Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Enhanced tumor-targeted gene delivery by bioreducible polyethylenimine tethering EGFR divalent ligands†
Duhwan
Lee‡
a,
Yeong Mi
Lee‡
a,
Jihoon
Kim
a,
Myung Kyu
Lee
b and
Won Jong
Kim
*a
aCenter for Self-assembly and Complexity, Institute for Basic Science and Department of Chemistry, Pohang University of Science and Technology (POSTECH) 77 Cheongam-ro, Nam-gu, Pohang 790-784, Republic of Korea. E-mail: wjkim@postech.ac.kr; Fax: +82-54-279-3399; Tel: +82-54-279-2104
bBionanotechnology Research Center, KRIBB, 125 Gwahak-ro, Yuseong-gu, Daejeon 305-806, Republic of Korea
First published on 17th February 2015
Abstract
This work demonstrates successful delivery of a gene to EGFR-overexpressed cancer cells by using a rationally designed branched GE11 peptide as a targeting ligand. In addition, we exploited the effect of the divalent structure of the branched GE11 peptide on the gene delivery and tumor targeting efficiency, compared to the monovalent GE11 peptide. The GE11 or branched GE11-tethered polymers were successfully synthesized. They are composed of a targeting peptide, disulfide crosslinked low molecular weight polyethylenimine and polyethylene glycol. Here, we evaluated the physicochemical properties, cytotoxicity and in vitro transfection efficiency and in vivo biodistribution of the GE11 and branched GE11 tethered polyplexes. Our results demonstrated that GE11 and bGE11-tethered gene delivery carriers showed efficient gene condensing ability, an enhanced transfection efficiency and targeting ability with low cytotoxicity. Interestingly, the branched GE11-tethered polymer showed the greater targeting ability to EGFR-overexpressed cancer cells in vivo than the GE11-tethered polymer. Therefore, this branched structure of targeting ligand has the potential for providing a novel strategy to design an efficient targeted delivery system.
Introduction
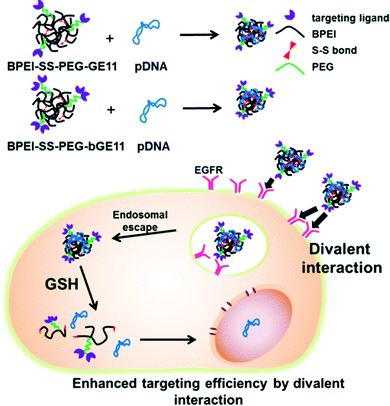
For an efficient polymeric delivery system, it is essential to precisely deliver the gene/polymer complex (polyplex) or drug into a targeted tissue or organ. To solve this issue, researchers have developed numerous targeting ligands which can bind to the specific receptor overexpressed on the target tissues or organs.1–3 Enhanced targeted delivery of payloads has been attained by using a combination of ligands and receptors; folate/folate receptor, RGD peptide/integrin αvβ5, NGR peptide/aminopeptidase N receptor and epidermal growth factor (EGF)/EGF receptor (EGFR).4–7 Among them, EGFR has been considered as one of the promising targets for gene and drug delivery systems because of its overexpression in lung, liver, head and neck cancers and a wide spectrum of human cancers from epithelial origin.8,9 Researchers have shown enhanced targeting and delivery efficiency of therapeutic genes or drugs by tethering EGF to polymers, carbon nanotubes, liposomes, and dendrimers.10–13 However, EGF itself is at risk of activating the EGFR, which leads to cell proliferation and reduces the antitumor effect of a therapeutic gene or drug.14 The GE11 peptide (YHWYGYTPQNVI), found by phage display library to be a substitute for EGF, binds to the receptor and has low mitogenic activity.15The structure of targeting ligands is an important issue in developing targeted delivery systems.16–18 For example, the cyclic form of NGR and RGD has shown better binding affinity to each respective receptor than the linear form because restriction of the structures enhances the receptor-ligand interactions by stabilizing the bent conformation of the peptide.19,20 In addition, multivalent interactions between ligand and target molecules are widely utilized in biological processes, such as in the adhesion of a virus to the surface of a cell, binding of transcription factors to multiple sites on DNA, and recognition of antigens by antibodies.21–24 These multivalent interactions are generally much stronger than monovalent interactions.21 However, a delivery system utilizing a multivalent form of targeting ligand has yet to be evaluated. Therefore, we used two types of targeting peptides which are structurally different. One is the GE11 peptide itself, and the other is branched GE11 (bGE11) which has two ligands in one peptide.
Recently, we reported an efficient gene delivery system composed of bioreducible branched polyethylenimine (BPEI-SS), polyethylene glycol (PEG) and targeting peptide.25–27 Branched polyethylenimine (BPEI), which is the representative cationic polymer for gene delivery, exhibits molecular weight (MW) dependency in transfection efficiency and cytotoxicity. It has been reported that BPEI with a high MW (>25 kDa) shows a high transfection efficiency and low biocompatibility, while BPEI with a low MW (<1.8 kDa) shows a low transfection efficiency and low cytotoxicity.28 The shortcomings of BPEI can be overcome by introducing the disulfide linkage to the low MW BPEI. BPEI-SS shows a high transfection efficiency as it is able to form a stable polyplex with pDNA in the extracellular compartments. Subsequently, BPEI-SS is degraded in reductive conditions in intracellular compartments and shows low cytotoxicity.29 In addition, the conjugation of a PEG moiety provides stability in blood and biocompatibility for the polyplex in vitro and in vivo.
In this study, we introduced two structurally different EGFR-targeting peptides to BPEI-SS-PEG and evaluated their physicochemical properties, in vitro transfection efficiency, cytotoxicity, cellular uptake efficiency and in vivo targeting efficiency (Fig. 1).
Experimental
Materials
BPEI with MW 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Da (BPEI25k), propylene sulfide and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) were purchased from Sigma-Aldrich (St. Louis, MO, USA). BPEI with MW 1200 Da (BPEI1.2k) was obtained from Polyscience, Inc. (Warrington, PA, USA). Methanol, diethyl ether and dimethyl sulfoxide (DMSO) were purchased from Samchun pure chemical (Pyeongtaek, Korea). A dialysis membrane with a molecular weight cut-off (MWCO) of 10 kDa was purchased from Spectrum Laboratories (Rancho Domingues, CA, USA). YOYO-1 iodide was obtained from Invitrogen (Eugene, OR, USA). Mounting medium for fluorescence with DAPI was purchased from VECTOR (Berlingame, CA, USA) and Cy5.5 NHS ester was obtained from GE healthcare (Little Chalfront, UK). Heterobifunctional polyethylene glycol with MW 5000 Da (α-maleimide-PEG-ω-N-hydroxysuccinimide ester polyethylene glycol, MAL-PEG-NHS) was purchased from NOF Corporation (White Plains, NY, USA). GE11 peptide and branched GE11 (bGE11) peptide were prepared by solid phase peptide synthesis (GE11 = YHWYGYTPQNVIGRC and bGE11 = (YHWYGYTPQNVI-GG)2-KGRC). Agarose powder and Tris-acetate-EDTA (TEA) buffer were purchased from bioneer Corp. (Daejeon, Korea). Dulbecco Modified Eagle's medium (DMEM) and Roswell Park Memorial Institute medium (RPMI 1640), penicillin-streptomycin, fetal bovine serum (FBS) and Dulbecco's phosphate buffered saline (DPBS) were obtained from Corning (Manassas, VA, USA). A luciferase assay system with reporter lysis buffer was purchased from Promega (Madison, WI, USA). Bradford protein assay reagent was purchased from Pierce Chemical Co. (Rockford, IL, USA). A549, Huh-7 and NIH3T3 cell lines were obtained from the Seoul National University Cell Bank (Seoul, Korea).
000 Da (BPEI25k), propylene sulfide and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) were purchased from Sigma-Aldrich (St. Louis, MO, USA). BPEI with MW 1200 Da (BPEI1.2k) was obtained from Polyscience, Inc. (Warrington, PA, USA). Methanol, diethyl ether and dimethyl sulfoxide (DMSO) were purchased from Samchun pure chemical (Pyeongtaek, Korea). A dialysis membrane with a molecular weight cut-off (MWCO) of 10 kDa was purchased from Spectrum Laboratories (Rancho Domingues, CA, USA). YOYO-1 iodide was obtained from Invitrogen (Eugene, OR, USA). Mounting medium for fluorescence with DAPI was purchased from VECTOR (Berlingame, CA, USA) and Cy5.5 NHS ester was obtained from GE healthcare (Little Chalfront, UK). Heterobifunctional polyethylene glycol with MW 5000 Da (α-maleimide-PEG-ω-N-hydroxysuccinimide ester polyethylene glycol, MAL-PEG-NHS) was purchased from NOF Corporation (White Plains, NY, USA). GE11 peptide and branched GE11 (bGE11) peptide were prepared by solid phase peptide synthesis (GE11 = YHWYGYTPQNVIGRC and bGE11 = (YHWYGYTPQNVI-GG)2-KGRC). Agarose powder and Tris-acetate-EDTA (TEA) buffer were purchased from bioneer Corp. (Daejeon, Korea). Dulbecco Modified Eagle's medium (DMEM) and Roswell Park Memorial Institute medium (RPMI 1640), penicillin-streptomycin, fetal bovine serum (FBS) and Dulbecco's phosphate buffered saline (DPBS) were obtained from Corning (Manassas, VA, USA). A luciferase assay system with reporter lysis buffer was purchased from Promega (Madison, WI, USA). Bradford protein assay reagent was purchased from Pierce Chemical Co. (Rockford, IL, USA). A549, Huh-7 and NIH3T3 cell lines were obtained from the Seoul National University Cell Bank (Seoul, Korea).
Purification of pDNA
Plasmids (pDNA) encoding luciferase were propagated in a chemically competent DH5α strain (GibcoBRL, Rockville, MD, USA) and prepared with a GeneJET Plasmid Maxiprep Kit (Thermo scientific, Waltham, MA, USA). The concentration of pDNA was determined by measuring absorbance at 260 nm, and we found the optical density at 260 to 280 nm to be in the range 1.8–1.9.Synthesis of bioreducible branched polyethylenimine (BPEI-SS)
BPEI-SS was synthesized by previously reported methods with some modification.29 Briefly, BPEI1.2k (1 g) in 20 mL vials was dissolved in 10 mL deionized water (DW) and the pH of the solution was adjusted to 7.2 by adding 1 M HCl. The solution was lyophilized for 2 days. The resultant solid was dissolved in methanol and the solution was purged with nitrogen for 15 min. Propylene sulfide (7 eq.) was added using a syringe, then the reaction mixture was stirred at 60 °C for 24 h. The product was purified by precipitation in cold diethyl ether twice, and the degree of thiolation was measured using 1H NMR in D2O (Bruker 300 MHz). To synthesize BPEI-SS, BPEI-SH was dissolved in DMSO and oxidative crosslinking was performed by stirring the solution at room temperature for 48 h. Finally, the product was obtained by dialysis (MWCO 10 kDa) against DW and following lyophilization.Synthesis of peptide-tethered bioreducible BPEI (BPEI-SS-PEG-GE11 and BPEI-SS-PEG-bGE11)
MAL-PEG-NHS (1 eq.) and GE11 or bGE11 peptides (1.2 eq. to maleimide groups) were dissolved in anhydrous DMSO and stirred at room temperature for 4 h. Next, BPEI-SS (19 eq. to NHS group) dissolved in DMSO was added to the solution. The reaction mixture was stirred at room temperature for 48 h. Finally, the product was obtained by dialysis (MWCO 10 kDa) against DW and following lyophilization. The chemical structure of BPEI-SS-PEG-GE11 (or bGE11) was analyzed by 1H NMR in D2O. The conjugation ratio of peptide to polymer was determined by fluorescence spectrometry (Shimadzu, Japan). Free peptide was used to draw a calibration curve for fluorescence intensity at known concentration. As a control, PEG-conjugated BPEI-SS (BPEI-SS-PEG) without targeting peptide was synthesized by using para-nitrophenylchloroformate (pNPC)-activated methoxy PEG.Preparation of polyplex
Polyplexes (polymer/pDNA complex) with 1 ≤ N/P ≤ 15, (where N is the molar amount of nitrogen in the polycation and P is the molar amount of phosphate in the pDNA) were prepared by adding polymer solution to the pDNA solution in PBS buffer. The polyplexes were incubated at room temperature for 30 min.Agarose gel retardation assay
Polyplexes with various N/P ratios were loaded onto a 1% (w/v) agarose gel containing ethidium bromide (EtBr, 0.5 μg mL−1) with a 6× loading dye. Electrophoresis was conducted at a constant voltage of 100 V for 20 min in 0.5× TAE buffer (Tris-acetate-EDTA). Naked pDNA (200 ng in 10 μL) was used as a control. The gel was analyzed on a UV illuminator to observe the position of the complexed pDNA relative to that of the naked pDNA.Size and zeta potential measurements
The polyplexes were prepared at various N/P ratios and diluted using PBS buffer (pH 7.4, 140 mM NaCl). The final pDNA concentration was adjusted to 3 μg mL−1. The particle size and zeta potential of each polyplex were measured by using a Zetasizer Nano S90 and Z (Malvern Instruments, Malvern, U.K.), respectively.Cell culture
The EGFR-overexpressed (EGFR-positive) adenocarcinomic human alveolar basal epithelial cell line (A549) and human hepato cellular carcinoma cell line (Huh-7) were cultured in RPMI164 medium and the EGFR-negative mouse embryo fibroblast (NIH3T3) cell line was cultured in DMEM medium. Both mediums contain 10% FBS and 1% antibiotics and the cells were grown and maintained in a humidified atmosphere with 5% CO2 at 37 °C.MTT assay
The cytotoxicity of the polyplexes was evaluated by the standard MTT assay protocol. Briefly, cells were seeded onto 96-well plates at a density of 5 × 103 cells per well and incubated for 24 h. pDNA (0.1 μg μL−1) was complexed with the polymer at predetermined N/P ratios in PBS. Cells were incubated with polyplex in 100 μL of serum free media for 4 h, then incubated for 20 h in 200 μL of fresh media containing 10% FBS. Next, the media was replaced with 200 μL of fresh media and 20 μL of 5 mg mL−1 MTT solution was added. After 4 h, the media was removed and 200 μL of DMSO was added to each well to dissolve the internalized purple formazan crystals. An aliquot of 100 μL was taken from each well and transferred to a fresh 96-well plate. The absorption was measured at 570 nm using a microplate spectrofluorometer (VICTOR3 V Multilabel Counter, Perkin-Elmer-Wellesley, MA, USA). The control cells which were not exposed to the transfection system were used to represent 100% cell viability. The results are presented as a mean and standard deviation (n = 3).Luciferase reporter gene assay
Cells were seeded on 24-well plates at an initial density of 6 × 104 cells per well and were incubated for 24 h in 500 μL of media containing 10% FBS at 37 °C in a humidified atmosphere with 5% CO2. pDNA (0.1 μg μL−1) was complexed with the polymer at predetermined N/P ratios in PBS. The cells were incubated with the polyplex in 250 μL of serum-free media for 4 h, then incubated for 20 h in 500 μL of media containing 10% FBS. The cells were washed twice with 500 μL of PBS and lysed by adding 200 μL of lysis buffer. Luciferase gene expression was measured by using a microplate spcetrofluorometer and a luciferase assay system. The results are presented as a mean and standard deviation (n = 3).Flow cytometry
Cells were seeded in 12-well plates at a density of 1 × 105 cells per well and incubated for 24 h. pDNA was labeled with YOYO-1 iodide for 12 h before use. YOYO-labeled pDNA (0.1 μg μL−1) was complexed with the polymer at a N/P ratio of 10 in PBS buffer and incubated for 30 min. The cells were incubated with polyplex for 4 h in serum-free media. After incubation, the cells were washed twice with PBS and then trypsinized. The harvested cells were centrifuged and the supernatant was removed, then the cells were fixed with 4% paraformaldehyde solution at 4 °C overnight. The paraformaldehyde solution was removed by centrifugation, then the cells were resuspended in PBS. The cells were analyzed using a FACS Caliber (Becton Dickinson, San Jose, CA, USA) and Becton Dickinson Cell Quest Software following the manufacturers’ protocol.In vivo imaging
The POSTECH Biotech Center Ethics Committee approved all of the animal experiments in this study. In vivo near-infrared (NIR) imaging was performed using an IVIS spectrum small-animal in vivo imaging system (Caliper Life Science) located at the Pohang Center of Evaluation of Biomaterials (Pohang Technopark). We evaluated the fluorescence intensity of the polyplexes labeled with Cy5.5 at an N/P ratio of 10 by using a Cy5.5 (ex = 675 nm, em = 740 nm) filter set. To investigate the targeting efficiency of GE11- and bGE11-conjugated and non-conjugated polyplexes, A549 cells were injected subcutaneously into the right flank of female BALB/c-nu/nu mice. After 4 weeks, the Cy5.5 labeled polyplexes (N/P = 20, 10 μg of pDNA) were injected into mice via the tail vein. The mice were anesthetized with 2–3% isoflurane and placed into the IVIS Spectrum system. Images were taken 4 h after injection. 24 h after injection the A549 xenografted mice were sacrificed for ex vivo imaging, and their major organs were collected. Each organ was rinsed with PBS three times, mounted on a board, and fluorescence images were taken using the IVIS Spectrum system.Results and discussion
Synthesis and characterization of bioreducible polymers with targeting ligands
Prior to synthesizing the bioreducible BPEI with targeting ligands, we firstly synthesized thiolated BPEI (BPEI-SH) by using propylene sulfide as a thiolation agent at 7 equiv. to low MW BPEI1.2k (Fig. S1†). Thiolation of BPEI was conducted by the ring opening reaction between propylene sulfide and a primary amine or secondary amine of BPEI. It was reported that during the thiolation reaction, the buffering capacity of BPEI was not changed significantly because primary and secondary amine groups are converted to higher order amines.29 Therefore, the endosomal escaping ability of BPEI is retained after thiolation. After purification of BPEI-SH by precipitation in diethyl ether, a sticky yellow product was obtained. The degree of thiolation was determined using 1H NMR (Fig. S2†) and the conjugation ratio was calculated as 5.32 (Table 1). The disulfide crosslinked BPEI (BPEI-SS) was synthesized through the oxidization of BPEI-SH with DMSO. After purification by dialysis (MWCO = 10 kDa) against deionized water (DW) and lyophilization by freezing drying, we could acquire a yellow solid, the BPEI-SS. The MW of BPEI1.2k and BPEI-SS was evaluated using gel permeation chromatography (GPC). By confirming that peaks of BPEI-SS appeared at earlier retention times compared to native BPEI1.2k, we could conclude that the synthesized BPEI-SS has a higher average MW than BPEI1.2k (Fig. S3†).| Denotation | Thiola | PEGb | Peptidec |
|---|---|---|---|
| a Measured by 1H NMR. Molar ratio of thiol groups to BPEI1.2k chain in polymers. b Confirmed by 1H NMR. Molar ratio of PEG to BPEI1.2k. c Confirmed by fluorescence spectrometry. Molar ratio of GE11 or bGE11 peptide to BPEI1.2k. | |||
| BPEI-SS-PEG | 5.32 | 0.16 | — |
| BPEI-SS-PEG-GE11 | 0.12 | 0.26 | |
| BPEI-SS-PEG-bGE11 | 0.12 | 0.11 | |
In order to introduce GE11 and bGE11 peptides to BPEI-SS, each type of GE11 peptide was firstly conjugated to the heterobifunctional polyethylene glycol (MAL-PEG-NHS). In each peptide, there is a free thiol group in a cysteine residue at the end of the peptide sequence. This free thiol group can react with maleimide of MAL-PEG-NHS by Michael reaction. BPEI-SS-PEG-GE11 (or bGE11) was synthesized via the coupling reaction between NHS in the resultant peptide-PEG-NHS and the primary amine in BPEI-SS. After dialysis (MWCO = 10 kDa) against DW and lyophilization, a slightly yellow sponge-like powder was obtained. Successful conjugation between BPEI and PEG was confirmed by 1H NMR (Fig. S2†). In the 1H NMR spectra, protons in BPEI and the PEG main chains were found at δ 2.6–3.1 and δ 3.6. Using the relative integral values of the respective peaks in the 1H NMR spectra, it was confirmed that the molar ratio of PEG to BPEI was 0.16, 0.12 and 0.12 for BPEI-SS-PEG, BPEI-SS-PEG-GE11 and BPEI-SS-PEG-bGE11, respectively. Each conjugated peptide was quantified by measuring the fluorescence spectrometry. In the sequence of the peptides there are several aromatic amino acids such as tryptophan and tyrosine which can emit intrinsic fluorescence (excitation: 280 nm, emission: 350 nm).30 Comparing the measured fluorescence intensity of GE11 or bGE11-tethered polymers with a calibration curve established by a predetermined concentration of free peptide (Fig. S4†), the molar ratio of peptide to BPEI1.2k was estimated to be 0.26 and 0.11 for BPEI-SS-PEG-GE11 and BPEI-SS-PEG-bGE11, respectively.
Physicochemical properties of polyplex
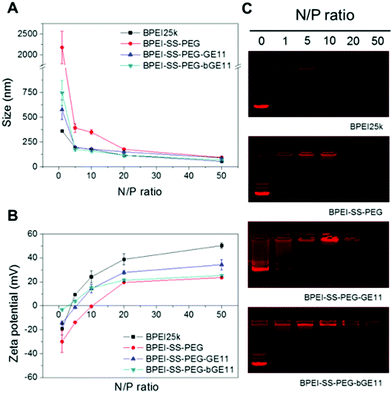
Cationic polymers can interact with negatively charged plasmid DNA (pDNA) through electrostatic interactions. The resultant polyplex protects the gene from enzymes in the blood flow as well as facilitates the cellular uptake of the gene by condensing the pDNA with micro-size to the nano scale. Therefore for successful gene delivery it is essential to demonstrate whether or not the synthesized polymer can condense genes effectively. After preparation of each polyplex at the various N/P ratios, the pDNA condensing ability of the polymers was evaluated by performing an agarose gel retardation assay. BPEI25k was used as a positive control. As shown in Fig. 2A, while BPEI25k could retard pDNA completely at N/P > 1, BPEI-SS-PEG and BPEI-SS-PEG-GE11 (or bGE11) could retard pDNA completely at N/P > 10 and N/P > 5, respectively. The lower pDNA condensing ability of PEG-decorated polymers compared with BPEI25k could be attributed to the hindrance of the PEG chain to the interaction between BPEI and the pDNA.31 These results indicate that BPEI-SS-PEG-GE11 (or bGE11) could successfully form the complex with pDNA.The particle size and surface charge of the polyplexes also affect the uptake and transfection in vitro and in vivo. To measure the size of the nanoparticles, polyplexes with various N/P ratios were analyzed by using dynamic light scattering (DLS). As shown in Fig. 2A, the particle size of all polyplexes decreased as the N/P ratios increased because of the high electrostatic interaction caused by the increased cationic charge. The size of BPEI-SS-PEG/pDNA and BPEI-SS-PEG-GE11/pDNA (or bGE11) changed to around 200 nm at a N/P ratio of 20 and 5, respectively, while these are above 2000 nm and 500 nm at a N/P ratio of 1. These results indicate that all polymers at each proper N/P ratio could form the condensed nano-complex with pDNA.
Positively charged polyplexes have the ability to adhere to the negatively charged plasma membrane of cells through electrostatic interactions. Therefore, it has been reported that positively charged polyplexes could be internalized by the cell effectively.32 As shown in Fig. 2B, the zeta potential increased with the increase in the N/P ratio. In the case of BPEI-SS-PEG, the polyplex showed a negative zeta potential at N/P < 10, while it was positive at a N/P ratio of 20. In addition, BPEI-SS-PEG-GE11/pDNA (or bGE11) at a N/P ratio of 5 showed a slightly positive zeta potential (±5 mV).
In summary, the results of the agarose gel retardation, DLS and zeta potential measurements are correlated with each other and demonstrate that the BPEI-SS-PEG-GE11 (or bGE11) polyplex has suitable properties for in vitro and in vivo gene delivery.
Cytotoxicity
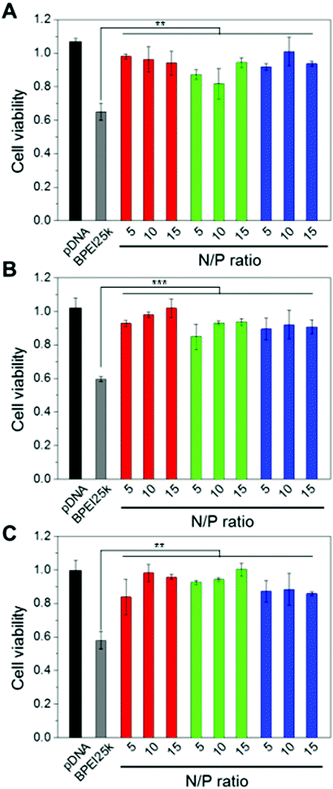
It is well-known that BPEI with a high MW displays toxicity to cells because the positively charged BPEI destabilizes the plasma membrane or interacts with cellular components, especially the membrane of mitochondria.33 To surmount these problems, a disulfide linkage and PEG chains were introduced to the low MW form of BPEI. The cytotoxicity of BPEI-SS-PEG-GE11 (or bGE11) polyplexes was evaluated by MTT assay using EGFR-positive A549 and Huh-7 cells and the EGFR-negative NIH3T3 cell line (Fig. 3). Naked pDNA and BPEI25k polyplexes at a N/P ratio of 10 as a gold standard were used as the control. The BPEI25k polyplex showed significant toxicity to all cell lines. However, no significant cytotoxicity was observed for pDNA, BPEI-SS-PEG, BPEI-SS-PEG-GE11 and BPEI-SS-PEG-bGE11 polyplexes regardless of the EGFR receptor expression profile of the cells. These results could be attributed to the degradation ability of BPEI and the outside exposure of PEG. BPEI-SS, which is dissociated from the polyplex, may be degraded into low MW BPEI with low cytotoxicity in the reductive conditions of the cytoplasm. In addition, BPEI and pDNA probably occupy the inner part of the polyplex and the PEG chain may be located at the outer part of the polyplex. Therefore, the PEG could hamper the interaction between positively-charged BPEI and the negatively-charged plasma membrane. These results confirmed the biocompatibility of the GE11- and bGE11-conjugated polymers.In vitro transfection efficiency
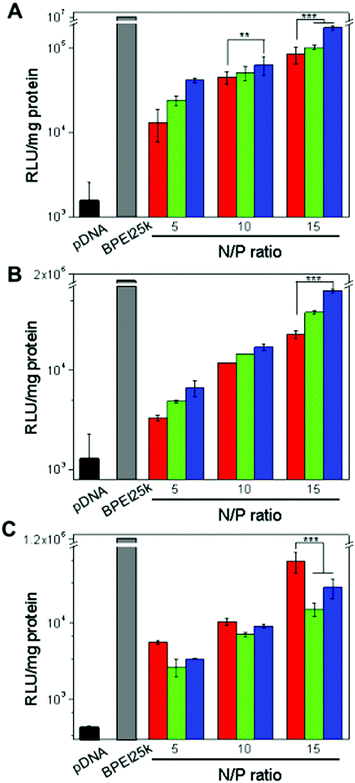
The in vitro gene transfection potential of the EGFR-targeted bioreducible polymers was investigated in EGFR-positive A549 and Huh-7 cell lines and the EGFR-negative NIH3T3 cell line (Fig. 4). Cells treated with naked pDNA and BPEI-SS-PEG polyplexes were used as controls. The pDNA showed a negligible transfection efficiency because cells do not internalize it with ease. BPEI25k exhibited the most effective transfection efficiency with high cytotoxicity owing to the high density of positive charge (Fig. 3 and 4). As the N/P ratio increased, the transfection efficiency of the polymers increased for all cell lines. In the EGFR-positive A549 and Huh-7 cell lines, GE11 and bGE11 tethered BPEI-SS polyplexes showed enhanced transfection efficiency compared to the non-targeted BPEI-SS-PEG polyplex at all N/P ratios. However, in the EGFR-negative NIH3T3 cell line, the non-targeted BPEI-SS-PEG polyplex showed a much higher transfection efficiency than the GE11 or bGE11 tethered BPEI-SS polyplexes. These results demonstrated that GE11- or bGE11-conjugated gene carriers have a higher affinity to EGFR-overexpressed cells than normal cells. Interestingly, BPEI-SS-PEG-bGE11 which has a branched peptide could deliver pDNA effectively to the target cells compared to BPEI-SS-PEG-GE11. These results can be explained by the hypothesis of EGFR dimerization.34 It has been supposed that proper EGFR dimerization seems to be necessary for EGFR internalization. Applying this hypothesis, the branched structure of bGE11 having two adjacent ligands can form a divalent interaction with EGFR, which facilitates the enhanced dimerization of the EGFR and receptor-mediated endocytosis, compared to GE11. Although the dimerization of EGFR was not directly confirmed, it could be concluded that the bGE11 tethered polyplex has a high transfection efficiency to EGFR-overexpressed cancer cells.Investigation of the interaction between polyplex and plasma membrane
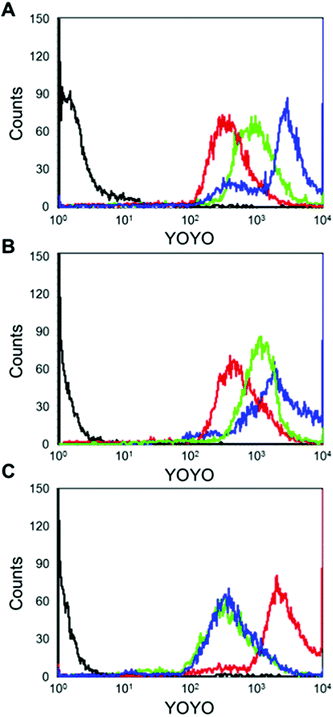
We hypothesized that the bGE11 tethered polyplex has more chance to interact with EGFR, enhancing trafficking of the polyplex into the cell. In order to confirm the strong interaction between the bGE11 tethered polyplexes and EGFR, we carried out fluorescence-activated cell sorting analysis (FACS) (Fig. 5). pDNA was labeled with YOYO-1 iodide before complexation with the polymers. BPEI-SS-PEG, BPEI-SS-PEG-GE11 and BPEI-SS-PEG-bGE11 polyplexes at a N/P ratio of 10 were transfected into EGFR-positive A549 and Huh-7 cell lines and the EGFR-negative NIH3T3 cell line. Non-treated cells were used as controls. In EGFR-positive A549 and Huh-7 cell lines, the GE11 and bGE11 tethered BPEI-SS polyplexes showed an enhanced interaction with the plasma membrane compared to the non-targeted polyplex due to the interaction between overexpressed EGFR and the peptides. Especially, the BPEI-SS-PEG-bGE11 polyplex showed a higher interaction with EGFR-positive cells than the BPEI-SS-PEG-GE11 polyplex. However, in the EGFR-negative NIH3T3 cell line, non-targeted BPEI-SS-PEG polyplex demonstrated a higher cellular uptake than the BPEI-SS-PEG-GE11 (or bGE11) polyplex. These results suggested that GE11 and bGE11 peptides enhance the cellular uptake of polyplexes by receptor-mediated endocytosis and the higher affinity of bGE11 for EGFR facilitates the higher transfection efficiency compared to the GE11 tethered polyplex.In vivo fluorescence bio-imaging and ex vivo biodistribution
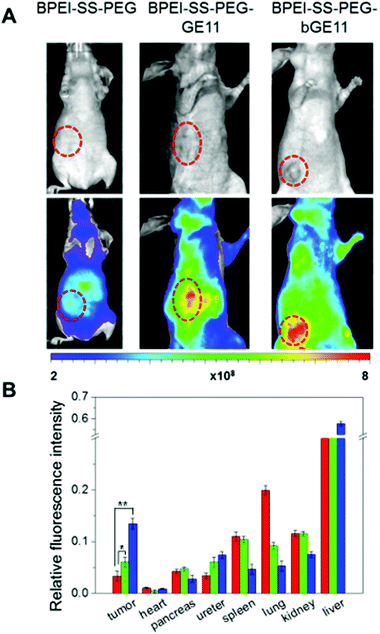
Although the targeting efficiency of the GE11 and bGE11 peptides tethered to BPEI-SS was successfully investigated in vitro, these results cannot guarantee the effective clinical efficacy of the developed polyplexes. Therefore, confirming the targeting ability of the polyplexes in vivo is a prerequisite for further clinical application. For in vivo fluorescence imaging, the polymers were labeled with Cy5.5 before complexation with pDNA. We xenografted EGFR-positive A549 cells to female nude mice. In vivo experiment was performed when the tumor had become an appropriate size (∼100 mm3) for imaging. BPEI-SS-PEG, BPEI-SS-PEG-GE11 and BPEI-SS-PEG-bGE11 polyplexes were injected into the tail vein of each nude mouse. 4 h after injection in vivo fluorescence images were acquired (Fig. 6A). The non-targeted BPEI-SS-PEG polyplex demonstrated a low fluorescence signal (yellow) in the tumor. However, the BPEI-SS-PEG-GE11 and BPEI-SS-PEG-bGE11 polyplexes showed an enhanced fluorescence signal at the tumor site compared to the BPEI-SS-PEG/pDNA polyplex. Especially, the BPEI-SS-PEG-bGE11 polyplex (red) showed the highest accumulation at the tumor site, which is consistent with the in vitro transfection and FACS results.In this gene delivery system, it is expected that the polyplexes should be mainly delivered into the target tumor without non-specific delivery to the normal organs including the spleen, lung and kidney, etc. Therefore, the biodistribution of the polyplexes was evaluated ex vivo to confirm the targeting ability (Fig. 6B). After 24 h post-injection, all mice were sacrificed and the fluorescence intensity of the major organs was measured. In the case of BPEI-SS-PEG, the fluorescence intensity of each organ was observed in the order of liver, lung, kidney, spleen and tumor. In the case of BPEI-SS-PEG-GE11, the polyplex accumulation was confirmed in the order of liver, kidney, spleen, lung and tumor. In the case of BPEI-SS-PEG-bGE11, polyplexes were present in organs in the order of liver, tumor, kidney, lung and spleen. As expected, the BPEI-SS-PEG-bGE11/pDNA polyplex showed the highest accumulation of polyplex at tumor sites. These results clearly indicate that the bGE11-conjugated polyplex can deliver a gene to the targeted tumor effectively and reduce non-specific delivery to the normal organs, showing the potential for use in treatments of EGFR-overexpressed cancer.
Conclusions
In this study, we have developed an EGFR targeted bioreducible BPEI for efficient targeted gene delivery. EGFR targeted BPEI-SS showed efficient gene complexation ability and a suitable size and zeta potential for gene delivery in vitro and in vivo. Compared to non-targeted polymers, GE11- or bGE11-conjugated BPEI-SS showed enhanced transfection efficiency in vitro and tumor-targeted biodistribution in vivo. In addition, the bGE11-conjugated polymers showed higher targeting ability than the GE11-conjugated polymers due to the multivalent interaction between ligand and receptor. Taken together, our results suggest that branched structure of the targeting ligand will be one of strategies used to design efficient targeted delivery systems.Acknowledgements
This work was supported by the Research Center Program of IBS (Institute for Basic Science) in Korea (CA1203-02) and Converging Research Center Program (2012K001474) through an NRF grant funded by the Korean government (MEST).Notes and references
- S. D. Li and L. Huang, Gene Ther., 2006, 13, 1313 CrossRef CAS PubMed.
- J. Sudimack and R. J. Lee, Adv. Drug Delivery Rev., 2000, 41, 147 CrossRef CAS.
- F. Danhier, O. Feron and V. Préat, J. Controlled Release, 2010, 148, 135 CrossRef CAS PubMed.
- A. C. Antony, Annu. Rev. Nutr., 1996, 16, 501 CrossRef CAS PubMed.
- E. Ruoslahti and M. D. Pierschbacher, Science, 1987, 238, 491 CAS.
- R. Pasqualini, E. Koivunen, R. Kain, J. Lahdenranta, M. Sakamoto, A. Stryhn, R. A. Ashmun, L. H. Shapiro, W. Arap and E. Ruoslahti, Cancer Res., 2000, 60, 722 CAS.
- J. Mendelsohn and J. Baselga, Oncogene, 2000, 19, 6550 CrossRef CAS PubMed.
- C. Yewale, D. Baradia, I. Vhora, S. Patil and A. Misra, Biomaterials, 2013, 34, 8690 CrossRef CAS PubMed.
- R. S. Herbst, Int. J. Radiat. Oncol., Biol., Phys., 2004, 59, S21 CrossRef PubMed.
- A. A. Bhirde, V. Patel, J. Gavard, G. Zhang, A. A. Sousa, A. Masedunskas, R. D. Leapman, R. Weigert, J. S. Gutkind and J. F. Rusling, ACS Nano, 2009, 3, 307 CrossRef CAS PubMed.
- E. Bohl Kullberg, N. Bergstrand, J. Carlsson, K. Edwards, M. Johnsson, S. Sjöberg and L. Gedda, Bioconjugate Chem., 2002, 13, 737 CrossRef PubMed.
- J. Capala, R. F. Barth, M. Bendayan, M. Lauzon, D. M. Adams, A. H. Soloway, R. A. Fenstermaker and J. Carlsson, Bioconjugate Chem., 1996, 7, 7 CrossRef CAS PubMed.
- H. Lee, T. H. Kim and T. G. Park, J. Controlled Release, 2002, 83, 109 CrossRef CAS.
- K. Oda, Y. Matsuoka, A. Funahashi and H. Kitano, Mol. Syst. Biol., 2005, 1, 2005 CrossRef PubMed.
- Z. Li, R. Zhao, X. Wu, Y. Sun, M. Yao, J. Li, Y. Xu and J. Gu, FASEB J., 2005, 19, 1978 CrossRef CAS PubMed.
- Y. Guo, H. Feinberg, E. Conroy, D. A. Mitchell, R. Alvarez, O. Blixt, M. E. Taylor, W. I. Weis and K. Drickamer, Nat. Struct. Mol. Biol., 2004, 11, 591 CAS.
- R. C. Ladner, Trends Biotechnol., 1995, 13, 426 CrossRef CAS.
- Á. Roxin and G. Zheng, Future Med. Chem., 2012, 4, 1601 CrossRef PubMed.
- (a) G. Colombo, F. Curnis, G. M. S. De Mori, A. Gasparri, C. Longoni, A. Sacchi, R. Longhi and A. Corti, J. Biol. Chem., 2002, 277, 47891 CrossRef CAS PubMed; (b) E. Koivunen, B. Wang and E. Ruoslahti, Nat. Biotechnol., 1995, 13, 265 CrossRef CAS PubMed.
- S. Verrier, S. Pallu, R. Bareille, A. Jonczyk, J. Meyer, M. Dard and J. Amédée, Biomaterials, 2002, 23, 585 CrossRef CAS.
- M. Mammen, S.-K. Choi and G. M. Whitesides, Angew. Chem., Int. Ed., 1998, 37, 2754 CrossRef.
- W. J. Lees, A. Spaltenstein, J. E. Kingery-Wood and G. M. Whitesides, J. Med. Chem., 1994, 37, 3419 CrossRef CAS.
- H. Chen and M. L. Privalsky, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 422 CrossRef CAS.
- J. M. Davie and W. E. Paul, J. Exp. Med., 1972, 135, 643 CrossRef CAS.
- S. Son, K. Singha and W. J. Kim, Biomaterials, 2010, 31, 6344 CrossRef CAS PubMed.
- S. Son, D. W. Hwang, K. Singha, J. H. Jeong, T. G. Park, D. S. Lee and W. J. Kim, J. Controlled Release, 2011, 155, 18 CrossRef CAS PubMed.
- D. W. Hwang, S. Son, J. Jang, H. Youn, S. Lee, D. Lee, Y.-S. Lee, J. M. Jeong, W. J. Kim and D. S. Lee, Biomaterials, 2011, 32, 4968 CrossRef CAS PubMed.
- D. Fischer, T. Bieber, Y. Li, H.-P. Elsässer and T. Kissel, Pharm. Res., 1999, 16, 1273 CrossRef CAS.
- Q. Peng, Z. Zhong and R. Zhuo, Bioconjugate Chem., 2008, 19, 499 CrossRef CAS PubMed.
- F. W. J. Teale and G. Weber, Biochem. J., 1957, 65, 476 CAS.
- J. H. Choi, J. S. Choi, H. Suh and J. S. Park, Bull. Korean Chem. Soc., 2001, 22, 46 CrossRef CAS.
- Z.-G. Yue, W. Wei, P.-P. Lv, H. Yue, L.-Y. Wang, Z.-G. Su and G.-H. Ma, Biomacromolecules, 2011, 12, 2440 CrossRef CAS PubMed.
- S. M. Moghimi, P. Symonds, J. C. Murray, A. C. Hunter, G. Debska and A. Szewczyk, Mol. Ther., 2005, 11, 990 CrossRef CAS PubMed.
- Q. Wang, G. Villeneuve and Z. Wang, EMBO Rep., 2005, 6, 942 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Synthetic scheme and figures of 1H NMR, GPC of delivery carriers and calibration curve of each peptide. See DOI: 10.1039/c5bm00004a |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2015 |