Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Anion binding by biotin[6]uril in water†
Micke
Lisbjerg
,
Bjarne E.
Nielsen
,
Birgitte O.
Milhøj
,
Stephan P. A.
Sauer
 and
Michael
Pittelkow
*
and
Michael
Pittelkow
*
Department of Chemistry, University of Copenhagen, Universitetsparken 5, DK-2100 Copenhagen Ø, Denmark. E-mail: pittel@kiku.dk; Web: http://www.pittelkow.kiku.dk
First published on 19th November 2014
Abstract
In this contribution we show that the newly discovered 6 + 6 biotin-formaldehyde macrocycle Biotin[6]uril binds a variety of anionic guest molecules in water. We discuss how and why the anions are bound based on data obtained using NMR spectroscopy, mass spectrometry, isothermal titration calorimetry (ITC), computational calculations and single crystal X-ray crystallography.
Molecular recognition in water is particularly difficult to achieve due to the competitive nature of water as a hydrogen bond donor and acceptor.1 One may argue that medicinal chemistry relies heavily on supramolecular chemistry in water, which highlights the importance of pursuing new fundamental understanding within this particular area.2 In this paper we address the challenge of recognising anions in water.3
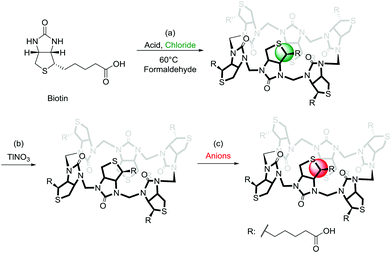
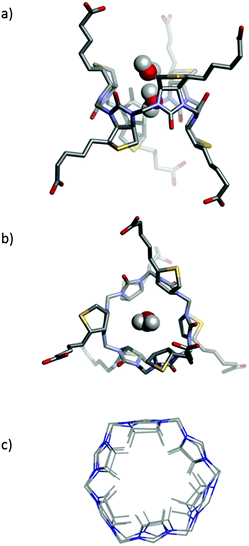
A large number of elegant and complex receptors have been prepared and studied for their molecular recognition properties. In supramolecular chemistry the most utilised receptors are those that are easily prepared, naturally occurring or commercially available. Among these are symmetrical macrocyclic structures such as calix[n]arenes,4 calix[n]pyrroles,5 and calix[n]resorcinarenes.6 Naturally occurring receptors such as the cyclodextrins7 and linear amyloses have also been studied.8 Recently a number of urea-based macrocycles,9 such as cucurbit[n]urils,10 hemicucurbit[n]urils11 and bambus[n]urils12 have been introduced and especially the family of cucurbit[n]urils have become popular due to the rich supramolecular chemistry offered by these structures in water.13 Common to many of the popular macrocycles is that they are prepared by simple condensation reactions using aldehydes as a condensation partner. To introduce attractive features such as chirality and enhanced water-solubility into these structures subsequent and sometimes elaborate synthetic efforts are necessary. We recently introduced a new type of receptor molecule that is easily prepared in a single synthetic step, is chiral, is water soluble and is capable of binding anions in water: the Biotin[6]uril (Fig. 1).14
The synthesis of this regiochemically well-defined macrocyclic structure was achieved in multigram quantities in one synthetic step, and the product precipitated directly from the reaction mixture, which made the purification of the product simple. The six biotin units of the Biotin[6]uril have twelve protons from the convex side of each biotin unit pointing into the cavity of the Biotin[6]uril. This makes the cavity distinctly hydrophobic (Fig. 1). In previous work we described how this hydrophobic pocket is the binding site for the halide anions in water at pH 10.8 in carbonate buffer.14
In this contribution we show how Biotin[6]uril is capable of binding a series of monovalent anions at pH 7.5 in phosphate buffer with binding constants ranging from log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K = 1.8 (Cl−) and log
K = 1.8 (Cl−) and log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K = 4.5 (−SCN). Initially we investigated a wide range of potential guest molecules and ions for their binding properties towards the Biotin[6]uril using 1H NMR spectroscopy. Binding was indicated by a change in the chemical shifts of the protons on the convex side of each biotin unit pointing into the cavity (protons b and f in Fig. 2c). We tested a series of aliphatic amines (e.g. 1,6-diaminohexane, 1,7-diaminoheptane, ethanolamine, and propargylamine), a series of cations (e.g. Na+, K+, and Cs+), a series of anions (e.g. CN−, ClO4−, PF6−, PhCO2− and CH3CO2− and the anions in Table 1) and a series of neutral guests (propionitrile, CO2, CS2 and propargyl-alcohol) for binding by Biotin[6]uril in water at pH 7.5 in 100 mM phosphate buffer.
K = 4.5 (−SCN). Initially we investigated a wide range of potential guest molecules and ions for their binding properties towards the Biotin[6]uril using 1H NMR spectroscopy. Binding was indicated by a change in the chemical shifts of the protons on the convex side of each biotin unit pointing into the cavity (protons b and f in Fig. 2c). We tested a series of aliphatic amines (e.g. 1,6-diaminohexane, 1,7-diaminoheptane, ethanolamine, and propargylamine), a series of cations (e.g. Na+, K+, and Cs+), a series of anions (e.g. CN−, ClO4−, PF6−, PhCO2− and CH3CO2− and the anions in Table 1) and a series of neutral guests (propionitrile, CO2, CS2 and propargyl-alcohol) for binding by Biotin[6]uril in water at pH 7.5 in 100 mM phosphate buffer.
| 1H-NMR log(Ka) | ITC log(Ka) | ΔHb (kJ mol−1) | TΔSb (kJ mol−1) | |
|---|---|---|---|---|
| a Data was obtained at pH 10.8 in carbonate buffer.14 b ΔH and ΔS are from the ITC data at 30 °C. All data obtained had less than 6% error. | ||||
| NaCl | 1.8 (1.7a) | 1.5 (1.0a) | −30.7 | −21.8 |
| NaBr | 3.0 | 2.7 | −37.5 | −21.6 |
| NaI | 3.7 | 3.4 | −42.8 | −23.0 |
| KIa | 3.3 | — | — | — |
| CsI | 3.7 | — | — | — |
| NaNO3 | 1.9 | 1.7 | −32.3 | −22.2 |
| NaN3 | 2.9 | 2.6 | −31.1 | −16.1 |
| NaClO4 | 2.7 | 2.4 | −33.3 | −19.5 |
| KCNO | 2.0 | 1.8 | −29.9 | −19.6 |
| KSeCN | 4.3 | 4.0 | −37.7 | −14.5 |
| NaSCN | 4.5 | 4.1 | −35.0 | −11.2 |
We were pleased to find that Biotin[6]uril interacts with a range of singly charged anions (Table 1). No binding to divalent anions was observed, as exemplified by experiments with SO42−, WO42−, CO32− and HPO42−.
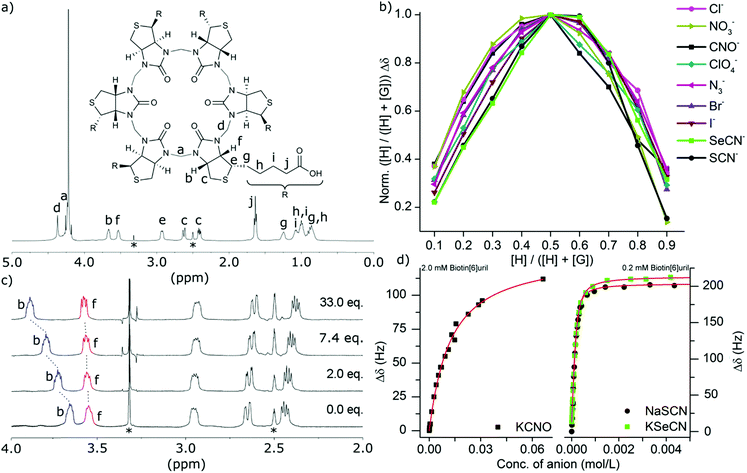
For the series of singly charged anions we measured the binding stoichiometries using the continuous variation method of Job by means of 1H NMR spectroscopy in water at pH 7.5 in 100 mM phosphate buffer (Fig. 2b). All the anions presented here showed a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
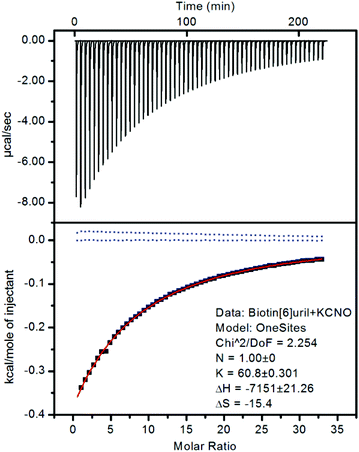
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding stoichiometry. To confirm that the chemical shift changes were not due to aggregation events we measured the 1H NMR spectra at different concentrations (ESI†) where no changes were observed as a function of concentration. To further evaluate the binding interactions of Biotin[6]uril with the anions we proceeded to measure the binding affinities using both 1H NMR titrations (Fig. 2c) and Isothermal Calorimetric Titration (ITC, Fig. 3). The binding affinity data are summarised in Table 1.
1 binding stoichiometry. To confirm that the chemical shift changes were not due to aggregation events we measured the 1H NMR spectra at different concentrations (ESI†) where no changes were observed as a function of concentration. To further evaluate the binding interactions of Biotin[6]uril with the anions we proceeded to measure the binding affinities using both 1H NMR titrations (Fig. 2c) and Isothermal Calorimetric Titration (ITC, Fig. 3). The binding affinity data are summarised in Table 1.
In the titration experiments using 1H NMR spectroscopy to monitor the binding event, the change in chemical shift values for the protons on the convex side of each biotin unit pointing into the cavity were monitored (Fig. 2c). When plotting the change in chemical shift value as a function of the anion concentration the characteristic graphs shown in Fig. 2d were obtained. By applying non-linear curve fitting the binding constants (Ka) and therefore binding energy (ΔG) can be found (Table 1 and ESI†). In Fig. 2d three such plots are shown for the three anions cyanate (OCN−), thiocyanate (SCN−) and selenocyanate (SeCN−). It can be seen that a lot more cyanate needs to be added to the Biotin[6]uril before saturation is achieved (Fig. 2d, left) than thiocyanate and selenocyanate (Fig. 2c, right). This immediately indicates that thiocyanate and selenocyanate bind Biotin[6]uril significantly more strongly than cyanate does.
The 1H NMR titrations revealed that Biotin[6]uril binds the halide anions in phosphate buffer at pH 7.5 in the order Cl− > Br− > I− which is the same as previously reported at pH 10.8 in carbonate buffer with comparable binding affinities at the two sets of conditions. To evaluate whether the cation plays a role in the binding of the anions we tested the Na+, K+ and Cs+ salts of iodide (Table 1). We found that the binding affinities were comparable. This indicates that any cation interaction is insignificant for the binding event in solution. This can also be seen by analysing the crystal structure of Biotin[6]uril with NaI which shows an interaction from a urea-carbonyl carbon to a Na+, but not to the anion binding cavity.
The binding of the halides show a trend where the larger anions fit better in the cavity. The same trend is observed for the cyanate anion family (Fig. 2 and 3). When increasing the thermodynamic radii of the anion going from CNO− (0.195 nm) to SeCN− and SCN− (0.213 nm) the binding gets significantly stronger.15 The larger binding affinities for the anions with larger thermodynamic radii can also be viewed as a consequence of the increased softness of the larger ions. Biotin[6]uril favours larger softer anions over smaller harder anions, until a certain size, as the anion needs to fit in the cavity of the receptor. The ITC data shows that all the binding interactions are enthalpically favorable and entropically unfavorable.
An interesting observation was that there is no binding to the ClO3− anion even though the receptor binds the ClO4− anion, which might be because of the trigonal pyramidal structure of the ClO3− anion which has a smaller thermodynamic radii than the ClO4− anion (0.171 nm vs. 0.240 nm).15 We did not observe any oxidative degradation of the Biotin[6]uril when exposing it to ClO3− or ClO4−. We speculate that the ClO3− anion binds more weakly due to the smaller size – and therefore its fit to the cavity within the Biotin[6]uril is simply less favourable. We have also observed that the F− anion binds very weakly which is possibly also due to the small size of this anion. Even though the ClO4− anion is only slightly larger than the SCN− anion (0.213 nm) and the I− (0.210 nm) anion its binding is much weaker. We hypothesise that this is due to the larger size that makes the ClO4− anion fit in the cavity less well.
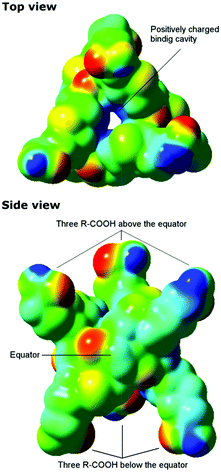
To gain understanding of why Biotin[6]uril, which is a hexa-carboxylic acid, and therefore presumably an anionic species at pH 7.5, binds anions we calculated a plot of the electrostatic potential for Biotin[6]uril using density functional theory (DFT) calculations. The calculations were performed directly for the crystal structures employing the long-range corrected CAM-B3LYP functional in combination with the 6-31G(d) basis set in the Gaussian 09 program.16–18 From the electrostatic potential plot it is clear that the macrocycle has a positive electrostatic potential in the cavity (blue, Fig. 4), whereas around the oxygen atoms of the urea-carbonyl groups the electrostatic potential is more negative (red, Fig. 4). The sulphur atoms lead also to a weakly negative electrostatic potential.
Biotin[6]uril has an alternating orientation of the negatively charged urea carbonyls relative to the equator of the macrocyclic receptor. Therefore the macrocycle does not bind cations at the peripheries like the cucurbit[n]urils do.13 The electrostatic potential is very similar to that reported for Bambus[6]urils, which also show a significant positive electrostatic potential in the cavity.12
In our previous work we have reported two single crystal X-ray structures of Biotin[6]uril.14 In one of those structures an iodide anion was bound in the binding cavity, and in the other structure a molecule of ethanol was situated in the cavity. Herein we report a new single crystal X-ray structure of Biotin[6]uril (Fig. 5a,b). In this structure the cavity contains two molecules of water that are hydrogen bonded to each. On the outside of the binding cavity we observe further molecules of water (disordered) to one side, and a carboxylic acid moiety from one of the side arms of the Biotin[6]uril to the other side. The two water molecules that reside within the cavity must be replaced by the anions upon binding of the anion to the cavity.
While the cavities of the Biotin[6]uril containing water, EtOH and iodide appear similar the cavities are actually subtly different. In Fig. 5c the urea-containing five membered rings of the each Biotin unit (including the H's that point into the binding cavity) and the connecting formaldehyde derived CH2-groups are shown (overlaid). This shows that the radius of the binding pocket is relatively unaffected by the different guest molecules. It is, however, possible for the biotin units to rotate slightly within the macrocyclic structure, thus changing slightly the directionality of the C–H bonds with respect to the centre of the cavity, and also the length of the binding cavity. The cavity size is similar to that of Bambus[6]uril reported by Sindelar and co-workers.12
One can view the centre of Biotin[6]uril as a cylinder shaped binding cavity defined by the 12 hydrogen atoms originating from the six C–H bonds from the biotin units. The 12 H-atoms define two offset circles with 6 H-atoms at the rim of each circle. By measuring the radius of each of these circles and the distance between them we get a cylinder shaped cavity with a volume of 86–102 Å3 (see ESI† for details) for the three X-ray structures. This internal volume is not constant for the three crystal structures, indicating that the binding cavity does have some flexibility. This iodide containing structure has a volume of 93 Å3, the EtOH containing structure a volume of 102 Å3 and the water containing cavity a volume of 86 Å3. This difference in cavity sizes are mainly due to the six biotin units of the macrocycle tilting slightly, giving a longer more narrow binding pocket.
Finally we studied the Biotin[6]uril-anion complexes using electrospray ionisation mass spectrometry. Solutions of Biotin[6]uril and an excess of the various anions were prepared in water, and these were analysed by direct injection ESI mass spectrometry. The spectra convincingly indicate the formation of complexes of the Biotin[6]uril and the anions. For the halide series of anions the mass spectra are shown in Fig. 6. In all three cases (Cl−, Br− and I−) clear molecular ions for the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complexes are observed.
1 complexes are observed.
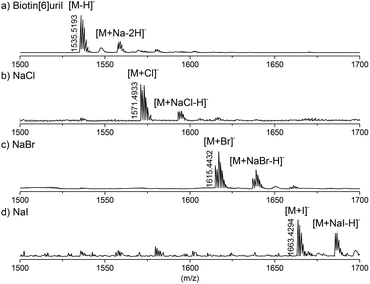 | ||
| Fig. 6 High resolution mass of the Biotin[6]uril without guest (a), with NaCl (b), NaBr (c), and NaI (d). | ||
Conclusions
In this communication we show how it is possible to bind a series of simple mono-charged anions to our recently discovered anion receptor Biotin[6]uril in water. We notice that the cavity of the Biotin[6]uril contains two water molecules, which upon release could contribute favourably to the enthalpically driven binding (non-classical hydrophobic effect).19 The enthalpically driven binding event is evident from the ITC data. The binding of anions, we speculate, is governed by a delicate balance between the anions size in order to fit in the cavity, and the hardness/softness of the anion.Acknowledgements
We acknowledge financial support from the Danish Research council (FNU) for a Steno Fellowship (MP) and the Lundbeck Foundation for a Young Group Leader Fellowship (MP) and a PhD scholarship (BMO). The support and sponsorship provided by COST Action CM1005 and MP1002 are acknowledged. We thank Dr Sophie R. Beeren for insightful discussions.Notes and references
- G. V. Oshovsky, D. N. Reinhoudt and W. Verboom, Angew. Chem., Int. Ed., 2007, 46, 2366–2393 CrossRef CAS PubMed.
- D. K. Smith, J. Chem. Educ., 2005, 82, 393–400 CrossRef CAS.
- (a) S. Kubik, Chem. Soc. Rev., 2010, 39, 3648–3663 RSC; (b) S. Kubik, Chem. Soc. Rev., 2009, 38, 585–605 RSC; (c) P. A. Gale and C. Caltagirone, Chem. Soc. Rev., 2014 10.1039/c4cs00179f; (d) J. L. Sessler, P. A. Gale and W.-S. Cho, Anion Receptor Chemistry, RSC, Cambridge, 2006 Search PubMed.
- L. Baldini, R. Sansone, A. Casnati and R. Ungaro, Supramolecular Chemistry: From Molecules to Nanomaterials, John Wiley & Sons Ltd, 2012 Search PubMed.
- P. Ballester, Isr. J. Chem., 2011, 51, 710–724 CrossRef CAS.
- V. K. Jain and P. H. Kanaiya, Russ. Chem. Rev., 2011, 82, 75–102 CrossRef PubMed.
- R. Breslow, Acc. Chem. Res., 1995, 28, 146–153 CrossRef CAS.
- (a) S. Meier and S. R. Beeren, J. Am. Chem. Soc., 2014, 136, 11284–11287 CrossRef CAS PubMed; (b) S. R. Beeren and O. Hindsgaul, Angew. Chem., Int. Ed., 2013, 52, 11265–11268 CrossRef CAS PubMed.
- N. Voiz and J. Clayden, Angew. Chem., Int. Ed., 2011, 50, 12148–12155 CrossRef PubMed.
- E. Masson, X. Ling, R. Joseph, L. Kyeremeh-Mensah and X. Lu, RSC Adv., 2012, 2, 1213–1247 RSC.
- Y. Miyahara, K. Goto, M. Oka and T. Inazu, Angew. Chem., Int. Ed., 2004, 43, 5019–5022 CrossRef CAS PubMed.
- (a) J. Svec, M. Necas and V. Sindelar, Angew. Chem., Int. Ed., 2010, 49, 2378–2381 CrossRef CAS PubMed; (b) V. Havel, J. Svec, M. Wimmerova, M. Dusek, M. Pojarova and V. Sindelar, Org. Lett., 2011, 13, 4000–4003 CrossRef CAS PubMed.
- L. Cao, M. Sekutor, P. Y. Zavalij, K. Mlinaric-Majerski, R. Glaser and L. Isaacs, Angew. Chem., Int. Ed., 2014, 53, 988–993 CrossRef CAS PubMed.
- M. Lisbjerg, B. M. Jessen, B. Rasmussen, B. E. Nielsen, A. Ø. Madsen and M. Pittelkow, Chem. Sci., 2014, 5, 2647–2650 RSC.
- H. D. B. Jenkins and K. P. Thakur, J. Chem. Educ., 1979, 56, 576–577 CrossRef CAS.
- W. J. Hehre, R. Ditchfield and J. A. Pople, J. Chem. Phys., 1972, 56(5), 2257–2261 CrossRef CAS PubMed.
- T. Yanai, D. P. Tew and N. C. Handy, Chem. Phys. Lett., 2004, 393, 51–57 CrossRef CAS PubMed.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, B. Mennucci, G. A. Petersson, H. Nakatsuji, M. Caricato, X. Li, H. P. Hratchian, A. F. Izmaylov, J. Bloino, G. Zheng, J. L. Sonnenberg, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, J. J. A. Montgomery, J. E. Peralta, F. Ogliaro, M. Bearpark, J. J. Heyd, E. Brothers, K. N. Kudin, V. N. Staroverov, T. Keith, R. Kobayashi, J. Normand, K. Raghavachari, A. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, N. Rega, J. M. Millam, M. Klene, J. E. Knox, J. B. Cross, V. Bakken, C. Adamo, J. Jaramillo, R. Gomperts, R. E. Stratmann, O. Yazyev, A. J. Austin, R. Cammi, C. Pomelli, J. W. Ochterski, R. L. Martin, K. Morokuma, V. G. Zakrzewski, G. A. Voth, P. Salvador, J. J. Dannenberg, S. Dapprich, A. D. Daniels, O. Farkas, J. B. Foresman, J. V. Ortiz, J. Cioslowski and D. J. Fox, Gaussian 09, Revision B.01, Gaussian, Inc., Wallingford CT, 2010 Search PubMed.
- F. Biedermann, W. M. Nau and H.-J. Schneider, Angew. Chem., Int. Ed., 2014, 53, 11158–11171 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Binding studies, X-ray crystal structure and Job plots. CCDC 1029718. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c4ob02211d |
| This journal is © The Royal Society of Chemistry 2015 |