. N. Zuckermann and F. E. Cohen, Folding Des., 1997, 2, 369–375 CrossRef CAS; (c) K. Kirshenbaum, et al. , Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 4303–4308 CrossRef CAS PubMed; (d) C. W. Wu, T. J. Sanborn, K. Huang, R. N. Zuckermann and A. E. Barron, J. Am. Chem. Soc., 2001, 123, 6778–6784 CrossRef CAS PubMed; (e) P. Armand, et al. , Proc. Natl. Acad. Sci. U. S. A., 1998, 14, 4309–4314 CrossRef.
- R. N. Zuckermann, J. M. Kerr, S. B. H. Kent and W. H. Moos, J. Am. Chem. Soc., 1992, 114, 10646 CrossRef CAS.
- (a) B. C. Lee, T. K. Chu, K. A. Dill and R. N. Zuckermann, J. Am. Chem. Soc., 2008, 130, 8847–8855 CrossRef CAS PubMed; (b) G. Maayan, M. D. Ward and K. Kirshenbaum, Chem. Commun., 2009, 56–58 RSC; (c) N. Maulucci, I. Izzo, G. Bifulco, A. Aliberti, C. De Cola, D. Comegna, C. Gaeta, A. Napolitano, C. Pizza, C. Tedesco, D. Flot and F. De Riccardis, Chem. Commun., 2008, 3927–3929 RSC; (d) C. De Cola, S. Licen, D. Comegna, E. Cafaro, G. Bifulco, I. Izzo, P. Tecilla and F. De Riccardis, Org. Biomol. Chem., 2009, 7, 2851 RSC; (e) G. Della Sala, B. Nardone, F. De Riccardis and I. Izzo, Org. Biomol. Chem., 2013, 11, 726–731 RSC; (f) I. Izzo, G. Ianniello, C. De Cola, B. Nardone, L. Erra, G. Vaughan, C. Tedesco and F. De Riccardis, Org. Lett., 2013, 15, 598–601 CrossRef CAS PubMed; (g) C. De Cola, G. Fiorillo, A. Meli, S. Aime, E. Gianolio, I. Izzo and F. De Riccardis, Org. Biomol. Chem., 2014, 12, 424 RSC; (h) C. Tedesco, L. Erra, I. Izzo and F. De Riccardis, CrystEngComm, 2014, 16, 3667–3687 RSC; (i) T. Zabrodski, M. Baskin, J. K. Prathap and G. Maayan, Synlett, 2014, A–F Search PubMed; (j) M. Baskin and G. Maayan, Biopolymers, 2015, 104, 577–584 CrossRef CAS PubMed; (k) A. S. Knight, E. Y. Zhou, J. G. Pelton and M. B. Francis, J. Am. Chem. Soc., 2013, 135, 17488–17493 CrossRef CAS PubMed; (l) A. S. Knight, E. Y. Zhou and M. B. Francis, Chem. Sci., 2015, 6, 4042–4048 RSC.
- G. Maayan, Y. Dayagi, R. Arad-Yellin, L. J. W. Shimon and A. Shanzer, Polyhedron, 2013, 64, 365–370 CrossRef CAS.
- T. S. Burkoth, A. T. Fafarman, D. H. Charych, M. D. Connolly and R. N. Zuckermann, J. Am. Chem. Soc., 2003, 125, 8841–8845 CrossRef CAS PubMed.
- G. Maayan, B. Yoo and K. Kirshenbaum, Tetrahedron Lett., 2008, 49, 335 CrossRef CAS PubMed.
- M. Baskin and G. Maayan, Chem. Sci., 2016, 7, 2809–2820 RSC.
- S. B. Y. Shin, B. Yoo, L. J. Todaro and K. Kirshenbaum, J. Am. Chem. Soc., 2007, 129, 3218–3225 CrossRef CAS PubMed.
- P. J. Kaniraj and G. Maayan, Org. Lett., 2015, 17, 2110–2113 CrossRef CAS PubMed.
- J. V. Caspar and T. J. Meyer, J. Am. Chem. Soc., 1983, 105, 5583–5590 CrossRef CAS.
- J. M. Holub, H. Jang and K. Kirshenbaum, Org. Lett., 2007, 17, 3275–3278 CrossRef PubMed.
- M. H. Filby, J. Muldoon, S. Dabb, N. C. Fletcher, A. E. Ashcroft and A. J. Wilson, Chem. Commun., 2011, 47, 559–561 RSC.
- X. Huanget, et al. , J. Am. Chem. Soc., 2002, 124, 10320–10335 CrossRef.
- V. Balzani, A. Juris and M. Venturi, Chem. Rev., 1996, 96, 759–833 CrossRef CAS PubMed and ref. 21 and 37 in that review.
Footnote |
| † Electronic supplementary information (ESI) available: Detailed synthetic procedures, HPLC and MS spectra, and complementary UV spectra. See DOI: 10.1039/c6cc04346a |
| This journal is © The Royal Society of Chemistry 2016 |