Open Access Article
Open Access ArticleSingle-cell mass cytometry reveals cell type- and cluster-specific heterogeneity in silver nanoparticle responses in a 3D alveolar tetra-culture model†
Eunseo
Lee
 a,
Seung-Geun
Park
a,
Seung-Geun
Park
 a,
Seung Min
Ha
a,
Seung Min
Ha
 a,
Minseop
Kim
a,
Minseop
Kim
 a,
Sehee
Park
a,
Sehee
Park
 b,
Aline
Chary
b,
Aline
Chary
 c,
Tommaso
Serchi
c,
Tommaso
Serchi
 c and
Tae Hyun
Yoon
c and
Tae Hyun
Yoon
 *adefg
*adefg
aDepartment of Chemistry, Hanyang University, Seoul 04763, Republic of Korea. E-mail: taeyoon@hanyang.ac.kr
bInfectious Diseases Therapeutic Research Center, Korea Research Institute of Chemical Technology, Daejeon 34114, Republic of Korea
cLuxembourg Institute of Science and Technology (LIST), 41 rue du Brill, L-4422 Belvaux, Luxembourg
dInstitute of Next Generation Material Design, Hanyang University, Seoul 04763, Republic of Korea
eYoon Idea Lab. Co. Ltd., Seoul 04763, Republic of Korea
fResearch Institute for Convergence of Basic Science, Hanyang University, Seoul 04763, Republic of Korea
gDepartment of Medical and Digital Engineering, Hanyang University, Seoul 04763, Republic of Korea
First published on 15th July 2025
Abstract
Silver nanoparticles (AgNPs) are widely used in medicine, environmental science, and industry. However, their heterogeneous interactions with complex biological systems, especially at the single-cell level, are not fully understood yet. Conventional toxicity assessment methods are typically conducted on oversimplified in vitro models that fail to replicate actual physiological conditions, and measure collective responses across cell populations, obscuring differences among individual cells. To overcome these limitations, we utilized single-cell mass cytometry (CyTOF) to investigate individual cell responses to AgNP-induced stress, combined with a 3D alveolar tetra-culture model designed to better reflect the complexity of biological systems. Single-cell mass cytometry of a 3D alveolar model revealed heterogeneous, cell type–specific responses to AgNP exposure. Specifically, PMA-differentiated THP-1, A549 and EA.hy926 cells exhibited high AgNP association but limited cytotoxicity, indicating activation of stress-mitigation pathways, while THP-1 cells showed early inflammatory activation despite minimal AgNP association, suggesting an indirect mechanism. Single-cell analysis and FlowSOM clustering revealed distinct subpopulations exhibiting diverse intracellular signaling profiles of inflammatory cytokines, anti-inflammatory mediators, and stress-response proteins, which unveiled common cellular responses and unique cell-type specific pathways determining cell fate (survival, transitional states, or apoptosis) upon AgNP exposure. This study introduces a novel framework for studying heterogeneous interactions of nanoparticles with complex biological systems by integrating a 3D alveolar tetra-culture model with single-cell mass cytometry analysis, enabling the dissection of nanoparticle-induced stress responses at an unprecedented level of detail. These insights have broad implications for nanotoxicology and nanomedicine, underscoring the need to account for cellular heterogeneity when evaluating nanoparticle-induced toxicity.
Environmental significanceConventional nanotoxicology often overlooks how different cell types and subpopulations respond within complex tissues, which may lead to underestimating health risks in realistic exposure scenarios. We addressed this by integrating a 3D alveolar tetra-culture model with single-cell mass cytometry to study silver nanoparticle (AgNP) effects in a physiologically relevant system. Our study suggests that toxicity is not solely determined by nanoparticle dose but also shaped by cell-intrinsic pathways and intercellular signals. Using hierarchical clustering of single-cell data, this approach provides a systematic framework to identify shared and cell-specific stress responses, capturing heterogeneity that bulk assays often miss. These insights emphasize the need to consider biological complexity when assessing the environmental and human health risks of nanomaterials. |
Introduction
Nanoparticles (NPs) have been extensively applied in biomedical applications, environmental systems, and industrial processes, including drug delivery, diagnostic imaging, and catalysis.1–3 Among them, silver nanoparticles (AgNPs) are widely studied because of their potent antimicrobial properties and broad applications in biomedical and consumer products.4–7 However, their interactions with biological systems are not fully understood, particularly in the context of cellular heterogeneity.8–10 While AgNPs have been reported to induce oxidative stress, inflammatory responses, and apoptosis,11,12 the extent to which these pathways vary at the single-cell level is often oversimplified. Most toxicity studies rely on bulk-level measurements that obscure cell-to-cell variability, failing to capture how individual cells respond to nanoparticle-induced stress.13,14 Given this inherent heterogeneity, single-cell approaches are needed to characterize nanoparticle-induced stress responses at the level of individual cells.Single-cell analysis techniques have emerged as powerful tools to address these challenges, enabling high-resolution characterization of cellular stress responses. While single-cell RNA sequencing (scRNA-seq) has been employed to characterize cellular heterogeneity, its ability to quantify nanoparticle association at the single-cell level is limited. Flow cytometry, another single-cell method, allows for multiparametric analysis of intracellular markers but is constrained by spectral overlap, which limits the number of markers that can be analyzed simultaneously.10,15–17 Whereas scRNA-seq profiles more than thousands of transcripts in each cell, mass cytometry (CyTOF) focus on proteome—simultaneously measuring roughly 40–60 metal-tagged surface and intracellular proteins, including diverse post-translational modifications—as well as the quantification of cellular dose of nanoparticles. Used together, these platforms provide complementary insight into how nanoparticle dosing shapes cell-type-specific outcomes such as survival or apoptosis.18,19
Despite the molecular-level insights offered by single-cell analysis, conventional in vitro models—particularly two-dimensional (2D) monolayer cultures—fail to capture the structural complexity and diffusion dynamics present in vivo.20–22 2D culture models often assume uniform nanoparticle exposure across a homogenous cell population, neglecting the influence of cell-type-specific differences on nanoparticle interactions with cells. To overcome these limitations, researchers have increasingly adopted three-dimensional (3D) culture models to better replicate physiological conditions and tissue architecture.20–22 However, many existing 3D culture studies have analyzed only a subset of cell types without employing gating strategies to distinguish and assess individual populations. While a few recent studies have begun to apply flow-cytometric gating in 3D culture models,23 these approaches have not yet, to our knowledge, been extended to high-dimensional mass cytometry. Similarly, most analyses assume uniform nanoparticle distribution, overlooking the heterogeneous exposure patterns present in multicellular systems.24
In this study, we used 3D alveolar tetra-culture model,21 to more accurately mimic the alveolar microenvironment, incorporating multiple cell types with distinct spatial distributions. Unlike prior approaches that treat the cellular environment as uniform, this model replicates in vivo-like exposure scenarios where epithelial, endothelial, and immune cells experience different nanoparticle interactions.12,25 By integrating nanoparticle diffusion, stress adaptation, inflammatory signaling, and apoptosis, this model provides a mechanistic framework for dissecting nanoparticle-induced cytotoxicity.
We employed mass cytometry combined with hierarchical clustering to profile subpopulation-level responses. Previous studies have identified immune cell populations that respond to nanoparticle exposure,9,26 but they have not systematically linked specific cellular subpopulations to distinct stress response pathways and fate decisions. This study bridges this gap, providing a more detailed, mechanistic view of AgNP-induced cellular heterogeneity at single-cell resolution. This approach enables us to determine how different cell subpopulations engage common and unique responses, uncovering how intracellular signaling dynamics dictate survival or apoptosis upon nanoparticle exposure.
By systematically mapping nanoparticle-induced heterogeneity at single-cell resolution, this study not only enhances our understanding of AgNP cytotoxicity but also introduces a novel framework for evaluating nanoparticle safety under physiologically relevant conditions. These findings have broad implications for nanotoxicology and nanomedicine, where accounting for inter- and intracellular variability is crucial for predicting real-world biological responses.
Experimental
Cell culture
Human umbilical vein-derived endothelial EA.hy926 cells (ATCC, Cat. No. CRL-2922, USA), human alveolar epithelial A549 cells (Korean Cell Line Bank, Cat. No. 10185, Korea), human monocytic THP-1 cells (Korea Cell Line Bank, Cat. No. 40202, Korea), and phorbol myristate acetate (PMA)-differentiated THP-1 cells were cultured to model the alveolar barrier. EA.hy926 and A549 cells were maintained in Dulbecco's modified Eagle medium (DMEM; Gibco, Cat. No. 61965) supplemented with 10% fetal bovine serum (FBS; Gibco, Cat. No. 16000-044, USA) and 1% penicillin–streptomycin (Pen-Strep; Gibco, Cat. No. 15140-122), while THP-1 cells were cultured in RPMI-1640 (GenDEPOT, Cat. No. CM058-050, USA) supplemented with 10% FBS and 1% Pen-Strep. To include both monocytic and macrophage-like THP-1 cells, a portion of the THP-1 cells were differentiated into macrophages by treatment with 100 nM PMA (Sigma Aldrich, Cat. No. P1585, USA). The culture medium was replaced with fresh medium for 2 days, and cells were incubated for an additional 4 days.3D alveolar tetra-culture model setup
The 3D alveolar model was established to mimic the human alveolar barrier.21 A 6-well insert (cellQART, SABEU GmbH & Co. KG, Germany; Cat. No. 9305012; 5 μm pore size, PET membrane) was used to separate the apical and basolateral compartments, enabling the precise seeding of distinct cell types. The cells positioned in the apical compartment simulated the airway-facing surface, whereas those in the basolateral compartment, which was exposed to culture medium, represented the underlying tissue environment. To establish the alveolar barrier, EA.hy926 cells were seeded onto inverted inserts at a density of 1.8 × 105 cells per mL and allowed to attach for 4 hours, ensuring their adhesion to the basolateral surface. The inserts were then returned to their upright orientation, and A549 cells were seeded onto the newly exposed surface at a density of 1.4 × 105 cells per mL, corresponding to the apical compartment. Following a 3-day incubation period to facilitate barrier formation, THP-1 cells were added to the basolateral compartment at 1 × 106 cells per mL, while PMA-differentiated THP-1 cells were seeded into the apical compartment at 1.6 × 105 cells per mL to simulate the presence of the immune system in the alveolar microenvironment. After a 4-hour attachment period, the medium in the apical compartment was removed to establish air–liquid interface (ALI) conditions. The final model setup, along with the sample preparation and data acquisition workflow, is illustrated in Fig. 1.Silver nanoparticles
The silver nanoparticles (AgNPs) used in this study were 10 nm silver nanospheres (nanoComposix, Cat. No. AGCB10-1 M, USA), stabilized with sodium citrate dihydrate. The stock suspension was provided in aqueous solution at a silver mass concentration of 1.05 mg mL−1. To ensure physiologically relevant interpretation of nanoparticle interactions, the AgNPs were comprehensively characterized in line with the GUIDEnano quality assessment framework27 and previous studies highlighting the importance of high-quality physicochemical data.28 Their physicochemical properties are presented in ESI† methods, Fig. S1, S2 and Tables S1, S4, S5.Silver nanoparticle exposure
To model inhalation exposure, AgNPs were applied directly onto the apical compartment, simulating nanoparticle deposition onto the alveolar epithelium. The stock suspension was sonicated in an ultrasonic bath (SD-300H, Seongdong Ultrasonic Co., Korea) for 1 minute, then diluted in culture medium to a final concentration of 2 μg mL−1. This corresponds to approximately 0.44 μg cm−2 on the insert surface, which lies within the physiologically relevant doses (0.05–5 μg cm−2) reported in prior models.29 This dose with an exposure time of 24 hours have been shown to induce oxidative stress and inflammatory gene responses (HMOX-1, NF-κB) without overt cytotoxicity.29 The suspension was vortexed to ensure uniform dispersion before application. Control wells received particle-free medium as a negative control. After 24 hours of exposure, THP-1 suspension cells were collected from the supernatant. Adherent cells (A549, EA.hy926, and PMA-differentiated THP-1) were washed 2 times with Dulbecco's phosphate-buffered saline (DPBS; Welgene, Cat. No. LB001-02, Korea) to remove residual nanoparticles before being detached with 0.25% Trypsin-EDTA (Gibco, Cat. No. 25200056, USA). The cells were incubated at 37 °C for 5 minutes with Trypsin-EDTA and collected by centrifugation at 200 × g for 5 minutes at room temperature (RT). The supernatant was discarded, and the resulting cell pellet was prepared for downstream analysis.Sample preparation for mass cytometry
The suspension and adherent cell populations were processed independently throughout all the preparation steps including fixation, staining, and antibody labeling. The cell pellet was resuspended in 2 mL of DPBS and washed twice to remove residual media and reagents. The cells were stained with cisplatin (Standard BioTools Inc., Cat. No. 201064, USA) to assess cell viability. The cells were then incubated with a cocktail of metal-tagged antibodies targeting surface markers for 30 minutes at RT. After staining, the cells were fixed with 1.6% paraformaldehyde (Electron Microscopy Sciences, Cat. No. 15710, USA) in DPBS for 20 minutes at RT. Phosphoprotein and cell cycle markers were then applied following the Maxpar Phosphoprotein Staining with Fresh Fix Protocol (Standard BioTools Inc., USA). The specific markers used, all purchased from Standard BioTools Inc., USA, are listed in Table S2.† Finally, the cells were stained with Cell-ID Intercalator-Ir (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 in DPBS; Standard BioTools Inc., USA) for nuclear DNA labeling and stored at 4 °C until acquisition.
1000 in DPBS; Standard BioTools Inc., USA) for nuclear DNA labeling and stored at 4 °C until acquisition.
Mass cytometry data acquisition
Mass cytometry was performed using a Helios™ mass cytometer (Standard BioTools Inc., USA), equipped with a high-throughput sample introduction system to optimize the data acquisition efficiency. Prior to sample acquisition, instrument calibration was performed using EQ™ calibration beads (Standard BioTools Inc., Cat. No. 201078, USA). Each sample was rewashed and diluted in cell acquisition solution (Standard BioTools Inc., Cat. No. 201240, USA) to a concentration of approximately 1 × 106 cells per mL. Immediately before analysis, the cells were filtered through strainer-capped tubes to prevent clogging. Finally, the samples were introduced to a Helios™ mass cytometer and data were acquired. Three independently prepared samples were analyzed to assess reproducibility across biological replicates.Data analysis
As outlined in Fig. 1, the data analysis pipeline consisted of preprocessing and downstream analysis to characterize AgNP-induced cellular responses. The mass cytometry data were normalized using CyTOF software (v7.0.5189, Standard BioTools Inc., USA) according to the manufacturer's recommendations. Preprocessing included variance-stabilizing transformation via the inverse hyperbolic sine (arcsinh) function, ensuring consistent marker intensity scaling. To ensure data quality, non-bead and singlet events were manually gated in FlowJo (v10.10.0, FlowJo LLC, USA). Manual gating was also used to distinguish four cell populations—PMA-differentiated THP-1, A549, EA.hy926, and THP-1 cells (Fig. S3†). Cell type proportions were calculated based on cell counts and normalized to 100% per sample (Table S3†). To investigate single-cell variability in AgNP responses, uniform manifold approximation and projection (UMAP) was applied to visualize subpopulation distributions and AgNP association trends across over 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 single-cell events. The quantification of AgNP association was performed using calibrated ion standards, with transport efficiency (TE) calculations ensuring accurate single-cell metal quantification. To identify functionally distinct subpopulations, flow self-organizing map (FlowSOM), a clustering algorithm that integrates hierarchical relationships, was employed to identify cellular subpopulations. While key phenotypic parameters such as AgNP association, cell viability, and cell-type distribution were evaluated across three independent biological replicates (Fig. S4†), the UMAP and FlowSOM analyses shown in Fig. 3 were derived from a single representative sample, selected based on its high cell yield. To compare marker expression patterns between FlowSOM clusters, heatmaps were generated in R (v4.4.0, R Core Team, Austria) using the ComplexHeatmap package (v2.20.0). These analytical methods collectively allow high-resolution profiling of shared and unique regulatory pathways governing the inflammation response, stress adaptation, and apoptosis across different cell types.
000 single-cell events. The quantification of AgNP association was performed using calibrated ion standards, with transport efficiency (TE) calculations ensuring accurate single-cell metal quantification. To identify functionally distinct subpopulations, flow self-organizing map (FlowSOM), a clustering algorithm that integrates hierarchical relationships, was employed to identify cellular subpopulations. While key phenotypic parameters such as AgNP association, cell viability, and cell-type distribution were evaluated across three independent biological replicates (Fig. S4†), the UMAP and FlowSOM analyses shown in Fig. 3 were derived from a single representative sample, selected based on its high cell yield. To compare marker expression patterns between FlowSOM clusters, heatmaps were generated in R (v4.4.0, R Core Team, Austria) using the ComplexHeatmap package (v2.20.0). These analytical methods collectively allow high-resolution profiling of shared and unique regulatory pathways governing the inflammation response, stress adaptation, and apoptosis across different cell types.
Statistical analysis
All analyses were conducted in R 4.4.0. Reproducibility of FlowSOM metacluster profiles across biological replicates was assessed by computing pairwise Spearman's rank correlation coefficients of Z-scored marker means in stats package (v4.4.0) and visualized with corrplot package (v0.95). Cell-type dependence of fate distributions was evaluated by Fisher's exact test to confirm significant differences in fate composition across cell types.Results and discussion
Cell type-specific AgNP association and cytotoxicity in a 3D alveolar model
To elucidate the distinct phenotypes of individual cells within the 3D alveolar model, UMAP was employed to visualize cellular heterogeneity at single-cell resolution. Using the manual-gating strategy described in Fig. S3,† four distinct cell types comprising the model—PMA-differentiated THP-1, A549, EA.hy926, and THP-1—were distinguished and overlaid in UMAP. To capture cell type-specific characteristics based on surface markers and cell states, we used UMAP with six surface markers, three cell cycle markers, and a viability marker (Table S2†). By grouping cells with similar characteristics, UMAP facilitates the visualization of high-dimensional mass cytometry data. Overlaying manually gated cells onto the UMAP plot showed four distinct cellular regions, where green, orange, blue, and red correspond to the manually gated cell types (A549, EA.hy926, THP-1, and PMA-differentiated THP-1, respectively) as shown in Fig. 2a. Notably, PMA-differentiated THP-1 and THP-1 cells formed spatially proximate clusters, reflecting their phenotypic similarity.30To explore the variability in AgNP association and cytotoxicity across distinct cell types within the 3D alveolar model, we performed qualitative comparisons between negative control (NC) and AgNP-treated (AgNPs) samples. Cytotoxicity was represented by cisplatin intensity. Given that cisplatin penetrates cells with compromised membrane integrity, higher cisplatin intensity indicates reduced cell viability.31,32 UMAP plots overlaid with 107Ag and cisplatin intensity revealed distinct patterns of AgNP association and cytotoxicity among different cell types as shown in Fig. 2b. High levels of 107Ag (represented by red regions) were observed in PMA-differentiated THP-1, A549, and EA.hy926 cells, which are in the apical compartment of the insert and are thus directly exposed to nanoparticles. In contrast, THP-1 cells, located in the basolateral compartment, showed relatively low AgNP association (blue regions), likely due to limited nanoparticle penetration through the epithelial–endothelial barrier formed on the insert.
Although THP-1 cells exhibited low AgNP association, they showed high cisplatin uptake, suggesting that their cytotoxicity may not solely depend on direct association of AgNPs. Instead, this increased toxicity could result from indirect inflammatory signaling, as THP-1 cells are highly sensitive to stress mediators released by neighboring cells,33,34 or from Ag+ ion-mediated toxicity. As AgNPs can release Ag+ ions upon dissolution, these ions may diffuse across compartments and induce cytotoxic effects in THP-1 cells independent of direct nanoparticle association. Indeed, previous studies have shown that AgNPs release Ag+ in serum-containing media and that silver can be detected in basolateral compartments in similar models, although the exact chemical form of the translocated species remains unsolved.35,36 To address this, it is necessary to conduct further investigations that measure Ag+ concentrations and evaluate their impacts within the apical and basolateral compartments. Within the THP-1 population, the subpopulations displayed varying degrees of sensitivity to AgNP-induced stress, as indicated by differential cisplatin uptake intensities. While some THP-1 cells were highly reactive and exhibited significant stress responses, others remained relatively resistant to AgNP-induced damage. In contrast, A549, EA.hy926, and PMA-differentiated THP-1 cells demonstrated a greater capacity to maintain viability despite AgNP exposure, exhibiting lower cisplatin uptake levels. This observation indicates that these cell types may engage in cellular repair or survival mechanisms, mitigating the cytotoxic effects of AgNPs.37–39
Box plots (shown in Fig. 2c) were generated to quantitatively compare AgNP association and cisplatin uptake among different cell types in the 3D alveolar model, complementing the qualitative data shown in Fig. 2b. The analysis revealed distinct variations in AgNP association and cytotoxicity among cell types, further supporting the observations from UMAP plots. THP-1 cells exhibited the lowest AgNP association, with a mean value of 32.1 femtograms per cell, which aligns with the qualitative findings from the UMAP plots. However, the interquartile range (25th to 75th percentile) of cisplatin intensities within THP-1 cells (116.5 to 1647.1) suggests that subpopulations have differential sensitivity to nanoparticle-induced stress, potentially driven by indirect inflammatory signaling.33,34
In contrast, A549 and EA.hy926 cells showed significantly higher AgNP association, with mean values of 421.9 and 392.1 fg per cell, respectively, approximately 12-fold greater than those of THP-1 cells. Despite this high AgNP association, most A549 and EA.hy926 cells maintained membrane integrity and exhibited relatively low cisplatin uptake, suggesting activation of stress mitigation pathways, such as ERK-mediated survival signaling or antioxidant defense mechanisms.40–42 PMA-differentiated THP-1 cells, although representing a relatively small proportion of the population, displayed the highest AgNP association, with a mean value of 901.5 fg per cell. This elevated association aligns with their phagocytic nature, which facilitates enhanced nanoparticle association, and may also activate intracellular processing mechanisms that help regulate cytotoxic effects.31
The cells positioned within the apical compartment, which were directly exposed to AgNPs, exhibited greater resilience and maintained viability. These findings suggest that these cells may have developed adaptive mechanisms, such as upregulation of stress–response proteins or efficient antioxidant pathways, to mitigate nanoparticle-induced cytotoxicity. In contrast, cells in the basolateral compartment, where direct exposure to nanoparticles was limited, exhibited higher cisplatin uptake, suggesting that indirect signaling mechanisms, rather than direct nanoparticle exposure, contributed to their cytotoxic responses.
Our findings reveal that cellular heterogeneity plays a critical role in understanding stress adaptation and apoptotic pathways in response to AgNP exposure. The extent of nanoparticle association and the cellular location within the 3D alveolar model influence these differential responses. This variability is driven by both intrinsic cellular mechanisms—such as activation of specific stress response pathways—and external signals from neighboring cells. These results underscore the importance of single-cell analysis in capturing the complexity of nanoparticle-induced cytotoxicity.
Subpopulation-specific AgNP association and cytotoxicity in a 3D alveolar model
To further investigate the heterogeneity of cellular responses to AgNP exposure at the single-cell level, biaxial plots were generated to visualize the relationship between AgNP association and cytotoxicity. The FlowSOM clustering algorithm was applied to the manually gated cell types to distinguish cellular subpopulations within each cell type, leveraging distinct marker expression profiles to capture nuanced differences in stress responses. This analysis incorporated surface markers, cell cycle markers, intracellular markers, and a viability marker as clustering parameters. FlowSOM, utilizing a minimum-spanning tree (MST) algorithm, revealed subpopulations of each cell type with distinct intracellular characteristics that were not distinguishable through manual gating.The number of metaclusters for each cell type was determined through iterative FlowSOM clustering, with the optimal number selected based on consensus clustering stability,43 as visualized in the delta area plot (Fig. S5†). The number of metaclusters for THP-1 cells was set higher than the optimal value because of the pronounced heterogeneity observed in AgNP association and cisplatin uptake, ensuring a more precise representation of subpopulations. The hierarchical relationships identified between these FlowSOM metaclusters are visualized on the MST (Fig. S6†). Although the FlowSOM clustering results were obtained from a single representative sample, high pairwise Spearman correlations demonstrate consistent cluster profiles across biological replicates (Fig. S7†). Each biaxial plot in Fig. 3 displays these FlowSOM clusters overlaid on the distribution of AgNP association and cisplatin uptake for PMA-differentiated THP-1, A549, EA.hy926, and THP-1 cells.
In PMA-differentiated THP-1 cells, four FlowSOM clusters (FS#P1 to FS#P4) were identified, each displaying distinct stress response patterns. Despite their high AgNP association (Fig. 3a), FS#P1 and FS#P2 exhibited low cisplatin uptake, suggesting that these subpopulations engage mechanisms to preserve membrane integrity despite elevated nanoparticle association. These results align with previous reports on macrophage resilience to metal nanoparticle-induced oxidative stress.39
Similarly, six FlowSOM clusters (FS#A1 to FS#A6) were identified within the A549 population, and four clusters (FS#E1 to FS#E4) were identified within the EA.hy926 population. Notably, FS#A3 and FS#E3 exhibited high cytotoxicity, whereas the other clusters displayed low cytotoxicity despite a wide range of AgNP association levels. These findings indicate that A549 and EA.hy926 subpopulations exhibit varying levels of susceptibility to AgNP-induced cytotoxicity, with some cells maintaining viability even at high levels of nanoparticle association.
In contrast, the THP-1 population demonstrated pronounced heterogeneity, with eight distinct clusters (FS#T1 to FS#T8). Among these, FS#T2, FS#T3, FS#T7, and FS#T8 exhibited high cytotoxicity, with cellular AgNP association ranging from low to high levels. This broad distribution suggests that THP-1 cells can exhibit significant stress responses even at relatively low nanoparticle association. This finding, when considered alongside the results from Fig. 2, suggests the possibility that THP-1 cells are more susceptible to indirect stress signaling, although further investigation is needed to confirm this hypothesis.
Interestingly, despite sharing similar cellular characteristics and spatial locations within the 3D alveolar model, different subpopulations within the same cell type exhibited distinct patterns of AgNP association and cisplatin uptake. This observation suggests that each subpopulation may follow different survival or apoptosis pathways, depending on its response to AgNP-induced stress. Notably, the high heterogeneity observed in THP-1 cells underscores their diverse stress responses, which can be revealed only through single-cell-level analysis or intracellular signaling, demonstrating the importance of this approach in elucidating cellular heterogeneity.
Dissecting cluster-specific intracellular signaling pathways governing cellular responses to AgNP exposure
Exposure to AgNPs often induces both inflammatory and cytotoxic responses, necessitating an integrated analysis of stress adaptation, inflammation, and apoptosis mechanisms.12 FlowSOM clustering was used to categorize cells based on intracellular marker expression, allowing for a high-resolution analysis of how different clusters respond to AgNP exposure. The heterogeneity of the subpopulations within FlowSOM metaclusters was visualized through a MST (Fig. S8†) showing the expression of stress, inflammatory, and apoptotic markers. The hierarchical relationships among these clusters were visualized using heatmaps (Fig. 4), which illustrate patterns of intracellular signaling and their varying levels of AgNP association. Heatmaps display the relative expression of key markers, including inflammatory cytokines (IL-6, TNF-α, and IFN-γ), an anti-inflammatory regulator (IL-4), an apoptotic marker (cleaved caspase-7), and intracellular stress response proteins (pERK1/2 and pBad), along with AgNP association (107Ag) and cytotoxicity (cisplatin). This analysis aimed to uncover key intracellular mechanisms that regulate cell fate decisions between survival and apoptosis following AgNP exposure.Among PMA-differentiated THP-1 cells, distinct clusters demonstrated contrasting responses depending on AgNP association and intracellular marker expression, highlighting the complex balance between inflammation and anti-inflammation. FS#P1 exhibited relatively high AgNP association but low expression of IL-6, TNF-α, IFN-γ, and pERK1/2, with elevated IL-4 levels. These findings suggest that anti-inflammatory signaling mitigated excessive cellular stress, enabling cells to maintain low cytotoxicity despite nanoparticle exposure. These findings align with previous findings that certain AgNP formulations selectively suppress TNF-α and IL-6 expression in macrophages.12 Additionally, previous studies have suggested that nanoparticle association may enhance macrophage survival by inhibiting apoptosis through lysosomal signaling, without triggering pro-inflammatory effects.12,39 FS#P2 exhibited a transitional apoptotic state, characterized by elevated TNF-α, IFN-γ, pERK1/2, and pBad expression while maintaining relatively low cisplatin uptake. The concurrent activation of cleaved caspase-7 suggested that these cells had entered the intrinsic apoptosis pathway while preserving membrane integrity, positioning them at the boundary between survival and apoptosis.44 Conversely, FS#P4 showed a complete transition to apoptosis, with high expression of IL-6, TNF-α, IFN-γ, pERK1/2, and pBad, ultimately driving cell death. These findings suggest that the combined activation of inflammatory signaling and intracellular stress responses may contribute to cell death. Similar to previous reports, AgNP exposure induces inflammatory cytokine upregulation and oxidative stress in macrophages, potentially leading to apoptosis through NF-κB and MAPK pathways.45,46
A549 cells, despite their epithelial barrier nature, exhibited varying responses to AgNP exposure. FS#A2 and FS#A6 maintained low expression of all the markers, including pERK1/2, despite high AgNP association, resulting in low cytotoxicity. These findings suggest that reduced ERK pathway activation may have allowed these cells to tolerate nanoparticle-induced stress without progressing to apoptosis, potentially reflecting their epithelial barrier function.12 FS#A3, however, displayed moderate pBad expression and increased cytotoxicity, suggesting that while pBad typically supports survival, excessive stress can override its protective function, leading to membrane damage.47 This observation suggests that cytotoxicity may have been driven by alternative, dominant cell-death pathways such as endoplasmic reticulum (ER) stress. For example, ER stress can induce transcriptional changes via the CHOP pathway that shift the balance of Bcl-2 family proteins toward apoptosis.48,49 These mechanisms offer an explanation for the observed membrane damage despite moderate pBad expression. FS#A5, which exhibited high IFN-γ and IL-4 expression, maintained low cytotoxicity, suggesting that the anti-inflammatory effect of IL-4 counterbalanced IFN-γ-induced inflammation. However, cleaved caspase-7 expression indicated that these cells had begun transitioning toward apoptosis. Notably, FS#A1 and FS#A4 showed similar marker profiles to FS#P2 (THP-1), all of which exhibited high cleaved caspase-7 expression while maintaining low cisplatin uptake. These findings suggest that although A549 and PMA-differentiated THP-1 cells differ in their biological functions, they can activate comparable apoptotic signaling pathways under specific AgNP exposure conditions.
Compared with other cell types, EA.hy926 cells showed unique inflammatory responses. FS#E3 displayed high IL-6 and TNF-α expression, leading to elevated cytotoxicity. Unlike A549 and THP-1 cells, where apoptotic pathways are influenced by intracellular stress responses, endothelial cells are more susceptible to inflammation-induced cell death. These findings align with previous findings showing that TNF-α-mediated endothelial activation leads to increased vascular permeability and programmed cell death under oxidative stress conditions.50 FS#E1 represented a mixed response characterized by the activation of both inflammatory and intracellular signaling pathways. The presence of high cleaved caspase-7 and low cytotoxicity indicate a transitional phase similar to that observed in other cell types.
THP-1 cells also exhibited distinct responses to AgNP exposure, ranging from stress adaptation and survival to progressive stages of cellular damage and death. FS#T6 displayed pBad activation with suppressed inflammatory responses, enabling cells to endure high AgNP association without entering apoptosis. This cluster may represent a stress-adapted phenotype capable of resisting nanoparticle-induced cytotoxicity. Notably, FS#T1 exhibited selective TNF-α expression with low expression of other markers and low cisplatin uptake, suggesting an early-stage inflammatory response unique to THP-1 cells. These findings suggest that monocyte-mediated immune activation precedes apoptosis,26 potentially triggering a paracrine signaling that modulates the response of neighboring cells. FS#T2 and FS#T3 represented the advanced stages of cell death, with high cisplatin uptake but low expression of stress adaptation markers, indicating that accumulated damage had surpassed the repair capacity of the cells. Moreover, FS#T8 displayed high IL-4 expression alongside increased cisplatin uptake, suggesting that while IL-4 initially helped suppress inflammation, prolonged stress ultimately led to membrane damage and cell death.
Unraveling common and distinct intracellular signaling pathways in AgNP-mediated cell fate decisions
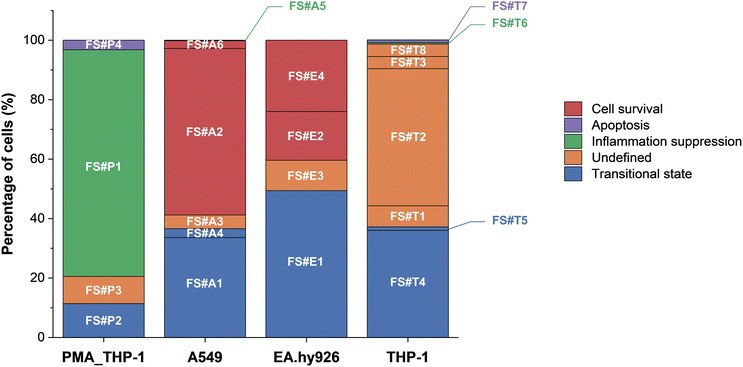
To systematically understand how AgNP exposure affects cell fate, we analyzed common and unique intracellular signaling patterns across all FlowSOM clusters. A general decision-making flow chart was constructed based on pathways that were shared by at least two different cell types, illustrating common responses rather than cell type-specific pathways, leading to survival, transitional states, or apoptosis (Fig. 5). In this context, we defined transitional states as cell clusters exhibiting apoptotic signals—such as caspase-7 activation or, under certain stress conditions, ERK pathway activation—while retaining membrane integrity, indicating that the cells remained in a reversible, pre-apoptotic state without being fully committed to apoptosis.The framework highlights key decision points, such as early inflammatory signaling and stress adaptation, which are associated with apoptotic or survival outcomes, although additional factors may influence final fate decisions. In addition to these common pathways, clusters that did not align with any commonly observed patterns were classified as undefined, indicating responses that appeared unique to a specific cell type. The presence of these undefined clusters suggests that additional regulatory factors, such as cell cycle state or alternative stress response pathways, could play a role in influencing cell fate.51,52 These undefined clusters, along with other clusters associated with cell survival, anti-inflammatory cell survival, transitional states, and apoptosis, are represented in the distribution of cell fate outcomes (Fig. 6).
A key observation from our analysis is that inflammatory signaling may play an important role in determining early cell fate following AgNP exposure. Clusters such as FS#A2, FS#A6, and FS#E4 exhibited minimal expression of pro-inflammatory markers (e.g., TNF-α and IFN-γ) despite relatively high AgNP association and maintained membrane integrity. Although further functional assays are needed to confirm whether reduced inflammation directly contributes to decreased cytotoxicity, this observation suggests that these subpopulations may resist nanoparticle-induced stress through intracellular adaptation mechanisms. In contrast, other clusters (e.g., FS#P2, FS#A4, FS#E1, and FS#T5) showed sustained inflammatory responses, which coincided with elevated levels of cleaved caspase-7. These results align with previous studies linking chronic inflammation and oxidative stress to activation of the intrinsic apoptotic pathway.36 Additionally, IL-4 expression was observed in clusters such as FS#P1 and FS#A5, where low cisplatin uptake was also noted, suggesting that anti-inflammatory signaling contributes to limiting inflammatory damage and promoting cell survival.
In other clusters, inflammation did not appear to be the dominant driver of cytotoxic outcomes. Instead, intracellular stress response pathways, particularly those involving pERK1/2 and pBad, have been implicated in regulating cellular fate.41 For example, FS#P2 displayed activation of both pERK1/2 and pBad alongside low levels of cisplatin uptake, suggesting a transitional state characterized by early apoptotic signaling without full loss of membrane integrity. In contrast, FS#P4 and FS#T7 exhibited elevated expression of these stress markers together with increased cisplatin uptake, indicative of progression toward late-stage apoptosis involving membrane disruption. Taken together, these findings highlight the multifaceted nature of AgNP-induced stress responses, in which both inflammatory signaling and intracellular stress adaptation collectively shape divergent cell fate trajectories.
We quantified the distribution of cell fate outcomes by classifying each FlowSOM cluster into inflammation suppression, survival, transitional, apoptosis, or undefined states. The resulting proportions are illustrated in the bar graph (Fig. 6). The results indicate statistically significant cell-type-specific differences in fate outcomes (Fisher's exact test, p = 0.0005). A549 and EA.hy926 cells exhibited a high proportion of transitional states, suggesting that these cells may enter a transient and reversible phase of stress signaling prior to committing to survival or apoptosis. This observation is consistent with previous findings showing that epithelial and endothelial cells exposed to nanoparticles often activate temporary survival mechanisms, such as ERK-mediated stress responses, before undergoing apoptosis.42 In contrast, THP-1 cells displayed a high proportion of undefined states, which may reflect greater variability in stress responses or enhanced sensitivity to intercellular signaling cues. Supporting this interpretation, cell cycle analysis (Fig. S9†) revealed that most undefined clusters remained in the G0/G1/S phase. This finding highlights the need for further investigations incorporating cell cycle-specific markers (e.g., Ki-67 and cyclins) and stress regulators (e.g., p53, p21, and autophagy markers) to improve the classification of these unresolved states.
Conclusions
By employing a physiologically relevant 3D alveolar tetra-culture model alongside multi-parametric single-cell mass cytometry, we revealed diverse cellular responses triggered by AgNPs. Our results indicate that nanoparticle-induced cytotoxicity is governed not only by the amount of cell associated nanoparticles but significantly by cell type and subpopulation dependent intracellular signaling pathways and intercellular communication. Notably, THP-1 cells exhibited inflammatory responses despite minimal direct nanoparticle uptake, suggesting that indirect or paracrine pathways play critical roles. Conversely, PMA-differentiated THP-1, A549 and EA.hy926 cells, despite higher nanoparticle association, showed notable resilience against AgNP-induced stress.Hierarchical clustering analysis enabled us to further identify distinct cellular subpopulations with unique expression patterns of stress-related, inflammatory, and apoptotic markers. This comprehensive single-cell analysis captured a spectrum of responses from survival and intermediate states to apoptosis, highlighting substantial heterogeneity often overlooked by conventional bulk methods. By enabling the identification of sensitive cell phenotypes and early molecular stress responses, single-cell profiling provides mechanistic insights essential for hazard characterization. These high-resolution data can support the development of adverse outcome pathways (AOPs), identification of cell-type-specific biomarkers. Such mechanistic profiling may facilitate grouping of nanomaterials by biological response similarity, thereby supporting their integration into regulatory frameworks—such as OECD read-across—by supplying direct evidence of cell-type-specific vulnerabilities.
However, further research is needed to validate these findings through longitudinal studies, including expanded antibody panels targeting a broader array of biological markers. Additionally, evaluating a wider variety of nanomaterials and standardizing data processing protocols will significantly enhance the practical applicability and adoption of this single-cell analytical approach in nanotoxicology and safety assessment.
Data availability
The datasets for this study are available in Zenodo (https://doi.org/10.5281/zenodo.15173760). Additional supporting information, including supplementary tables and figures, is provided as part of the ESI.†Author contributions
All authors contributed to the article and approved the submitted version. Conceptualization: T. H. Y.; methodology: E. L., S. P. and A. C.; investigation: E. L., S. G. P., S. M. H. and M. K.; validation: E. L.; formal analysis: E. L.; writing – original draft preparation: E. L. and T. H. Y.; writing – review and editing: E. L., A. C., T. S. and T. H. Y.; visualization: E. L.; supervision: T. H. Y.; funding acquisition: T. H. Y.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by Nano Material Technology Development Program (Grant number RS-2024-00452934), Basic Science Research Program (Grant number 2020R1A6A1A06046728) and International Cooperative R&D Program (Grant number 2022K1A3A1A78097929) through the National Research Foundation of Korea (NRF). This work was also conducted as part of the European Commission Horizon 2020 program project CompSafeNano (Grant agreement number 101008099), and the Horizon Europe program projects INSIGHT (Grant agreement number 101137742), CHIASMA (Grant agreement number 101137613), and MACRAMÉ (Grant agreement number 101092686).References
- O. Afzal, A. S. A. Altamimi, M. S. Nadeem, S. I. Alzarea, W. H. Almalki, A. Tariq, B. Mubeen, B. N. Murtaza, S. Iftikhar, N. Riaz and I. Kazmi, Nanoparticles in Drug Delivery: From History to Therapeutic Applications, Nanomaterials, 2022, 12, 4494 CrossRef CAS PubMed.
- T. M. Joseph, D. Kar Mahapatra, A. Esmaeili, Ł. Piszczyk, M. S. Hasanin, M. Kattali, J. Haponiuk and S. Thomas, Nanoparticles: Taking a Unique Position in Medicine, Nanomaterials, 2023, 13, 574 CrossRef CAS PubMed.
- S. Yu, Y. Yin and J. Liu, Silver nanoparticles in the environment, Environ. Sci.: Processes Impacts, 2013, 15, 78–92 RSC.
- A. C. Burduşel, O. Gherasim, A. M. Grumezescu, L. Mogoantă, A. Ficai and E. Andronescu, Biomedical Applications of Silver Nanoparticles: An Up-to-Date Overview, Nanomaterials, 2018, 8, 681 CrossRef PubMed.
- T. Bruna, F. Maldonado-Bravo, P. Jara and N. Caro, Silver Nanoparticles and Their Antibacterial Applications, Int. J. Mol. Sci., 2021, 22, 7202 CrossRef CAS PubMed.
- H. A. Hussein and M. A. Abdullah, Novel drug delivery systems based on silver nanoparticles, hyaluronic acid, lipid nanoparticles and liposomes for cancer treatment, Appl. Nanosci., 2022, 12, 3071–3096 CrossRef CAS.
- H. Jangid, S. Singh, P. Kashyap, A. Singh and G. Kumar, Advancing biomedical applications: an in-depth analysis of silver nanoparticles in antimicrobial, anticancer, and wound healing roles, Front. Pharmacol., 2024, 15, 1438227 CrossRef CAS PubMed.
- M. K. Ha, K. H. Chung and T. H. Yoon, Heterogeneity in Biodistribution and Cytotoxicity of Silver Nanoparticles in Pulmonary Adenocarcinoma Human Cells, Nanomaterials, 2019, 10, 36 CrossRef PubMed.
- M. K. Ha, J. S. Choi, S. J. Kwon, J. Song, Y. Lee, Y. E. Kim and T. H. Yoon, Mass cytometric study on the heterogeneity in cellular association and cytotoxicity of silver nanoparticles in primary human immune cells, Environ. Sci.: Nano, 2020, 7, 1102–1114 RSC.
- H. Perumalsamy, X. Xiao, H. Y. Kim and T. H. Yoon, scRNA-seq analysis discovered suppression of immunomodulatory dependent inflammatory response in PMBCs exposed to silver nanoparticles, J. Nanobiotechnol., 2024, 22, 118 CrossRef CAS PubMed.
- R. P. Singh and P. Ramarao, Cellular uptake, intracellular trafficking and cytotoxicity of silver nanoparticles, Toxicol. Lett., 2012, 213, 249–259 CrossRef CAS PubMed.
- X. F. Zhang, W. Shen and S. Gurunathan, Silver Nanoparticle-Mediated Cellular Responses in Various Cell Lines: An in Vitro Model, Int. J. Mol. Sci., 2016, 17, 1603 CrossRef PubMed.
- M. Ghasemi, T. Turnbull, S. Sebastian and I. Kempson, The MTT Assay: Utility, Limitations, Pitfalls, and Interpretation in Bulk and Single-Cell Analysis, Int. J. Mol. Sci., 2021, 22, 12827 CrossRef CAS PubMed.
- S. He, C. Li, M. Lu, F. Lin, S. Hu, J. Zhang, L. Peng and L. Li, Comprehensive analysis of scRNA-seq and bulk RNA-seq reveals the non-cardiomyocytes heterogeneity and novel cell populations in dilated cardiomyopathy, J. Transl. Med., 2025, 23, 17 CrossRef CAS PubMed.
- Z. Zhou, B. Xu, A. Minn and N. R. Zhang, DENDRO: genetic heterogeneity profiling and subclone detection by single-cell RNA sequencing, Genome Biol., 2020, 21, 10 CrossRef CAS PubMed.
- S. K. Maden, S. H. Kwon, L. A. Huuki-Myers, L. Collado-Torres, S. C. Hicks and K. R. Maynard, Challenges and opportunities to computationally deconvolve heterogeneous tissue with varying cell sizes using single-cell RNA-sequencing datasets, Genome Biol., 2023, 24, 288 CrossRef CAS PubMed.
- H. T. Maecker and A. Harari, Immune monitoring technology primer: flow and mass cytometry, J. ImmunoTher. Cancer, 2015, 3, 44 CrossRef PubMed.
- J. Lee, D. Y. Hyeon and D. Hwang, Single-cell multiomics: technologies and data analysis methods, Exp. Mol. Med., 2020, 52, 1428–1442 CrossRef CAS PubMed.
- M. K. Ha, S. J. Kwon, J. Choi, N. T. Nguyen, J. Song, Y. Lee, Y. Kim, I. Shin, J. Nam and T. H. Yoon, Mass Cytometry and Single-Cell RNA-seq Profiling of the Heterogeneity in Human Peripheral Blood Mononuclear Cells Interacting with Silver Nanoparticles, Small, 2020, 16, 1907674 CrossRef CAS PubMed.
- I. Fizesan, C. Iacovita, A. Pop, B. Kiss, R. Dudric, R. Stiufiuc, C. M. Lucaciu and F. Loghin, The Effect of Zn-Substitution on the Morphological, Magnetic, Cytotoxic, and In Vitro Hyperthermia Properties of Polyhedral Ferrite Magnetic Nanoparticles, Pharmaceutics, 2021, 13, 2148 CrossRef CAS PubMed.
- A. Chary, An in vitro coculture system for the detection of sensitization following aerosol exposure, Altex, 2019, 36, 403–418 Search PubMed.
- M. Saibene, T. Serchi, P. Bonfanti, A. Colombo, I. Nelissen, R. Halder, J.-N. Audinot, B. Pelaz, M. G. Soliman, W. J. Parak, P. Mantecca, A. C. Gutleb and S. Cambier, The use of a complex tetra-culture alveolar model to study the biological effects induced by gold nanoparticles with different physicochemical properties, Environ. Toxicol. Pharmacol., 2024, 106, 104353 CrossRef CAS PubMed.
- S. Mayer, T. Milo, A. Isaacson, C. Halperin, S. Miyara, Y. Stein, C. Lior, M. Pevsner-Fischer, E. Tzahor, A. Mayo, U. Alon and R. Scherz-Shouval, The tumor microenvironment shows a hierarchy of cell-cell interactions dominated by fibroblasts, Nat. Commun., 2023, 14, 5810 CrossRef CAS PubMed.
- N. V. S. Vallabani and H. L. Karlsson, Primary and Secondary Genotoxicity of Nanoparticles: Establishing a Co-Culture Protocol for Assessing Micronucleus Using Flow Cytometry, Front. Toxicol., 2022, 4, 845987 CrossRef PubMed.
- P. J. Shiny, A. Mukherjee and N. Chandrasekaran, DNA damage and mitochondria-mediated apoptosis of A549 lung carcinoma cells induced by biosynthesised silver and platinum nanoparticles, RSC Adv., 2016, 6, 27775–27787 RSC.
- J. Bae, M. K. Ha, H. Perumalsamy, Y. Lee, J. Song and T. H. Yoon, Mass Cytometry Exploration of Immunomodulatory Responses of Human Immune Cells Exposed to Silver Nanoparticles, Pharmaceutics, 2022, 14, 630 CrossRef CAS PubMed.
- M. L. Fernández-Cruz, D. Hernández-Moreno, J. Catalán, R. K. Cross, H. Stockmann-Juvala, J. Cabellos, V. R. Lopes, M. Matzke, N. Ferraz, J. J. Izquierdo, J. M. Navas, M. Park, C. Svendsen and G. Janer, Quality evaluation of human and environmental toxicity studies performed with nanomaterials – the GUIDEnano approach, Environ. Sci.: Nano, 2018, 5, 381–397 RSC.
- T. X. Trinh, M. K. Ha, J. S. Choi, H. G. Byun and T. H. Yoon, Curation of datasets, assessment of their quality and completeness, and nanoSAR classification model development for metallic nanoparticles, Environ. Sci.: Nano, 2018, 5, 1902–1910 RSC.
- I. Fizeşan, S. Cambier, E. Moschini, A. Chary, I. Nelissen, J. Ziebel, J.-N. Audinot, T. Wirtz, M. Kruszewski, A. Pop, B. Kiss, T. Serchi, F. Loghin and A. C. Gutleb, In vitro exposure of a 3D-tetraculture representative for the alveolar barrier at the air-liquid interface to silver particles and nanowires, Part. Fibre Toxicol., 2019, 16, 14 CrossRef PubMed.
- M. A. Forrester, H. J. Wassall, L. S. Hall, H. Cao, H. M. Wilson, R. N. Barker and M. A. Vickers, Similarities and differences in surface receptor expression by THP-1 monocytes and differentiated macrophages polarized using seven different conditioning regimens, Cell. Immunol., 2018, 332, 58–76 CrossRef CAS PubMed.
- H. G. Fienberg, E. F. Simonds, W. J. Fantl, G. P. Nolan and B. Bodenmiller, A platinum-based covalent viability reagent for single-cell mass cytometry, Cytom. A, 2012, 81, 467–475 CrossRef PubMed.
- N. Martinho, J. M. T. Marquês, I. Todoriko, M. Prieto, R. F. M. De Almeida and L. C. Silva, Effect of Cisplatin and Its Cationic Analogues in the Phase Behavior and Permeability of Model Lipid Bilayers, Mol. Pharmaceutics, 2023, 20, 918–928 CrossRef CAS PubMed.
- J. Song, M. Guan, Z. Zhao and J. Zhang, Type I Interferons Function as Autocrine and Paracrine Factors to Induce Autotaxin in Response to TLR Activation, PLoS One, 2015, 10, e0136629 CrossRef PubMed.
- E. Panzarini, C. Vergallo, F. P. Fanizzi, S. Mariano, A. M. Tata and L. Dini, The dialogue between died and viable cells: in vitro and in vivo bystander effects and 1H-NMR-based metabolic profiling of soluble factors, Pure Appl. Chem., 2020, 92, 399–411 CrossRef CAS.
- A. R. Gliga, S. Skoglund, I. Odnevall Wallinder, B. Fadeel and H. L. Karlsson, Size-dependent cytotoxicity of silver nanoparticles in human lung cells: the role of cellular uptake, agglomeration and Ag release, Part. Fibre Toxicol., 2014, 11, 11 CrossRef PubMed.
- F. Zhang, G. V. Aquino, A. Dabi and E. D. Bruce, Assessing the translocation of silver nanoparticles using an in vitro co-culture model of human airway barrier, Toxicol. In Vitro, 2019, 56, 1–9 CrossRef CAS PubMed.
- R. Niranjan, K. P. Mishra, S. N. Tripathi and A. K. Thakur, Proliferation of Lung Epithelial Cells Is Regulated by the Mechanisms of Autophagy Upon Exposure of Soots, Front. Cell Dev. Biol., 2021, 9, 662597 CrossRef PubMed.
- J. G. González-Vega, J. C. García-Ramos, R. A. Chavez-Santoscoy, J. E. Castillo-Quiñones, M. E. Arellano-Garcia and Y. Toledano-Magaña, Lung Models to Evaluate Silver Nanoparticles' Toxicity and Their Impact on Human Health, Nanomaterials, 2022, 12, 2316 CrossRef PubMed.
- B. M. Jarai and C. A. Fromen, Nanoparticle Internalization Promotes the Survival of Primary Macrophages, Adv. NanoBiomed Res., 2022, 2, 2100127 CrossRef CAS PubMed.
- D. Redolfi-Bristol, K. Yamamoto, E. Marin, W. Zhu, O. Mazda, P. Riello and G. Pezzotti, Exploring the cellular antioxidant mechanism against cytotoxic silver nanoparticles: a Raman spectroscopic analysis, Nanoscale, 2024, 16, 9985–9997 RSC.
- X. Sun, Y. Yang, J. Shi, C. Wang, Z. Yu and H. Zhang, NOX4- and Nrf2-mediated oxidative stress induced by silver nanoparticles in vascular endothelial cells, J. Appl. Toxicol., 2017, 37, 1428–1437 CrossRef CAS PubMed.
- M. I. Saad, S. Alhayyani, L. McLeod, L. Yu, M. Alanazi, V. Deswaerte, K. Tang, T. Jarde, J. A. Smith, Z. Prodanovic, M. D. Tate, J. J. Balic, D. N. Watkins, J. E. Cain, S. Bozinovski, E. Algar, T. Kohmoto, H. Ebi, W. Ferlin, C. Garbers, S. Ruwanpura, I. Sagi, S. Rose-John and B. J. Jenkins, ADAM17 selectively activates the IL-6 trans-signaling/ERK MAPK axis in KRAS-addicted lung cancer, EMBO Mol. Med., 2019, 11, e9976 CrossRef PubMed.
- S. Monti, P. Tamayo, J. Mesirov and T. Golub, Consensus Clustering: A Resampling-Based Method for Class Discovery and Visualization of Gene Expression Microarray Data, Mach. Learn., 2003, 52, 91–118 CrossRef.
- M. Lamkanfi and T. D. Kanneganti, Caspase-7: A protease involved in apoptosis and inflammation, Int. J. Biochem. Cell Biol., 2010, 42, 21–24 CrossRef CAS PubMed.
- L. Müller, M. Riediker, P. Wick, M. Mohr, P. Gehr and B. Rothen-Rutishauser, Oxidative stress and inflammation response after nanoparticle exposure: differences between human lung cell monocultures and an advanced three-dimensional model of the human epithelial airways, J. R. Soc., Interface, 2010, 7, S27–S40 CrossRef PubMed.
- T. Brzicova, E. Javorkova, K. Vrbova, A. Zajicova, V. Holan, D. Pinkas, V. Philimonenko, J. Sikorova, J. Klema, J. Topinka and P. Rossner, Molecular Responses in THP-1 Macrophage-Like Cells Exposed to Diverse Nanoparticles, Nanomaterials, 2019, 9, 687 CrossRef CAS PubMed.
- M. M. J. P. E. Sthijns, W. Thongkam, C. Albrecht, B. Hellack, A. Bast, G. R. M. M. Haenen and R. P. F. Schins, Silver nanoparticles induce hormesis in A549 human epithelial cells, Toxicol. In Vitro, 2017, 40, 223–233 CrossRef CAS PubMed.
- L. Xu, Y. Bi, Y. Xu, Y. Wu, X. Du, Y. Mou and J. Chen, Suppression of CHOP Reduces Neuronal Apoptosis and Rescues Cognitive Impairment Induced by Intermittent Hypoxia by Inhibiting Bax and Bak Activation, Neural Plast., 2021, 2021, 1–14 Search PubMed.
- K. Liu, C. Zhao, R. C. Adajar, D. DeZwaan-McCabe and D. T. Rutkowski, A beneficial adaptive role for CHOP in driving cell fate selection during ER stress, EMBO Rep., 2024, 25, 228–253 CrossRef CAS PubMed.
- B. Drabarek, D. Dymkowska, J. Szczepanowska and K. Zabłocki, TNFα affects energy metabolism and stimulates biogenesis of mitochondria in EA.hy926 endothelial cells, Int. J. Biochem. Cell Biol., 2012, 44, 1390–1397 CrossRef CAS PubMed.
- R. J. Holmila, S. A. Vance, S. B. King, A. W. Tsang, R. Singh and C. M. Furdui, Silver Nanoparticles Induce Mitochondrial Protein Oxidation in Lung Cells Impacting Cell Cycle and Proliferation, Antioxidants, 2019, 8, 552 CrossRef CAS PubMed.
- J. A. Kim, C. Åberg, A. Salvati and K. A. Dawson, Role of cell cycle on the cellular uptake and dilution of nanoparticles in a cell population, Nat. Nanotechnol., 2012, 7, 62–68 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5en00439j |
| This journal is © The Royal Society of Chemistry 2025 |