Molecular beacon-based NAND logic gate for sensing triplex DNA binders†
Kai-Cheng Lina,
Chia-Yin Kuob,
Chih-Chun Niehb and
Wei-Lung Tseng*bc
aDepartment of Emergency Medicine, Division of Traumatology, Kaohsiung Veterans General Hospital, Taiwan
bDepartment of Chemistry, National Sun Yat-sen University, Kaohsiung, Taiwan. E-mail: tsengwl@mail.nsysu.edu.tw; Fax: +886-7-3684046
cSchool of Pharmacy, College of Pharmacy, Kaohsiung Medical University, Taiwan. E-mail: tsengwl@mail.nsysu.edu.tw; Fax: +886-7-3684046
First published on 6th August 2014
Abstract
This study developed a thymine-based molecular beacon (MB), which can be used for screening triplex DNA binders and quantifying coralyne and sanguinarine. Triplex DNA binder triggered the formation of triple-stranded DNA between MB and 7-repeat adenosine (A7), switching off the fluorescence of MB. Based on the use of triplex DNA binder and A7 as inputs and the fluorescence change of MB as the output, the NAND gate was constructed to discriminate triplex DNA binders from duplex DNA binders.
Developing molecular beacon (MB)-related techniques for highly sensitive DNA detection has been of great interest to many researchers because of their advantages of simplicity, rapidity, specificity, and sensitivity. An MB is a single-strand oligonucleotide that consists of a stem-loop structure, a reported fluorophore, and a non-fluorescent quencher/another fluorophore at its 5′- and 3′-ends.1 Watson–Crick hydrogen bonding forms a stem by joining 5–7 nucleotides at the 5′-end and 5–7 nucleotides complementing nucleotides at the 3′-end. The resultant stem reduces the fluorescence of a reported fluorophore through fluorescence resonance energy transfer, collisional quenching, and contact quenching between the two organic dyes.2 When the central loop hybridizes with the complementary target DNA or RNA, MB can change from a stem-loop to an open-chain form. This conformational change subsequently restores the fluorescence of the reported fluorophore. In addition to single-stranded DNA or RNA, the MBs were shown to be useful for detecting a wide variety of analytes, including Hg2+,3 Ag+,4 coralyne,5 heparin,5 thiol-containing molecules,6 adenosine triphosphate,7 and thrombin.8 However, no study has reported the use of MBs for screening triplex DNA binders.
Triplex DNA binders, such as benzo[e]pyrido indole derivatives and coralyne, can stabilize triple-stranded DNA, which is implicated by the modulation of gene expression and DNA transcription.9 Typical methods for screening triplex DNA binders have included competitive dialysis,10 mass spectroscopy,11 gold nanoparticle-based sensors,12 gold nanoparticle-based microarrays,13 and UV-vis melting experiments.14 However, most of these methods have high equipment cost, a complex synthesis procedure, time-consuming sample preparation, and/or low-throughput screening.
To overcome these problems, this study developed a rapid, convenient, sensitive and selective MB probe for probing triplex DNA binders. Grossmann et al. reported that a triplex-based molecular beacon, consisting of a stem of a pair of polythymine and polyadenine peptide nucleic acid, was selective and sensitive for detecting target DNA.15 The triplex stem region was formed through the binding of two polythymine to polyadenine peptide nucleic acid. Inspired by this result, Scheme 1 illustrates the use of a DNA MB probe (T8-MB-T8) for screening triplex DNA binders. The T8-MB-T8 probe contained a 15-mer loop, a stem of a pair of 8-mer T bases, a reporter of carboxyfluorescein (FAM) at the 5′-end, and a quencher of 4-([4-(dimethylamino)phenyl]azo)-benzoic acid (DABCYL) at the 3′-end. Without a triplex DNA binder, a mixture of T8-MB-T8 and 7-repeat adenosine (A7) exhibits weak fluorescence because FAM is separated far from DABCYL. A7 could only hybridize to a stem of seven T bases in MB, because a triple-stranded DNA complex is considerably less stable than that of double-stranded DNA.16 The presence of a triplex DNA binder converts the formed duplex DNA to triplex DNA.17 The random-coil structure of T8-MB-T8 is transformed into a hairpin structure because of the formation of triplex DNA in the MB stem, bringing terminal labels into close proximity. Consequently, collisional quenching of fluorescence occurs between FAM and DABCYL.
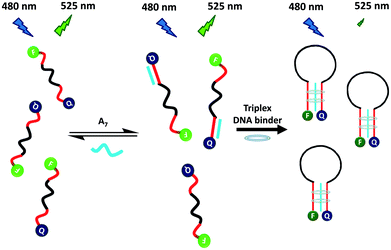 | ||
| Scheme 1 Turn-off fluorescence for the detection of a triplex DNA binder based on triplex DNA binder-induced conformation change of a random-coil MB to a hairpin-shaped MB in the presence of A7. | ||
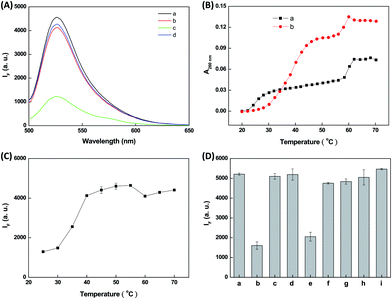
To test our hypothesis, we initially monitored the fluorescence spectra of a solution containing 10 nM T8-MB-T8, 10 mM HEPES (pH 7.0), and 300 mM NaCl in the absence and presence of 1 μM coralyne and 100 nM A7. The MB probe was incubated with a mixture of A7 and coralyne at ambient temperature for 10 min. Coralyne, a synthetic protoberberine alkaloid, has been reported as a stabilizer of triple-stranded DNA.12,13,17 Compared with the fluorescence spectrum of T8-MB-T8 (curve a in Fig. 1A), the presence of A7 induced a slight decrease in the fluorescence of T8-MB-T8 (curve b in Fig. 1A). This result indicates the formation of small amounts of triple-stranded DNA when incubating T8-MB-T8 with A7. This is likely because the triplex DNA structure is unstable at room temperature.12 Similarly, in the absence of A7, the addition of coralyne to a solution of T8-MB-T8 cannot cause the quenching of FAM fluorescence (curve c in Fig. 1A). When coralyne was present in a solution containing T8-MB-T8 and A7, more than a 70% quenching of FAM fluorescence signified the formation of stable triple-stranded DNA (curve d in Fig. 1A). To confirm coralyne-induced stabilization of triple-stranded DNA, the 260 nm UV absorbance of a solution containing T8-MB-T8, A7, and coralyne was monitored as a function of temperature. Tm1 and Tm2 represent the melting points of the dissociation of triple-stranded DNA (the complex of 2 × T8 and A7) into double-stranded DNA (the complex of T8 and A7) and the dissociation of the resultant double-stranded DNA into single-stranded DNA, respectively. When T8-MB-T8 and A7 were present in an aqueous solution, the values of Tm1 and Tm2 were determined to be 25 °C and 60 °C, respectively (curve a in Fig. 1B). Upon the addition of coralyne, the value of Tm1 increased to 40 °C and the value of Tm2 remained almost constant (curve b in Fig. 1B). Xue et al. reported a similar UV melting profile of triple-stranded DNA in the presence of the triplex DNA binder, neomycin.18 Because double-stranded DNA is more stable than triple-stranded DNA, the melting point of double-stranded DNA is higher than that of triple-stranded DNA. Similarly, Lytton-Jean et al. reported that the melting points of triple- and double-stranded DNA were 17.1 and 60.1 °C in the presence of coralyne.13 These results signified that coralyne efficiently stabilized triple-stranded DNA and exerted little effect on double-stranded DNA. We also monitored fluorescence intensity at 520 nm of a solution containing T8-MB-T8, A7, and coralyne as a function of temperature. The melting point representing the triplex to duplex transition was observed to be approximately 45 °C (Fig. 1C), which was in agreement with the value obtained from the monitoring of the UV melting profile of T8-MB-T8·A7 in the presence of coralyne.
We consequently investigated the effects of the polyadenosine length, the A7 concentration, the NaCl concentration, and the solution pH on the stability of the complexes of the T8-MB-T8·A7 and coralyne. Fig. S1 (ESI†) shows that the fluorescence intensity at 520 nm of the T8-MB-T8·An–coralyne complexes were nearly unchanged after increasing the polyadenosine length. Because a previous study demonstrated that coralyne was capable of disproportionating the duplex DNA into triplex DNA,17 we rule out that polyadenosine forms a self-structure with coralyne with an increase in the polyadenosine length. As exhibited in Fig. S2 (ESI†), an increase in the concentration of A7 caused a progressive reduction in the fluorescence intensity at 520 nm of the T8-MB-T8·A7–coralyne complexes, indicating that the stability of triple-stranded DNA relied on the concentration of A7. When the concentration of NaCl varied from 100 to 400 mM NaCl, the fluorescence at 520 nm of the T8-MB-T8·A7–coralyne complexes progressively decreased (Fig. S3, ESI†). Obviously, the binding affinity of the T8-MB-T8·A7 to coralyne increased upon increasing the NaCl concentration. Similarly, Lytton-Jean et al. reported that the stability of the triplex DNA–coralyne complexes increased upon increasing the NaCl concentration.13 Because the fluorescence intensity of FAM is highly dependent on the solution pH,19 we monitored the fluorescence intensity of the T8-MB-T8·A7 in the absence and presence of coralyne. Fig. S4 (ESI†) shows that the maximum difference in fluorescence intensity between T8-MB-T8·A7 and the T8-MB-T8·A7 complexes was observed at pH 7.0, signifying that the T8-MB-T8·A7–coralyne complexes are stable at a neutral pH.
To further demonstrate the feasibility of the proposed method and to rule out fluorescence quenching of the T8-MB-T8 probe through duplex formation, we chose several known duplex DNA binders, including ethidium bromide, 4′,6-diamidino-2-phenylindole (DAPI), anthraquinone-2-carboxylic acid (AQ2A), and 9-aminoacridine, and triplex binders, including berberine, palmatine, sanguinarine, and coralyne. The binding of the chosen duplex binders to DNA is classified as groove binders (DAPI) and intercalators (ethidium bromide, AQ2A, and 9-aminoacridine). Coralyne analogues, including berberine, palmatine, and sanguinarine, were also tested in this study. The incubation time was fixed at 10 min. Fig. 1D shows that duplex DNA binder-induced fluorescence quenching of the T8-MB-T8·A7 probe was not observed, whereas only coralyne and sanguinarine, triplex DNA binders, can trigger fluorescence quenching of the T8-MB-T8·A7–coralyne complexes through triplex stabilization. Their corresponding fluorescence spectra are shown in Fig. S5 and S6 (ESI†). This result provides clear evidence that the proposed method is effective in screening triplex DNA binders. More importantly, the degree of triplex DNA binder-induced fluorescence quenching of the T8-MB-T8·A7 probe provides useful information to determine the relative binding strengths of triplex DNA binders. No reduction in the fluorescence of the T8-MB-T8·A7 probe was observed in the case of berberine and palmatine because they exhibited weak binding to triple-stranded DNA. Sinha and Kumar reported that the binding of coralyne to triplex-stranded RNA was remarkably high in comparison to berberine and palmatine.20 Das et al. demonstrated that the complexation of triplex-stranded DNA was more efficient for sanguinarine than bertaine.21 These results clearly indicate that coralyne and sanguinarine are stronger triplex DNA binders than berberine and palmatine.
Coralyne has been reported to be a promising drug for cancer therapy because of its relatively low toxicity.22 The plant alkaloid, sanguinarine, has been shown to be capable of inhibiting the activity of Na+, K+-ATPase in different organs, causing epidemic dropsy.23 Thus, the T8-MB-T8·A7 probe was applied for the quantitative detection of coralyne and sanguinarine at 10 min incubation time. As the concentration of coralyne and sanguinarine increased at a fixed concentration of T8-MB-T8 and A7, the fluorescence of the T8-MB-T8·A7 was progressively decreased (Fig. S7 and S8, ESI†). The linear relationship of the fluorescence intensity (IF) at 520 nm versus the concentrations of coralyne and sanguinarine, as shown in the inset of Fig. S7 and S8,† was from 2 to 100 nM (R2 = 0.9880) and from 10 to 100 nM (R2 = 0.9900), respectively. This probe enabled the detection of coralyne with limit of detection (LOD; signal-to-noise ratio of 3) corresponding to 0.6 nM, which is lower than the LOD values measured from the reported methods (Table S1, ESI†). The LOD of sanguinarine was estimated to be 3 nM, which is lower than or comparable the LOD values obtained from the reported methods (Table S2, ESI†). Compared to these reported methods, we suggest that this probe offers the advantages of sensitivity, speed and simplicity for detecting coralyne and sanguinarine.
To make a judgment about whether or not a triplex DNA binder was present in the input samples, the NAND gate was constructed using A7 and triplex DNA binder as inputs and the fluorescence change of the T8-MB-T8 probe as the output (Fig. 2). The fluorescence intensity at 520 nm of the T8-MB-T8 probe did not change after adding either A7 (i1 = 1 and i2 = 0) or triplex DNA binder (i1 = 0 and i2 = 1). When the MB probe reacted with the mixture (i1 = 1 and i2 = 1) of A7 and triplex DNA binder, the formation of triplex-stranded DNA disrupted the conformation of the MB probe and quenched their fluorescence. Fig. 2 shows that the fluorescence intensity at 520 nm of the T8-MB-T8 probe displayed different combinations of input signals corresponding to the true table of the NAND gate.
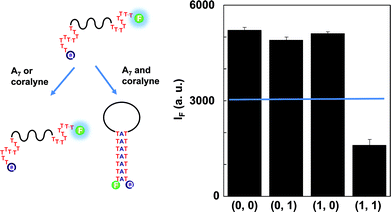 | ||
| Fig. 2 Schematic illustration and truth table of the designed probe for the operation of the two input NAND gate. | ||
In summary, the present study introduced the T8-MB-T8·A7 probe for screening triplex DNA binders. The relative binding strength among triplex DNA binders was simultaneously determined by monitoring the degree of triplex DNA binder-induced fluorescence quenching of the T8-MB-T8·A7 probe. The integrated logic circuit probe was adapted to screen triplex DNA binders. These findings facilitate the implementation of MB as an optical probe to screen other triplex DNA binders.
Acknowledgements
We would like to thank National Science Council (NSC 100-2628-M-110-001-MY 4) for the financial support of this study.Notes and references
- W. H. Tan, L. Tan, Y. Li, T. J. Drake, L. Moroz, K. M. Wang, J. Li, A. Munteanu, C. Y. J. Yang and K. Martinez, Analyst, 2005, 130, 1002–1005 RSC.
- S. A. Marras, Mol. Biotechnol., 2008, 38, 247–255 CrossRef CAS PubMed.
- A. Ono and H. Togashi, Angew. Chem., Int. Ed., 2004, 43, 4300–4302 CrossRef CAS PubMed.
- Y. Wang, J. Li, H. Wang, J. Jin, J. Liu, K. Wang, W. Tan and R. Yang, Anal. Chem., 2010, 82, 6607–6612 CrossRef CAS PubMed.
- C. Y. Kuo and W. L. Tseng, Chem. Commun., 2013, 49, 4607–4609 RSC.
- H. Xu and M. Hepel, Anal. Chem., 2011, 83, 813–819 CrossRef CAS PubMed.
- L. M. Lu, X. B. Zhang, R. M. Kong, B. Yang and W. Tan, J. Am. Chem. Soc., 2011, 133, 11686–11691 CrossRef CAS PubMed.
- E. Heyduk and T. Heyduk, Anal. Chem., 2005, 77, 1147–1156 CrossRef CAS.
- S. Buchini and C. J. Leumann, Curr. Opin. Chem. Biol., 2003, 7, 717–726 CrossRef CAS PubMed.
- J. B. Chaires, J. Ren, D. Hamelberg, A. Kumar, V. Pandya, D. W. Boykin and W. D. Wilson, J. Med. Chem., 2004, 47, 5729–5742 CrossRef CAS PubMed.
- X. Niusheng, Y. Hongmei, C. Meng, W. Cuihong and L. Shuying, Anal. Chem., 2012, 84, 2562–2568 CrossRef PubMed.
- M. S. Han, A. K. R. Lytton-Jean and C. A. Mirkin, J. Am. Chem. Soc., 2006, 128, 4954–4955 CrossRef CAS PubMed.
- A. K. Lytton-Jean, M. S. Han and C. A. Mirkin, Anal. Chem., 2007, 79, 6037–6041 CrossRef CAS PubMed.
- G. C. Silver, J. S. Sun, C. H. Nguyen, A. S. Boutorine, E. Bisagni and C. Helene, J. Am. Chem. Soc., 1997, 119, 263–268 CrossRef CAS.
- T. N. Grossmann, L. Roglin and O. Seitz, Angew. Chem., Int. Ed., 2007, 46, 5223–5225 CrossRef CAS PubMed.
- G. E. Plum, Biopolymers, 1997, 44, 241–256 CrossRef CAS.
- M. Polak and N. V. Hud, Nucleic Acids Res., 2002, 30, 983–992 CrossRef CAS PubMed.
- L. Xue, H. Xi, S. Kumar, D. Gray, E. Davis, P. Hamilton, M. Skriba and D. P. Arya, Biochemistry, 2010, 49, 5540–5552 CrossRef CAS PubMed.
- J. Y. Han and K. Burgess, Chem. Rev., 2010, 110, 2709–2728 CrossRef CAS PubMed.
- R. Sinha and G. S. Kumar, J. Phys. Chem. B, 2009, 113, 13410–13420 CrossRef CAS PubMed.
- S. Das, G. S. Kumar, A. Ray and M. Maiti, J. Biomol. Struct. Dyn., 2003, 20, 703–714 CAS.
- W. D. Wilson, A. N. Gough, J. J. Doyle and M. W. Davidson, J. Med. Chem., 1976, 19, 1261–1263 CrossRef CAS.
- M. Das and S. K. Khanna, Crit. Rev. Toxicol., 1997, 27, 273–297 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Fig. S1–S8 and Tables S1–S3. See DOI: 10.1039/c4ra06158f |
| This journal is © The Royal Society of Chemistry 2014 |