Open Access Article
Open Access ArticleStructural elucidation of HIV-1 G-quadruplexes in a cellular environment and their ligand binding using responsive 19F-labeled nucleoside probes†
Sarupa
Roy
a,
Priyasha
Majee
b,
Sruthi
Sudhakar
b,
Satyajit
Mishra
c,
Jeet
Kalia
 cd,
P. I.
Pradeepkumar
cd,
P. I.
Pradeepkumar
 *b and
Seergazhi G.
Srivatsan
*b and
Seergazhi G.
Srivatsan
 *a
*a
aDepartment of Chemistry, Indian Institute of Science Education and Research (IISER), Pune, Dr Homi Bhabha Road, Pune 411008, India. E-mail: srivatsan@iiserpune.ac.in
bDepartment of Chemistry, Indian Institute of Technology Bombay, Mumbai 400076, India. E-mail: pradeep@chem.iitb.ac.in
cDepartment of Biological Sciences, Indian Institute of Science Education and Research (IISER) Bhopal, Bhopal Bypass Road, Bhauri, Bhopal 462066, India
dDepartment of Chemistry, Indian Institute of Science Education and Research (IISER) Bhopal, Bhopal Bypass Road, Bhauri, Bhopal 462066, India
First published on 25th April 2024
Abstract
Understanding the structure and recognition of highly conserved regulatory segments of the integrated viral DNA genome that forms unique topologies can greatly aid in devising novel therapeutic strategies to counter chronic infections. In this study, we configured a probe system using highly environment-sensitive nucleoside analogs, 5-fluoro-2′-deoxyuridine (FdU) and 5-fluorobenzofuran-2′-deoxyuridine (FBFdU), to investigate the structural polymorphism of HIV-1 long terminal repeat (LTR) G-quadruplexes (GQs) by fluorescence and 19F NMR. FdU and FBFdU, serving as hairpin and GQ sensors, produced distinct spectral signatures for different GQ topologies adopted by LTR G-rich oligonucleotides. Importantly, systematic 19F NMR analysis in Xenopus laevis oocytes gave unprecedented information on the structure adopted by the LTR G-rich region in the cellular environment. The results indicate that it forms a unique GQ-hairpin hybrid architecture, a potent hotspot for selective targeting. Furthermore, structural models generated using MD simulations provided insights on how the probe system senses different GQs. Using the responsiveness of the probes and Taq DNA polymerase stop assay, we monitored GQ- and hairpin-specific ligand interactions and their synergistic inhibitory effect on the replication process. Our findings suggest that targeting GQ and hairpin motifs simultaneously using bimodal ligands could be a new strategy to selectively block the viral replication.
Introduction
HIV-1 is one of the most lethal retroviruses, which induces a chronic infection by etching the host cell genome with a proviral DNA that is reverse transcribed from its RNA genome. Established treatments use a cocktail of drugs having different modes of action to control the disease progression.1 However, due to persistence of latent reservoirs, drug-resistance and promiscuity of the viral polymerase, it is very hard to eradicate the virus completely from the host system.2,3 One of the current ways to counter latency involves awakening dormant viruses and simultaneously inhibiting viral replication with antiviral agents.4,5 It is also hypothesized that targeting certain structural and functional segments of the integrated viral DNA genome could complement the above strategy and help in curing the disease.6 An important and a highly conserved gene segment that could be suitable for this purpose is the long terminal repeat (LTR) of the HIV-1 promoter region.7,8The initiation of HIV-1 transcription is navigated by the promoter region 5′-LTR, which is composed of U3, R and U5 regions.9 The U3 region consists of three functional segments including the highly conserved core-binding site of NF-κB and Sp1 transcription factors,10 which harbors contiguous G-rich tracts capable of forming G-quadruplex (GQ) structures namely LTR-II, LTR-III and LTR-IV, and HIVpro1 and HIVpro2 (Fig. 1A).11–13 Notably, LTR-III and LTR-IV form GQs in vitro in a mutually exclusive manner. While LTR-III adopts a unique architecture made of a hybrid-type GQ juxtaposed with a three G-C paired hairpin motif, LTR-IV attains a parallel GQ topology with a T-bulge (Fig. 1B).14,15 However, the entire G-rich region majorly forms the GQ-hairpin form like the LTR-III motif, and the parallel form of LTR-IV is induced when it binds to ligands or protein factors.14–16 Importantly, the LTR GQ region represents an evolutionary conserved element across all primate lentiviruses, and the balance between different GQ structures is implicated in the propagation and latency of the virus.17,18 Therefore, we envision that the virus status can be selectively controlled in the host cell by using structure-specific binders. In this direction, it is important to gain a comprehensive understanding of the structural polymorphism, dynamics and druggable space of LTR GQs in a cellular environment to advocate a viable therapeutic strategy.
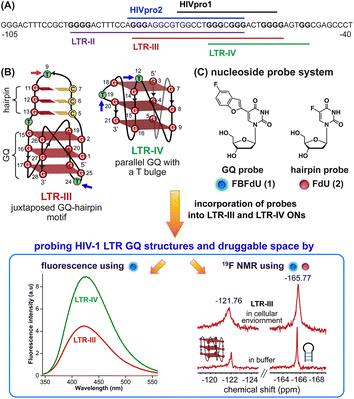 | ||
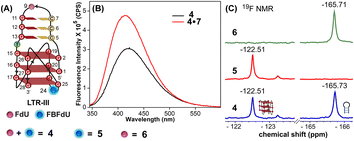
| Fig. 1 (A) Schematic representation of the HIV-1 LTR G-rich region. (B) Secondary structures of LTR-III and LTR-IV GQs are depicted using the respective NMR structures (PDB: 6H1K and 2N4Y). (C) Environment-sensitive nucleoside system designed to probe the structural polymorphism and druggable space of LTR GQs by fluorescence and 19F NMR techniques in cell-free and cellular environments. Potential sites for incorporation of the GQ probe (FBFdU 1) and hairpin probe (FdU 2) into the loop region of LTR-III and LTR-IV G-rich sequences are shown with blue and magenta arrows, respectively. | ||
A multitude of biophysical and biochemical techniques including CD, UV thermal melting, fluorescence, NMR and X-ray crystallography add pieces of valuable information to characterize GQs in vitro.19–25 Recently, antibodies and chemical probes have been developed to detect ensembles of DNA GQs in cells.26–30 However, the majority of tools fall short when evaluating co-existing structures, as the challenges are twofold: (i) inability to differentiate different GQ topologies (exceptions are few)31–34 and (ii) limited capability to assess structures in a cellular environment. Also, sequences with multiple G tracts exhibit high structural polymorphism and dynamics, which can vary between cell-free and cellular environments.35,36 Therefore, we sought to devise a probe platform that would (i) provide spectral signatures for different GQs formed by the LTR G-rich region, which could help identify the preferred GQ in cells and (ii) allow us to survey the chemical space of LTR GQs to design structure-specific binders.
In this context, we developed microenvironment-sensitive dual-functional nucleoside probes that immensely aid in studying nucleic acid conformations and topology-specific ligand/drug interaction.37,38 In particular, 5-fluorobenzofuran-modified 2′-deoxyuridine (FBFdU) serves as an excellent two-channel readout system to detect different GQ conformations of the human telomeric repeat in vitro and in cellular milieu by using fluorescence and 19F NMR techniques.37 However, LTR-III forms a juxtaposed GQ-hairpin motif that is unique for the HIV-1 virus. Hence, we realized that by using only a GQ sensing probe (FBFdU) it would not be possible to survey the landscape of the LTR. In this regard, here we report the development of a probe system that uses FBFdU as a GQ sensor and 5-fluoro-2′-deoxyuridine (FdU) as a hairpin sensor (Fig. 1C). The probes judiciously placed in loop positions of the LTR G-rich region are minimally perturbing, and importantly, produce distinct and resolved spectral signatures for LTR-III and LTR-IV GQs. Rewardingly, we deduced the GQ structure adopted by the LTR promoter region in an ex vivo model (Xenopus laevis oocyte extract) by using 19F NMR signatures obtained in vitro. Furthermore, the probe platform and Taq DNA polymerase assay helped in mapping ligand interactions and their influence on the LTR replication process.
Results and discussion
Structural investigation of the modified HIV-1 LTR by fluorescence and 19F NMR in vitro
| G-rich domain | ONa | 5′---------------------------------------3’ |
|---|---|---|
| a ONs 3, 8 and 12 are native unmodified ONs of LTR-III, LTR-IV and LTR-(III + IV), respectively. ONs 4–6 are native LTR-III modified with FBFdU (1) and or FdU (2) at T24 and T9, respectively. ONs 9 and 10 are native LTR-IV modified with FBFdU (1) at T12 and T19, respectively. ONs 13–16 are native LTR-(III + IV) modified with FBFdU (1) and or FdU (2) at T25 or T32 and T10, respectively. 7 and 11 are complementary ONs of 3 and 8, respectively. | ||
| LTR-III | 3 |
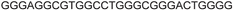
|
| 4 |
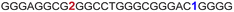
|
|
| 5 |
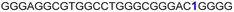
|
|
| 6 |
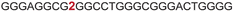
|
|
| 7 |
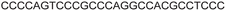
|
|
| LTR-IV | 8 |
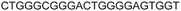
|
| 9 |
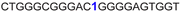
|
|
| 10 |
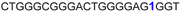
|
|
| 11 |
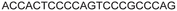
|
|
| LTR-III + IV | 12 |
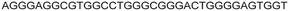
|
| 13 |
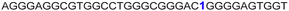
|
|
| 14 |
|
|
| 15 |
|
|
| 16 |
|
|
The ability of FBFdU to serve as a GQ reporter was evaluated by recording fluorescence of LTR-III ON 4 and its corresponding duplex (4·7) in a buffer containing K+ ions (Fig. 2A and B). The GQ form of 4 displayed a discernibly lower fluorescence intensity and a slightly red-shifted emission band (417 nm) as compared to its perfect duplex (4·7, 414 nm). In support of our probe system design, the 19F NMR spectrum of ON 4 exhibited two distinct peaks at −122.51 ppm and −165.73 ppm arising from FBFdU and FdU, respectively (Fig. 2C, blue line). 1H NMR spectra of ON 4 and 3 revealed imino proton signals for both GQ and hairpin domains (Fig. S4†). To assign signals in the 19F NMR spectrum, two singly modified ONs 5, containing FBFdU at T24 (GQ domain) and 6, containing FdU at T9 (hairpin domain), were synthesized (Table 1, Fig. 2A, S1 and S2†). CD profiles and Tm values indicated the formation of a stable hybrid GQ structure like the native 3 and modified 4 ONs (Fig. S3A and B, Table S2†). While ON 5 produced a single peak at −122.51 ppm from the GQ sensor (Fig. 2C, red line), ON 6 gave a signal at −165.71 ppm from the hairpin sensor similar to ON 4 (Fig. 2C, green line). Hence, signals emanating from FBFdU (−122.51 ppm) and FdU (−165.73 ppm) of ON 4 are assigned to GQ and hairpin domains, respectively. When ON 4 was annealed to its complementary ON 7, the duplex structure produced new peaks at −121.54 ppm associated with FBFdU and −165.62 ppm associated with FdU (Fig. S5†). 1H NMR also validated the formation of a duplex structure where characteristic peaks for Watson–Crick H-bonded imino protons appeared between 12 and 14 ppm, with no peaks in the GQ region (Fig. S5†). Henceforth, the probe system provides a simplified 19F NMR spectrum to detect the two domains of LTR-III ON 4 simultaneously.
Probing the GQ structure of the LTR in a physiological environment by NMR
The LTR G-rich promoter region encompassing both LTR-III and IV segments was recently characterized by Richter and Phan groups in vitro using 1H NMR. Their results suggest that it largely forms a juxtaposed GQ-hairpin motif like LTR-III.14 As an important step forward, we decided to use the spectral properties of our nucleoside analogs to systematically determine the GQ structure adopted by the LTR region in a cellular environment and probe its druggable space. In-cell 19F NMR has become a powerful tool to study nucleic acid structures in cellular milieu,37,47–49 as fluorine is 100% abundant, highly sensitive and importantly, absent in cellular systems (no background signal).50–55 Furthermore, its signal does not undergo significant line broadening in the heterogeneous cellular environment, which is very severe in the case of a proton signal.56 To determine 19F signatures of the longer LTR region and survey GQ and hairpin structures, ONs 13–15 labeled with FBFdU in the GQ domain or FdU in the hairpin domain were synthesized (Table 1, Fig. 4A, S1 and S2†). Importantly, the modification position was maintained as in the individual LTR G-rich segments. CD spectra of modified (13–15) and control unmodified (12) ONs exhibited bands similar to the hybrid topology adopted by the LTR-III region (Fig. S8A†). While ON 13 (FBFdU at T25) displayed a nearly 4 °C higher Tm compared to the native ON 12, ON 14 (FBFdU at T32) and 15 (FdU at T10) displayed similar Tm values (Fig. S8B and Table S2†). ON 13 showed a single broad 19F peak at −123.20 ppm and ON 14 exhibited a sharper peak at −122.10 ppm for the GQ domain (Fig. 4B). ON 15 produced a distinct peak (−165.72 ppm) for the hairpin domain. 1H NMR spectra of ONs clearly revealed the presence of imino protons for GQ and hairpin motifs (Fig. 4B). Furthermore, the absence of multiple 19F peaks suggests that the parallel topology of the LTR-IV region is possibly not formed by the longer promoter region (compare with Fig. 3C). To detect both GQ and hairpin motifs simultaneously, we synthesized ON 16 containing FdU at T10 and FBFdU at T32 positions. Modification at T25 (like in ON 13) was avoided as it led to a broader peak and poor base line.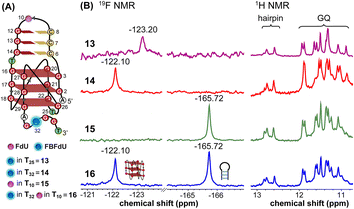 | ||
| Fig. 4 (A) Schematic representation of the juxtaposed GQ-hairpin structure of the LTR-III + IV region. (B) 19F NMR and partial 1H NMR spectra of ONs 13–16. | ||
Rewardingly, ON 16 displayed two distinct signals, one each for GQ (−122.10 ppm) and hairpin (−165.72 ppm) structures with the same chemical shifts as that of ONs 14 and 15 designed to detect the structures independently (Fig. 4B). Also, ON 16 depicted a 1H NMR spectrum revealing the presence of GQ and hairpin structures. Collectively, the probe combination provides distinct and simplified 19F signatures for GQ and hairpin structures, and our results demonstrate that the LTR G-rich region predominantly folds into a GQ-hairpin motif similar to LTR-III in vitro.
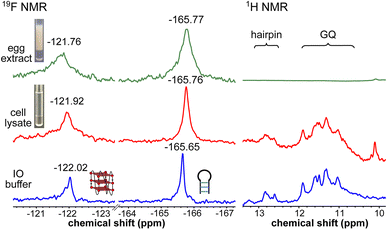
To obtain a progressive understanding of the LTR GQs in cell-free and cellular environments, we performed a systematic NMR analysis using Xenopus oocytes, a commonly used cellular model.47–49,57 While a buffer mimicking intraoocyte (IO) ionic conditions serves as a cell-free system, frog egg lysate and extract serve as very good ex vivo systems to carry out NMR experiments to determine the structure of nucleic acids. First, we recorded the 19F NMR spectrum of ON 16 in an IO buffer (25 mM HEPES pH 7.5, 110 mM KCl, 10.5 mM NaCl, 130 nM CaCl2, 1 mM MgCl2, 0.1 mM EDTA). The ON acquired a conformation like LTR-III, reflecting peaks at −122.02 ppm for the GQ and −165.65 ppm for the hairpin motifs (Fig. 5, blue line and Fig. S9†). The formation of the GQ-hairpin structure was confirmed by 1H NMR and CD experiments (Fig. 5 and S10†). ON 16 incubated in the lysate supported the formation of a hybrid architecture like in the IO buffer (Fig. 5, red line). The inter-phase egg extract obtained by simply centrifuging crushed eggs maintains metabolite and protein contents mimicking those in the biological environment.46 Interestingly, the 19F NMR spectrum of ON 16 incubated in the egg extract revealed the presence of GQ and hairpin motifs, albeit with a slight broadening and shift in the signal (green line). In contrast, due to extensive line broadening, the 1H NMR spectrum fails to provide structural information in egg extract (Fig. 5).46,57 Therefore, our fluorine-labeled nucleoside probes outweigh the applicability of proton NMR in cell-based analysis. To confirm if the signal is originating from the intact ON in cellular samples, after NMR acquisition, the samples were analyzed by HPLC and ESI-MS. The results indicated that the ON is not degraded in the cellular environment (Fig. S11 and S12†). Taken together, these results provide clear evidence for the presence of a monomeric architecture preserving juxtaposed GQ and hairpin domains both in vitro and under cellular conditions underscoring the potential of LTR GQ as a target of selective therapeutic intervention.
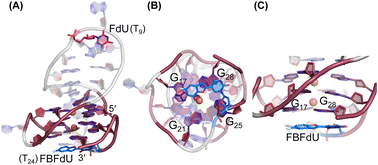 | ||
| Fig. 6 Representative images of major clusters of LTR-III ON 4. (A) Overall structure with FdU and FBFdU in ON 4. (B) Axial view showing the stacking of FBFdU over the bottom quartet. (C) Zoomed-in image showing the perpendicular orientation of FBFdU stacked with G17 and G28. GQ bases are represented in maroon, FdU in magenta and FBFdU in blue. K+ ions are represented as orange spheres. The clusters have been obtained from the 500 ns MD simulation. Details of MD simulations are provided in the ESI.† | ||
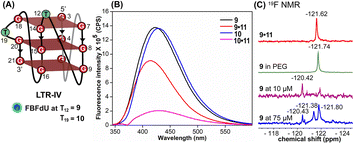
Simulations of LTR-IV ON 9 revealed 3 clusters of ∼30, 30, and 26% each, and ON 10 revealed 2 clusters accounting for ∼85% of the population (Fig. S19 and S20†). The modified base adopts an alternate conformation without affecting the GQ topology (Fig. S21 and S22†). Models of 9 and 10 revealed that the probe placed at T12 and T19 positions, respectively, is flipped out (solvent exposed) and is away from the G-tetrad core (Fig. S21 and S22†). Hence, these ONs exhibit high fluorescence intensity with emission maxima similar to the emission wavelength of the nucleoside analog 1 in water, indicating a polar environment around the probe (423 nm and 428 nm, Table S3†). In the case of duplex 9·11, FBFdU flanked between C11 and G13 would experience partial stacking interaction, and hence, shows lower fluorescence intensity at λem = 418 nm. FBFdU in duplex 10·11 exhibits significant reduction in fluorescence intensity due to partial stacking interaction followed by more quenching from two adjacent Gs (G18 and G20). Although the 19F component of nucleoside probes exhibits distinct chemical shifts for different GQ and duplex structures, rationalizing peak positions is not trivial. This is because the environment around the probes and their interaction with neighbouring bases as mentioned above can have varying shielding-deshielding effects on the 19F atom,62–64 which are difficult to predict and are also evident from an obscure trend in the chemical shift of nucleosides in different solvents (Table S4†).
Probing ligand binding to HIV-1 LTR GQs by fluorescence, 19F NMR and Taq DNA polymerase stop assay
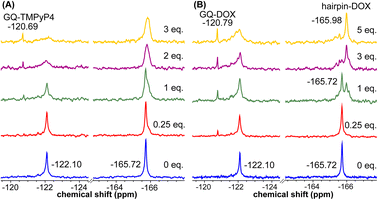
Using the spectral properties of the probes, we evaluated ligand recognition of LTR ONs using two structurally different GQ binders namely, TMPyP4 and BRACO19 (Fig. S23A†). Upon ligand binding to the GQ structure, the fluorescence of FBFdU placed in the GQ domain is known to diminish significantly because of its proximity to the polyaromatic ligands.37 LTR-III ON 4 was titrated with increasing concentrations of the ligands and changes in fluorescence were recorded. Titration with TMPyP4 and BRACO19 resulted in a dose-dependent quenching in fluorescence intensity with minimum changes in the emission maximum (Fig. S24A and B†). A plot of normalized fluorescence intensity versus ligand concentration fitted to the Hill equation gave an apparent Kd value of 0.28 ± 0.05 μM and 0.56 ± 0.09 μM, respectively (Fig. S23B†). Similarly the parallel GQ structure of LTR-IV ON 9 titrated with the ligands gave Kd values of 0.33 ± 0.02 μM and 0.39 ± 0.05 μM, respectively (Fig. S23C, S25A and B†). The 19F label of FBFdU efficiently reported the formation of different GQ-ligand complexes with distinct chemical shifts. TMPyP4 and BRACO19 binding to the GQ domain of LTR-III ON 4 produced a new peak at −120.75 ppm and −120.78 ppm, respectively, with a concomitant decrease in the GQ signal (Fig. S23D and S24C†). Gratifyingly, as these ligands bind preferentially to the G-tetrad, they did not exhibit detectable interaction with the hairpin structure. This is evident from the chemical shift of FdU (−165.73 ppm), placed in the hairpin domain, which remains mostly unchanged throughout the titration experiment. Similarly, ligand binding to the parallel topology of LTR-IV ON 9 exhibited a distinct peak for each complex (TMPyP4: −120.77 ppm and BRACO19: −120.60 ppm, Fig. S23E and S25C†). Interestingly, upon ligand binding, multiple GQs formed by 9 coalesce into one ligand-bound form.Bioinformatics and biophysical studies reveal the prevalence of quadruplex-hairpin/duplex junctions in genomes, characterized by varying loop sizes and conformations.65–69 These adjacently placed structures offer unique scaffolds to target the junction or both GQ and duplex elements simultaneously.70–72 Hence, bimodal ligands capable of doing the same can significantly enhance the specific targeting of hybrid GQs as opposed to autonomous GQ structures. In this direction, we evaluated the recognition properties of the GQ-hairpin motif of the longer LTR promoter ON 16 using TMPyP4 (GQ binder) and doxorubicin (DOX, duplex binder) by fluorescence and 19F NMR (Fig. S26A†). Addition of increasing concentrations of TMPyP4 (30 nM–2.5 μM) to ON 16 (0.5 μM) resulted in a progressive quenching in fluorescence intensity as before and gave an apparent Kd value of 0.52 ± 0.03 μM for the formation of the ligand-GQ complex (Fig. S26B and S27A†). Preferential binding of the ligand to the GQ region was ascertained by 19F NMR. FBFdU placed at the GQ domain responded to increasing concentrations of the ligand, giving rise to a new peak at −120.69 ppm for the complex (Fig. 7A). Notably, the chemical shift of FdU (−165.72 ppm) placed in the hairpin domain remained practically unaltered, indicating that TMPyP4 interacts specifically with the GQ structure.
DOX is intrinsically fluorescent and it shows changes in fluorescence upon binding to DNA. To avoid interference from FBFdU, DOX (2 μM) was titrated with a control unmodified ON 12 (2.5 nM–2 μM). We observed a sigmoidal quenching behavior, which gave a Kd value of 0.10 ± 0.02 μM (Fig. 26B and S27B†). 19F NMR using ON 16 gave better insights into the recognition process. Addition of DOX (1 equiv.) to the ON resulted in the emergence of two new peaks–(i) −120.79 ppm associated with GQ-DOX and (ii) −165.98 ppm associated with hairpin-DOX (Fig. 7B). Notably, at a higher equivalent of DOX, the ligand largely occupies the hairpin domain and to some extent the GQ domain. These results highlight the advantage of 19F-labeled nucleoside analogs in probing structure-specific ligand interactions. Based on these observations, we designed a polymerase stop assay to study the inhibitory effect of the GQ structure and the ligands independently and in a combination.
The effect of GQ structures on DNA polymerase activity was evaluated by Taq DNA polymerase stop assay using a native LTR template T1 encompassing III and IV regions and a mutated template T2 (does not fold into a GQ, Fig. 8A and Table S5†). GQ forming template T1 significantly halted the polymerization process yielding largely stalled products near the GQ site (Fig. S28,† lanes 2–6, 8B). Longer reaction times (30 min) produced only ∼33% of the full-length product. In contrast, reactions in the presence of a non-GQ forming template T2 produced significant amounts of the full-length product in only 2 min (∼45%), which progressively increased to ∼80% at 30 min (Fig. S28,† lanes 7–11, 8B). These observations indicate that the stalling of the primer extension reaction is due to the formation of a stable LTR GQ structure by T1.
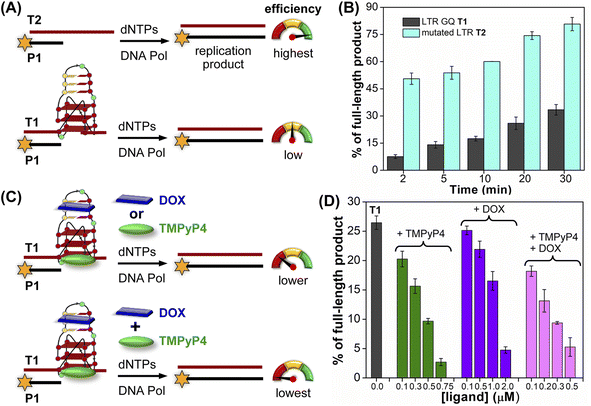 | ||
| Fig. 8 Schematic representation of primer extension reactions using Taq DNA polymerase (A) with non-GQ forming template T2 and GQ forming template T1, (C) in the presence of ligands TMPyP4 or DOX and TMPyP4 + DOX. (B) and (D) Percentage of the full-length product obtained from Taq DNA polymerase reactions. (B) Reactions performed using templates T1 and T2 at different time intervals. (D) Reactions performed using T1 with increasing concentrations of ligands TMPyP4, DOX and TMPyP4 + DOX at 20 min. For gel images, see Fig. S28 and S29.† Values are denoted as mean ± s.d for 2 independent experiments. | ||
Next, we studied the effect of ligand binding to the GQ structure on the polymerase activity using T1 (Fig. 8C). For this purpose, a reaction time of 20 min was chosen as it gave reasonable amounts of the full-length product (∼25%, Fig. S28,† lane 5). Varying concentrations of TMPyP4 and DOX were independently added and allowed to bind with LTR GQ, and then primer extension reactions were carried out as before. Upon addition of TMPyP4 there was a noticeable decrease in the formation of the full-length product, accompanied by a simultaneous increase in stalled products (Fig. S29,† lanes 2–5, Fig. 8D). At 0.75 μM of the ligand (7.5 equiv. w.r.t T1) no detectable full-length product was observed (Fig. S29,† lane 6). Similarly, increasing amounts of DOX resulted in a progressive reduction in the formation of the full-length product (Fig. S29,† lane 7–10, Fig. 8D). Although DOX inhibited the polymerase activity, it required a higher amount (20 equiv.) to produce an effect comparable to TMPyP4 (Fig. S29,† compare lane 6 and 10). This may be due to DOX competing for the primer-template duplex, hairpin and GQ regions. These results prompted us to study the combined effect of ligands, wherein different concentrations of TMPyP4 and DOX at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio were added to the reaction mixture. The gel image revealed a synergistic effect of ligands as the formation of the full-length product considerably decreased with a concomitant increase in truncated products in comparison to reactions in which only one ligand was added at an equivalent concentration (Fig. 8D, S29,† lanes 11–14). The effect is noticeable when we compare TMPyP4 (0.3 μM, ∼16%), DOX (1 μM, ∼17%), and TMPyP4+DOX (0.3 μM, ∼9%, Fig. 8D). These results suggest that the GQ-hairpin motif of the LTR G-rich region serves as a highly conserved regulatory element to potentially block the viral replication process by targeting both GQ and hairpin domains simultaneously.
1 ratio were added to the reaction mixture. The gel image revealed a synergistic effect of ligands as the formation of the full-length product considerably decreased with a concomitant increase in truncated products in comparison to reactions in which only one ligand was added at an equivalent concentration (Fig. 8D, S29,† lanes 11–14). The effect is noticeable when we compare TMPyP4 (0.3 μM, ∼16%), DOX (1 μM, ∼17%), and TMPyP4+DOX (0.3 μM, ∼9%, Fig. 8D). These results suggest that the GQ-hairpin motif of the LTR G-rich region serves as a highly conserved regulatory element to potentially block the viral replication process by targeting both GQ and hairpin domains simultaneously.
Based on our results and the formation of a unique GQ-hairpin architecture in a cellular environment, we propose that bimodal ligand scaffolds composed of GQ and duplex binders, clamped using an appropriate linker, could selectively target the HIV-1 LTR and profoundly attenuate its pathogenesis (Fig. S30†). Needless to say, careful consideration should be exercised when optimizing the ligand design. The choice of GQ and hairpin binders from available examples and linker length, flexibility and point of attachment to the ligands will be very crucial.
Conclusion
We have devised a probe platform using two highly environment-sensitive nucleoside analogs (FBFdU and FdU) to study the structural polymorphism of a conserved HIV-1 LTR G-rich region in cell-free and cellular environments. These minimally invasive analogs produced a very simplified spectrum with distinct fluorescence and 19F NMR signatures for different LTR GQ architectures. Importantly, using 19F signatures of FBFdU and FdU we successfully identified that the LTR G-rich region adopts a GQ-hairpin architecture in a cellular environment. MD simulations gave insights on the structural basis by which FBFdU fluorescently senses different GQ topologies and distinguishes them from the duplex form. Furthermore, the nucleoside probes facilitated the detection and estimation of structure-specific ligand interactions by fluorescence and 19F NMR techniques. Polymerase stop assay confirmed the regulatory function of LTR GQ structures. While TMPyP4 (GQ binder) and DOX (duplex binder) individually decreased polymerase activity, an equimolar mixture exhibited a synergistic inhibitory effect on LTR replication. Taken together, these findings suggest that simultaneous targeting of the juxtaposed GQ-hairpin motif using bimodal ligands could be a rational plan to selectively and efficiently inhibit the pathogenesis of the virus. In this direction, the FBFdU and FdU combination offers a versatile platform to study the structure as well as devise screening assays to identify hybrid ligands targeting GQ-hairpin/duplex motifs.Data availability
The datasets supporting this article have been uploaded as part of the ESI.† Deposited MD simulation models are available in https://www.modelarchive.org/. Details are provided in the ESI (Fig. S16, S19 and S20).†Author contributions
S. G. S. designed and supervised this project. S. R. designed and performed the experiments. P. M., S. S. and P. I. P. performed MD simulations to build models of labeled GQ structures. S. M. and J. K. helped in performing experiments in frog egg lysate and extract. All authors analysed the results, and S. R. and S. G. S. wrote the manuscript in consultation with all the authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
S. R. and S. M. acknowledge CSIR, India, for their graduate research fellowships. P. M. thanks IIT Bombay and S. S. thanks Prime Ministers Research Fellowship (PMRF) for graduate research fellowships. We thank Spacetime-IIT Bombay for HPC facilities. This work was supported by SERB (CRG/2022/000284) grant to S. G. S and IIT Bombay-Institute of Eminence (IOE) funds to P. I. P.References
- Y. Peng, Y. Zong, D. Wang, J. Chen, Z.-S. Chen, F. Peng and Z. Liu, Front. Pharmacol, 2023, 26, 1294966 CrossRef PubMed.
- R. J. Pomerantz, Clin. Infect. Dis., 2002, 34, 91–97 CrossRef CAS PubMed.
- R. K. Gupta, J. Gregson, N. Parkin, H. Haile-Selassie, A. Tanuri, F. L. Andrade, P. Kaleebu, C. Watera, A. Aghokeng, N. Mutenda, J. Dzangare, S. Hone, Z. Z. Hang, J. Garcia, Z. Garcia, P. Marchorro, E. Beteta, A. Giron, R. Hamers, S. Inzaule, L. M. Frenkel, M. H. Chung, T. de Oliveira, D. Pillay, K. Naidoo, A. Kharsany, R. Kugathasan, T. Cutino, G. Hunt, S. A. Rios, M. Doherty, M. R. Jordan and S. Bertagnolio, Lancet Infect. Dis., 2018, 18, 346–355 CrossRef PubMed.
- A. Archin, L. Liberty, A. D. Kashuba, S. K. Choudhary, J. D. Kuruc, A. M. Crooks, D. C. Parker, E. M. Anderson, M. F. Kearney, M. C. Strain, D. D. Richman, M. G. Hudgens, R. J. Bosch, J. M. Coffin, J. J Eron, D. J. Hazuda and D. M. Margolis, Nature, 2012, 487, 482–485 CrossRef PubMed.
- W. Nguyen, J. Jacobson, K. E. Jarman, H. Jousset Sabroux, L. Harty, J. McMahon, S. R. Lewin, D. F. Purcell and B. E. Sleebs, J. Med. Chem., 2019, 62, 5148–5175 CrossRef CAS PubMed.
- J. Zhang and C. Crumpacker, Viruses, 2022, 14, 1084 CrossRef CAS PubMed.
- K. A. Roebuck and M. Saifuddin, Gene Expression, 1999, 8, 67–84 CAS.
- T. van Opijnen, R. E. Jeeninga, M. C Boerlijst, G. P. Pollakis, V. Zetterberg, M. Salminen and B. Berkhout, J. Virol., 2004, 78, 3675–3683 CrossRef CAS PubMed.
- L. A. Pereira, K. Bentley, A. Peeters, M. J. Churchill and N. J. Deacon, Nucleic Acids Res., 2000, 28, 663–668 CrossRef CAS PubMed.
- R. E. Jeeninga, M. Hoogenkamp, M. Armand-Ugon, M. de Baar, K. Verhoef and B. Berkhout, J. Virol., 2000, 74, 3740–3751 CrossRef CAS PubMed.
- R. Perrone, M. Nadai, I. Frasson, J. A. Poe, E. Butovskaya, T. E. Smithgall, M. Palumbo, G. Palu and S. N. Richter, J. Med. Chem., 2013, 56, 6521–6530 CrossRef CAS PubMed.
- S. Amrane, A. Kerkour, A. Bedrat, B. Vialet, M.-L. Andreola and J.-L. Mergny, J. Am. Chem. Soc., 2014, 136, 5249–5252 CrossRef CAS PubMed.
- A. De Rache, J. Marquevielle, S. Bouaziz, B. Vialet, M. L. Andreola, J.-L. Mergny and S. Amrane, J. Mol. Biol., 2023, 436, 168359 CrossRef PubMed.
- E. Butovskaya, B. Heddi, B. Bakalar, S. N. Richter and A. T. Phan, J. Am. Chem. Soc., 2018, 140, 13654–13662 CrossRef CAS PubMed.
- B. De Nicola, C. J. Lech, B. Heddi, S. Regmi, I. Frasson, R. Perrone, S. N. Richter and A. T. Phan, Nucleic Acids Res., 2016, 44, 6442–6451 CrossRef CAS PubMed.
- E. Ruggiero, I. Frasson, E. Tosoni, M. Scalabrin, R. Perrone, M. Marušič, J. Plavec and S. N. Richter, ACS Infect. Dis., 2022, 8, 958–968 CrossRef CAS PubMed.
- Y. Sheng, B. Cao, M. X. Ou, Y. Wang, S. M. Yuan, N. Zhang, T. Zou and Y. Liu, Chem. Commun., 2021, 57, 5298–5301 RSC.
- R. Perrone, E. Lavezzo, G. Palù and S. N. Richter, Sci. Rep., 2017, 7, 2018 CrossRef PubMed.
- M. Vorlíčková, I. Kejnovská, J. Sagi, D. Renčiuk, K. Bednářová, J. Motlová and J. Kypr, Methods, 2012, 57, 64–75 CrossRef PubMed.
- R. A. Darby, M. Sollogoub, C. McKeen, L. Brown, A. Risitano, N. Brown, C. Barton, T. Brown and K. R. Fox, Nucleic Acids Res., 2002, 30, e39 CrossRef PubMed.
- B. R. Vummidi, J. Alzeer and N. W. Luedtke, ChemBioChem, 2013, 14, 540–558 CrossRef CAS PubMed.
- J. Mohanty, N. Barooah, V. Dhamodharan, S. Harikrishna, P. I Pradeepkumar and A. C. Bhasikuttan, J. Am. Chem. Soc., 2013, 135, 367–376 CrossRef CAS PubMed.
- D. Sun and L. H. Hurley, Methods Mol. Biol., 2010, 608, 65–79 CrossRef CAS PubMed.
- S. M. Haider, G. N. Parkinson and S. Neidle, J. Mol. Biol., 2003, 326, 117–125 CrossRef CAS PubMed.
- T. Santos, G. F. Salgado, E. J. Cabrita and C. S. Cruz, Pharmaceuticals, 2021, 14, 769 CrossRef CAS PubMed.
- G. Biffi, D. Tannahill, J. McCafferty and S. Balasubramanian, Nat. Chem., 2013, 5, 182–186 CrossRef CAS PubMed.
- A. Henderson, Y. Wu, Y. C. Huang, E. A. Chavez, J. Platt, F. B. Johnson, R. M. Brosh Jr, D. Sen and P. M. Lansdrop, Nucleic Acids Res., 2014, 42, 860–869 CrossRef CAS PubMed.
- S. Galli, L. Melidis, S. M. Flynn, D. Varshney, A. Simeone, J. Spiegel, S. K. Madden, D. Tannahill and S. Balasubramanian, J. Am. Chem. Soc., 2022, 144, 23096–23103 CrossRef CAS PubMed.
- F. Doria, M. Nadai, M. Zuffo, R. Perrone, M. Freccero and S. N. Richter, Chem. Commun., 2017, 53, 2268–2271 RSC.
- J. Robinson, S. G. Stenspil, K. Maleckaite, M. Bartlett, M. D. Antonio, R. Vilar and M. K. Kuimova, J. Am. Chem. Soc., 2024, 146, 1009–1018 CrossRef CAS PubMed.
- J.-H. Yuan, W. Shao, S.-B. Chen, Z.-S. Huang and J.-H. Tan, Biochem. Biophys. Res. Commun., 2020, 531, 18–24 CrossRef CAS PubMed.
- T. Vo, S. Oxenford, R. Angell, C. Marchetti, S. A. Ohnmacht, W. D. Wilson and S. Neidle, ACS Med. Chem. Lett., 2020, 11, 991–999 CrossRef CAS PubMed.
- S. Kumar, S. P. P. Pany, S. Sudhakar, S. B. Singh, C. S. Todankar and P. I. Pradeepkumar, Biochemistry, 2022, 61, 2546–2559 CrossRef CAS PubMed.
- S. Takahashi, A. Kotar, H. Tateishi-Karimata, S. Bhowmik, Z. F. Wang, T. C. Chang, S. Sato, S. Takenaka, J. Plavec and N. Sugimoto, J. Am. Chem. Soc., 2021, 143, 16458–16469 CrossRef CAS PubMed.
- R. Rigo, E. Groaz and C. Sissi, Pharmaceuticals, 2022, 15, 373 CrossRef CAS PubMed.
- J. T. Grün and H. Schwalbe, Biopolymers, 2022, 113, e23477 CrossRef PubMed.
- S. Manna, D. Sarkar and S. G. Srivatsan, J. Am. Chem. Soc., 2018, 140, 12622–12633 CrossRef CAS PubMed.
- A. Nuthanakanti, I. Ahmed, S. Y. Khatik, K. Saikrishnan and S. G. Srivatsan, Nucleic Acids Res., 2019, 47, 6059–6072 CrossRef CAS PubMed.
- M. Cheng, Yu. Cheng, J. Hao, G. Jia, J. Zhou, J.-L. Mergny and C. Li, Nucleic Acids Res., 2018, 46, 9264–9275 CrossRef CAS PubMed.
- J. Jana, Y. M. Vianney, N. Schröder and K. Weisz, Nucleic Acids Res., 2022, 50, 7161–7175 CrossRef CAS PubMed.
- M. Sproviero, K. L. Fadock, A. A. Witham and R. A. Manderville, ACS Chem. Biol., 2015, 10, 1311–1318 CrossRef CAS PubMed.
- S. Manna and S. G. Srivatsan, RSC Adv., 2018, 8, 25673–25694 RSC.
- M. Olejniczak, Z. Gdaniec, A. Fischer, T. Grabarkiewicz, L. Bielecki and R. W. Adamiak, Nucleic Acids Res., 2002, 30, 4241–4249 CrossRef CAS PubMed.
- K. Tanabe, M. Sugiura and S. Nishimoto, Bioorg. Med. Chem., 2010, 18, 6690–6694 CrossRef CAS PubMed.
- N. Q. Do and A. T. Phan, Chem.–Eur. J., 2012, 18, 14752–14759 CrossRef CAS PubMed.
- R. Hänsel, F. Löhr, S. Foldynová-Trantírková, E. Bamberg, L. Trantírek and V. Dötsch, Nucleic Acids Res., 2011, 39, 5768–5775 CrossRef PubMed.
- H.-L. Bao, T. Ishizuka, T. Sakamoto, K. Fujimoto, T. Uechi, N. Kenmochi and Y. Xu, Nucleic Acids Res., 2017, 45, 5501–5511 CrossRef CAS PubMed.
- H.-L. Bao, T. Masuzawa, T. Oyoshi and Y. Xu, Nucleic Acids Res., 2020, 48, 7041–7051 CAS.
- C. Wang, G. Xu, X. Liu, L. Jiang, X. Zhou, M. Liu and C. Li, J. Am. Chem. Soc., 2024, 146, 4741–4751 CrossRef CAS PubMed.
- H. Chen, S. Viel, F. Ziarelli and L. Peng, Chem. Soc. Rev., 2013, 42, 7971–7982 RSC.
- D. Gimenez, A. Phelan, C. D. Murphy and S. L. Cobb, Beilstein J. Org. Chem., 2021, 17, 293–318 CrossRef CAS PubMed.
- K. Fauster, C. Kreutz and R. Micura, Angew. Chem., Int. Ed., 2012, 51, 13080–13084 CrossRef CAS PubMed.
- M. R. Baranowski, M. Warminski, J. Jemielity and J. Kowalska, Nucleic Acids Res., 2020, 48, 8209–8224 CrossRef CAS PubMed.
- Q. Li, M. Trajkovski, C. Fan, J. Chen, Y. Zhou, K. Lu, H. Li, X. Su, Z. Xi, J. Plavec and C. Zhou, Angew. Chem., Int. Ed., 2022, 61, e2022018 Search PubMed.
- M. Wang, M. Lu, M. P. Fritz, C. M. Quinn, I. L. Byeon, C.-H. Byeon, J. Struppe, W. Maas, A. M. Gronenborn and T. Polenova, Angew. Chem., Int. Ed., 2018, 57, 16375–16379 CrossRef CAS PubMed.
- L. B. T. Pham, A. Costantino, L. Barbieri, V. Calderone, E. Luchinat and L. Banci, J. Am. Chem. Soc., 2023, 145, 1389–1399 CrossRef CAS PubMed.
- R. Hänsel, S. Foldynová-Trantírková, F. Löhr, J. Buck, E. Bongartz, E. Bamberg, H. Schwalbe, V. Dötsch and L. Trantírek, J. Am. Chem. Soc., 2009, 131, 15761–15768 CrossRef PubMed.
- S. Doose, H. Neuweiler and M. Sauer, ChemPhysChem, 2009, 10, 1389–1398 CrossRef CAS PubMed.
- H. S. Hayatshahi, N. M. Henriksen and T. E. Cheatham, J. Chem. Theory Comput., 2018, 14, 1456–1470 CrossRef CAS.
- N. J. Greco and Y. Tor, J. Am. Chem. Soc., 2005, 127, 10784–10785 CrossRef CAS.
- J. Riedl, R. Pohl, L. Rulíšek and M. Hocek, J. Org. Chem., 2012, 77, 1026–1044 CrossRef CAS PubMed.
- H. Sapper and W. Lohmann, Biophys. Struct. Mech., 1978, 4, 327–335 CrossRef CAS PubMed.
- C. S. Giam and J. L. Lyle, J. Am. Chem. Soc., 1973, 95, 3235–3239 CrossRef CAS.
- J. N. Dahanayake, C. Kasireddy, J. P. Karnes, R. Verma, R. M. Steinert, D. Hildebrandt, O. A. Hull, J. M. Ellis and K. R. Mitchell-Koch, Annu. Rep. NMR Spectrosc., 2018, 93, 281–365 CrossRef CAS.
- Y. M. Vianney and K. Weisz, Nucleic Acids Res., 2022, 50, 11948–11964 CrossRef CAS PubMed.
- M. L. Greco, A. Kotar, R. Rigo, C. Cristofari, J. Plavec and C. Sissi, Nucleic Acids Res., 2017, 45, 10132–10142 CrossRef CAS PubMed.
- M. Yang, S. Carter, S. Parmar, D. D. Bume, D. R. Calabrese, X. Liang, K. Yazdani, M. Xu, Z. Liu, C. J. Thiele and J. S. Schneekloth, Nucleic Acids Res., 2021, 49, 7856–7869 CrossRef CAS PubMed.
- S. Y. Khatik, S. Sudhakar, S. Mishra, J. Kalia, P. I. Pradeepkumar and S. G. Srivatsan, Chem. Sci., 2023, 14, 5627–5637 RSC.
- R. K. R. Sannapureddi, M. K. Mohanty, L. Salmon and B. Sathyamoorthy, J. Am. Chem. Soc., 2023, 145, 15370–15380 CrossRef CAS PubMed.
- L. Díaz-Casado, I. Serrano-Chacón, L. Montalvillo-Jiménez, F. Corzana, A. Bastida, A. G. Santana, C. González and J. L. Asensio, Chem.–Eur. J., 2021, 27, 6204–6212 CrossRef.
- T. Q. N. Nguyen, K. W. Lim and A. T. Phan, Sci. Rep., 2017, 7, 11969 CrossRef PubMed.
- S. Mandal, Y. Kawamoto, Z. Yue, K. Hashiya, Y. Cui, T. Bando, S. Pandey, M. E. Hoque, M. A. Hossain, H. Sugiyama and H. Mao, Nucleic Acids Res., 2019, 47, 3295–3305 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details, CD, Tm, fluorescence, MD simulation, mass, NMR spectra and gel images. Experimental procedure for NMR measurements of ONs in a cellular environment, and ligand interaction with LTR GQs and their effects on the Taq DNA polymerase activity are described. See DOI: https://doi.org/10.1039/d4sc01755b |
| This journal is © The Royal Society of Chemistry 2024 |