Open Access Article
Open Access ArticleA non-peptide-based fluorescent probe capable of sensitively visualizing asparagine endopeptidase†
Kang
Li
a,
Yanxian
Hou
a,
Jinliang
Han
a,
Chengyuan
Lv
a,
Wenkai
Liu
a,
Jianjun
Du
 a,
Wen
Sun
a,
Wen
Sun
 a,
Jiangli
Fan
a,
Jiangli
Fan
 *ab and
Xiaojun
Peng
*ab and
Xiaojun
Peng
 a
a
aState Key Laboratory of Fine Chemicals, Frontiers Science Centre for Smart Materials Oriented Chemical Engineering, Dalian University of Technology, Dalian 116024, China. E-mail: fanjl@dlut.edu.cn
bNingbo Institute of Dalian University of Technology, Ningbo 315016, China
First published on 23rd February 2024
Abstract
The non-peptide-based fluorescent probe QMC11 is capable of specifically targeting asparagine endopeptidase (AEP) and imaging cellular endogenous AEP. The motion of the probe can be restricted by AEP to activate fluorescence while keeping a low background signal.
Asparagine endopeptidase (AEP) is a member of the C13 cysteine protease family, predominantly found in cellular lysosomes.1 It specifically cleaves peptide bonds in the carboxyl terminus of aspartic acid (Asn) or aspartic acid (Asp) residues. The abnormal upregulation of AEP, compared with other biomarkers of Alzheimer's disease (AD) such as amyloid-β (Aβ),2,3 β-secretase (BACE1), tau protein, monoamine oxidases (MAOs), and methionine sulfoxide reductase (Msrs), is present in the early stage of the entire pathological process.4,5 At the same time, the upregulation of AEP leads to accumulation of Aβ protein, cleavage of SET (protein phosphatase 2A inhibitor 2) and abnormal aggregation of Tau, all of which contribute to the progression of both elderly and AD patients.6–11 This cascade effect is not seen with other biomarkers. Thus, the elevated AEP activation is considered an important marker in the regulation of pathological pathways and pre-monitoring in Alzheimer's disease.12,13
In recent years, various methods, such as mass spectrometry,14 immunoproteomic analysis,15 and magnetic resonance imaging (MRI),16 have been employed to detect AEP. However, these methods possess certain drawbacks including their complex technology, high costs, and the requirement of large instruments and equipment. Comparatively, fluorescence imaging exhibits several advantages such as non-invasiveness, real-time monitoring, and high sensitivity. Due to its specificity for substrates containing aspartate (Asp), AEP can cut certain specific polypeptide sequences.17 Consequently, a few fluorescent probes targeting AEP have been designed based on peptide substrates containing aspartic acid (Asp).18–23 But the synthesis and purification of substrates based on proteins or peptides entail multiple steps, thereby increasing the preparation difficulties of the probes.21,22 Also, some of them could not only be cleaved by AEP, but also by caspases, which means that the specificity to AEP was weak. To overcome these limitations, alternative strategies for AEP detection should be explored to avoid introducing peptide chains.
We chose the fluorophore sulfonate-substituted quinoline-malononitrile (QM-SO3) as the fluorescence reporting unit,24 in which the oxygen atom in the fluorophore DCM was replaced by a nitrogen atom to overcome the quenching effect and a sulfonate group was introduced to enhance the water solubility. C11 has been screened as a non-toxic δ-secretase (AEP) inhibitor through high throughput, specifically targeting AEP without affecting other related cysteine proteases. Additionally, it was found that the presence of an amino group on C11 did not impact the binding to AEP.25 Therefore, the amino group could serve as the active site for attaching the fluorophore. Combining these ideas, we herein presented QMC11, a new small molecule fluorescent probe (Fig. 1A) by connecting QMN3 and C11 through click reaction. QMC11 was not fluorescent in the physiological environment unless it specifically bound to AEP. The binding mode was mainly hydrogen bonding, and the binding sites included benzoxadiazole of C11 and the sulfonic acid group part. The binding restricted the molecular rotation and conformational change, thereby turning on the fluorescence. The probe molecule exhibited high SNR (∼35-fold) and greater specificity for AEP compared to other substances. As illustrated in Fig. 1B, the targeted portion of QMC11 contained a morpholine ring, which facilitated its arrival to the lysosomal region, where it combined with the highly expressed AEP to enable the fluorescence signal to be turned on.
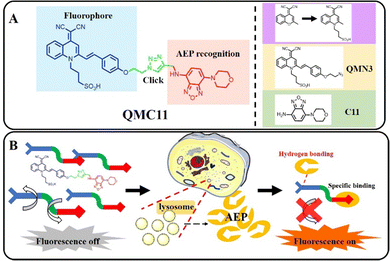 | ||
| Fig. 1 (A) The structure and design principle of QMC11. (B) Specific identification of the AEP mechanism. | ||
The details of the synthetic procedure are presented in Scheme S1 (ESI†). Key intermediates and the final product were characterized with 1H-NMR, 13C-NMR, and ESI-HRMS (Fig. S11–S13, ESI†).
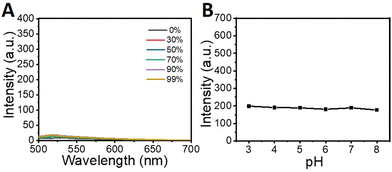
Firstly, the fluorescence properties of QMC11 in different solvent systems were conducted. QMC11 demonstrated good solubility in water, and it was not emissive in aqueous solution, ethanol, or tetrahydrofuran (Fig. 2A and Fig. S1A, ESI†). However, in the water–glycerol system, the fluorescence at 550 nm was continuously enhanced as the volume fraction of glycerol in the mixed solvent reached 99% (Fig. S1B, ESI†). These observations indicated a correlation between QMC11 and the viscosity of the system. To determine whether AEP is sufficient to turn on fluorescence by binding the inhibitor (C11), QMN3, which lacks the target group C11 (Fig. 1), was synthesized. It also remained non-emissive in aqueous solution, ethanol, and tetrahydrofuran and responded to the viscosity (Fig. S2, ESI†). It was reported that the maximal activity of AEP was at pH 4–6 under normal assay conditions, and the enzyme was irreversibly denatured at pH 7 and above.26 To determine the stability of QMC11 over a specific pH range, the effects of pH on QMC11 were tested using a glycerin–PBS solvent system (glycerin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PBS = 9
PBS = 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and the fluorescence behavior was investigated at pH levels ranging from 3 to 8 (Fig. 2B). It was observed that the fluorescence signal of QMC11 was minimally affected by pH, indicating its good stability.
1) and the fluorescence behavior was investigated at pH levels ranging from 3 to 8 (Fig. 2B). It was observed that the fluorescence signal of QMC11 was minimally affected by pH, indicating its good stability.
The binding behavior of QMC11 was investigated by performing an enzyme binding experiment in vitro using the buffer (pH = 4.5). Upon association with AEP, a significant fluorescence enhancement was observed from QMC11, as shown in Fig. 3A. It took approximately 90 minutes for the fluorescence to reach a plateau, as shown in Fig. 3B. In contrast, Fig. S3 (ESI†) illustrated that QMN3, without C11, did not produce a fluorescent signal upon the addition of AEP. Additionally, it was observed that with the increase of the AEP concentration, the fluorescence intensity of QMC11 gradually increased (Fig. 3C). This might be due to the fact that as QMC11 binds to AEP, while the AEP concentration increased, their conformational change and rotation were more restricted, resulting in enhanced fluorescence. To confirm the specific binding of the inhibitor part to AEP, we further added the mother liquor of inhibitor C11 to the incubated probe molecule and observed a gradual weakening and eventual disappearance of the fluorescence signal (Fig. 3D). To evaluate the specificity of QMC11 for AEP, potentially competitive species including amino acids, anions, cations and protein macromolecules were tested. Fig. 3G illustrates that QMC11 did not exhibit any fluorescence response to these species, indicating its fine specificity for AEP.
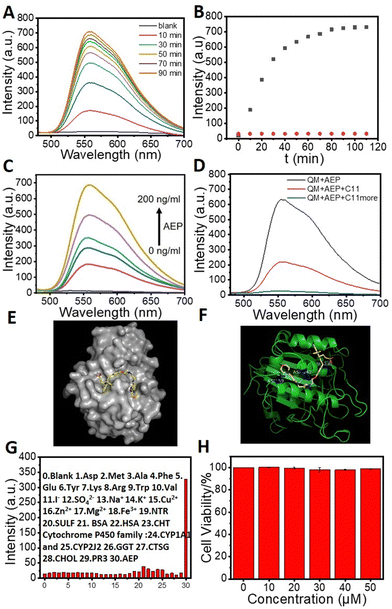 | ||
| Fig. 3 Response tests of QMC11 (20 μM except Fig. 3G, λex: 478 nm). (A) Fluorescence spectra of QMC11 with incubation with AEP (200 ng mL−1). (B) Kinetics of QMC11 (black) and QMN3 (20 μM; red) fluorescence response to AEP. (C) The fluorescence spectroscopy of various concentrations of AEP (0–200 ng mL−1) incubated with QMC11 for 90 min. (D) Inhibition of fluorescence response of QMC11 and AEP by addition of C11 (20 μM). (E) Simulated docking model of QMC11 with AEP. (F) The action sites in the simulation docking model. (G) Selectivity of QMC11 (10 μM) toward different analytes (0–30: blank, Asp, Met, Ala, Phe, Glu, Tyr, Lys, Arg, Trp, Val, I−, SO42−, Na+, K+, Cu2+, Zn2+, Mg2+, Fe3+, NTR, SULF, BSA, HAS, CHT Cytochrome P450 family, CYP1A1 and CYP2J2, GGT, CTSG, CHOL, PR3, AEP). Values are means ± SD (n = 3). (H) Cytotoxicity test of QMC11 (MTT). | ||
The docking study showed that the five-membered ring portion of benzoxadiazole of C11 would nicely bind to amino acid residue regions such as ASN-159 and ASP-242 via hydrogen bonding. Surprisingly, additional hydrogen bonds were also found between the sulfonic acid group portion of QMC11 and ASN-236 (Fig. 3E and F). Moreover, the binding energy of QMC11 and AEP was −8.3 kcal mol−1, indicating a strong affinity and potential for stable combination. This suggested that AEP had the ability to restrict the motion of the probe molecule, such as rotation and conformational change.
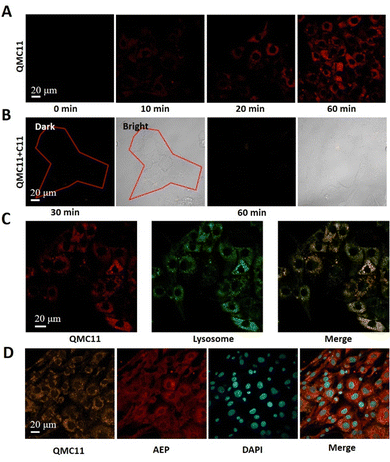
Based on the above properties we explored the application of QMC11 in living cells. Prior to bioimaging, the cell cytotoxicity of QMC11 was assessed using an MTT assay. We chose C6 cells (rat glioma cells), which express a significant level of AEP.27 As shown in Fig. 3H and Fig. S4 (ESI†), when incubated with QMC11 at the concentration of 10, 20, 40, 80 or 120 μM for 24 h, the viability of C6 cells was 99.2%, 99.7%, 98.2%, 97.8%, and 98.8%, respectively. Meanwhile, when incubated with QMN3 for 24 h, 96.0%, 94.5%, 93.0%, 93.1%, and 95.2% of C6 cells survived, respectively. The results indicated that over 95.0% of C6 cells retained viability in the presence of 40 μM of QMC11 over 24 h, demonstrating its low cytotoxicity and safety for cell imaging. Next, in order to determine whether QMC11 can enter cells and combine with cellular AEP, we incubated this probe with C6 cells. Meanwhile, we also incubated the control probe QMN3 alone with C6 cells. After a 1-h incubation, QMC11 permeated into the cytoplasm and emitted fluorescence (Fig. 4A), indicating that rotation of QMC11 was inhibited due to AEP specifically binding to C11. In contrast, C6 cells incubated with QMN3 showed little fluorescence (Fig. S5, ESI†). Moreover, when C6 cells were pre-treated with C11 (40 μM), the fluorescence of QMC11 was suppressed (Fig. 4B). These results were consistent with in vitro experiments, which showed that the specific binding of QMC11 and AEP was indeed due to the C11 part. In order to determine whether viscosity has an effect on QMC11 in cells, we incubated QMC11 with 4T1 cells (mouse breast cancer cell 4T1, no AEP expression27) for 1 h. Fortunately, no fluorescent signal appeared, indicating that the viscosity of cancer cells had no effect on QMC11 (Fig. S8, ESI†). Thus, it could be concluded that QMC11 was able to combine with cellular AEP and emitted fluorescence.
We verified the AEP expression levels of C6 and U251 through Western Blot experiments, indicating that C6 would overexpress AEP (Fig. S7, ESI†). In addition, we confirmed the high intracellular expression of AEP by immunofluorescence staining, further revealing the relationship between QMC11 and AEP in C6 cells. Fluorescence signals in the orange, red, and blue channels were observed in Fig. 4D. We found that QMC11 (orange) was co-localized with AEP (red) in the cytoplasm of C6 cells. Given that cellular AEP becomes activated in the acidic environment of lysosomes,28QMC11 must at least enter lysosomes to combine with AEP. To investigate the subcellular localization of QMC11, we incubated C6 cells with QMC11 for 1 h and employed a lysosome-targeted fluorescent dye Lysotracker Red. By imaging the red and green channels, we found that the red channel image overlapped well with the green channel image, indicating that the probe got into lysosomes where it played a significant role (Fig. 4C). Simultaneously, we found that it exhibits poor colocalization with mitochondria and the endoplasmic reticulum (Fig. S9 and S10, ESI†). Therefore, based on the endogenous AEP activity, QMC11 could real-time monitor AEP.
In summary, a new small molecule fluorescent probe QMC11 was designed by connecting a water-soluble fluorophore and a non-toxic AEP inhibitor C11 through click reaction. QMC11 did not exhibit fluorescence in the physiological environment, but was selectively turned on by AEP with high sensitivity. Importantly, QMC11 could stain lysosomal AEP in live cells. In contrast to other probes that require numerous steps for the synthesis and purification of substrates relying on proteins or peptides, QMC11 provided a simpler synthesis process and avoided the interference of caspases. We thought it might be a different way to identify early abnormal biomarkers of AD.
This work was financially supported by the National Natural Science Foundation of China (21925802, 22338005), Liaoning Binhai Laboratory (LBLB-2023-03) and the Fundamental Research Funds for the Central Universities (DUT22LAB601).
Conflicts of interest
There are no conflicts to declare.Notes and references
- L. Calugi, E. Lenci, F. Bianchini, A. Contini and A. Trabocchi, Bioorg. Med. Chem., 2022, 63, 116746 CrossRef CAS PubMed.
- X. Wang, A. Iyaswamy, D. Xu, S. Krishnamoorthi, S. G. Sreenivasmurthy, Y. Yang, Y. Li, C. Chen, M. Li, H.-W. Li and M. S. Wong, ACS Appl. Mater. Interfaces, 2022, 15, 39–47 CrossRef PubMed.
- J. An, P. Verwilst, H. Aziz, J. Shin, S. Lim, I. Kim, Y. K. Kim and J. S. Kim, Bioact. Mater., 2022, 13, 239–248 CAS.
- S. Ma, G. Chen, J. Xu, Y. Liu, G. Li, T. Chen, Y. Li and T. D. James, Coord. Chem. Rev., 2021, 427, 213553 CrossRef CAS.
- R. Ni, Z. Chen, X. L. Deán-Ben, F. F. Voigt, D. Kirschenbaum, G. Shi, A. Villois, Q. Zhou, A. Crimi, P. Arosio, R. M. Nitsch, K. P. R. Nilsson, A. Aguzzi, F. Helmchen, J. Klohs and D. Razansky, Nat. Biomed. Eng., 2022, 6, 1031–1044 CrossRef CAS PubMed.
- C. Kaether, C. Haass and H. Steiner, Neurodegener. Dis., 2006, 3, 275–283 CrossRef CAS PubMed.
- W. A. Campbell, M. L. Reed, J. Strahle, M. S. Wolfe and W. Xia, J. Neurochem., 2003, 85, 1563–1574 CrossRef CAS PubMed.
- K. Blennow, C. Chen, C. Cicognola, K. R. Wildsmith, P. T. Manser, S. M. S. Bohorquez, Z. Zhang, B. Xie, J. Peng, O. Hansson, H. Kvartsberg, E. Portelius, H. Zetterberg, T. Lashley, G. Brinkmalm, G. A. Kerchner, R. M. Weimer, K. Ye and K. Hoglund, Brain, 2020, 143, 650–660 CrossRef PubMed.
- G. Basurto-Islas, I. Grundke-Iqbal, Y. C. Tung, F. Liu and K. Iqbal, J. Biol. Chem., 2013, 288, 17495–17507 CrossRef CAS PubMed.
- C. M. Yates, John Buttenvorth, T. Mara C. Tennant and A. Gordon, J. Neurochem., 1990, 55, 1624–1630 CrossRef CAS PubMed.
- K. Wang, H. Gao, Y. Zhang, H. Yan, J. Si, X. Mi, S. Xia, X. Feng, D. Liu, D. Kong, T. Wang and D. Ding, Adv. Mater., 2022, 34, 2106994 CrossRef CAS PubMed.
- M. Song, Int. J. Mol. Sci., 2022, 23, 10223–10236 CrossRef CAS PubMed.
- P. Griffin, L. Apostolova, B. C. Dickerson, G. Rabinovici, S. Salloway, K. Brandt, J. Masdeu, D. Hammers, S. Raghuram, S. Hall and M. C. Carrillo, Alzheimer’s Dementia, 2023, 19, S126–S131 Search PubMed.
- A. Dutta, D. Potier, M. Walker, O. Gray and A. D. Whetton, Oncotarget, 2016, 7, 70822–70831 CrossRef PubMed.
- J.-W. Ju, H.-N. Joo, M.-R. Lee, S.-H. Cho, H.-I. Cheun, J.-Y. Kim, Y.-H. Lee, K.-J. Lee, W.-M. Sohn, D.-M. Kim, I.-C. Kim, B. C. Park and T.-S. Kim, Proteomics, 2009, 9, 3066–3078 CrossRef CAS PubMed.
- Y. Yuan, S. Ge, H. Sun, X. Dong and H. Zhao, ACS Nano, 2015, 9, 5117–5124 CrossRef CAS PubMed.
- M. Poreba, R. Solberg, W. Rut, N. N. Lunde, P. Kasperkiewicz, S. J. Snipas, M. Mihelic, D. Turk, B. Turk, G. S. Salvesen and M. Drag, Cell Chem. Biol., 2016, 23, 1023–1035 CrossRef CAS PubMed.
- X. Li, Q. Liu, S. Ye, S. Wang, K. Li, G. Lv, Y. Peng, L. Qiu and J. Lin, Chem. Biol. Drug Des., 2019, 94, 1494–1503 CrossRef CAS PubMed.
- J. Lee and M. Bogyo, ACS Chem. Biol., 2010, 5, 233–243 CrossRef CAS PubMed.
- J. A. Hong, N. E. Choi, Y. K. La, H. Y. Nam, J. Seo and J. Lee, Org. Biomol. Chem., 2017, 15, 8018–8022 RSC.
- L. E. Edgington, M. Verdoes, A. Ortega, N. P. Withana, J. Lee, S. Syed, M. H. Bachmann, G. Blum and M. Bogyo, J. Am. Chem. Soc., 2013, 135, 174–182 CrossRef CAS PubMed.
- Y. Zhao, Z. Hai, H. Wang, L. Su and G. Liang, Anal. Chem., 2018, 90, 8732–8735 CrossRef CAS PubMed.
- K. B. Sexton, M. D. Witte, G. Blum and M. Bogyo, Bioorg. Med. Chem. Lett., 2007, 17, 649–653 CrossRef CAS PubMed.
- W. Fu, C. Yan, Z. Guo, J. Zhang, H. Zhang, H. Tian and W. H. Zhu, J. Am. Chem. Soc., 2019, 141, 3171–3177 CrossRef CAS PubMed.
- Z. Zhang, O. Obianyo, E. Dall, Y. Du, H. Fu, X. Liu, S. S. Kang, M. Song, S. P. Yu, C. Cabrele, M. Schubert, X. Li, J. Z. Wang, H. Brandstetter and K. Ye, Nat. Commun., 2017, 8, 14740 CrossRef CAS PubMed.
- J.-M. Chen, P. M. Dando, N. D. Rawlings, M. A. Brown, N. E. Young, R. A. Stevens, E. Hewitt, C. Watts and A. J. Barrett, J. Biol. Chem., 1997, 272, 8090–8098 CrossRef CAS PubMed.
- S. Ruan, C. Hu, X. Tang, X. Cun, W. Xiao, K. Shi, Q. He and H. Gao, ACS Nano, 2016, 10, 10086–10098 CrossRef CAS PubMed.
- E. Dall and H. Brandstetter, Biochimie, 2016, 122, 126–150 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3cc05419e |
| This journal is © The Royal Society of Chemistry 2024 |