Open Access Article
Open Access ArticleGrowth, replication and division enable evolution of coacervate protocells
Annemiek D.
Slootbeek†
 ,
Merlijn H. I.
van Haren†
,
Merlijn H. I.
van Haren†
 ,
Iris B. A.
Smokers†
,
Iris B. A.
Smokers†
 and
Evan
Spruijt
and
Evan
Spruijt
 *
*
Institute for Molecules and Materials, Radboud University, Heyendaalseweg 135, 6525 AJ Nijmegen, The Netherlands. E-mail: e.spruijt@science.ru.nl
First published on 13th September 2022
Abstract
Living and proliferating cells undergo repeated cycles of growth, replication and division, all orchestrated by complex molecular networks. How a minimal cell cycle emerged and helped primitive cells to evolve remains one of the biggest mysteries in modern science, and is an active area of research in chemistry. Protocells are cell-like compartments that recapitulate features of living cells and may be seen as the chemical ancestors of modern life. While compartmentalization is not strictly required for primitive, open-ended evolution of self-replicating systems, it gives such systems a clear identity by setting the boundaries and it can help them overcome three major obstacles of dilution, parasitism and compatibility. Compartmentalization is therefore widely considered to be a central hallmark of primitive life, and various types of protocells are actively investigated, with the ultimate goal of developing a protocell capable of autonomous proliferation by mimicking the well-known cell cycle of growth, replication and division. We and others have found that coacervates are promising protocell candidates in which chemical building blocks required for life are naturally concentrated, and chemical reactions can be selectively enhanced or suppressed. This feature article provides an overview of how growth, replication and division can be realized with coacervates as protocells and what the bottlenecks are. Considerations are given for designing chemical networks in coacervates that can lead to sustained growth, selective replication and controlled division, in a way that they are linked together like in the cell cycle. Ultimately, such a system may undergo evolution by natural selection of certain phenotypes, leading to adaptation and the gain of new functions, and we end with a brief discussion of the opportunities for coacervates to facilitate this.
Introduction
The division of a cell in two virtually identical copies is one of the most captivating processes in biology and a distinctive sign of living organisms.1 Without reproduction, Darwinian evolution would not be possible and life could never have reached the degree of complexity we see today.2–4 In modern cells reproduction consists of copying (replication) of the cell's genetic code concurring with an increase in size (growth), followed by actual division of the cell into two. These steps are intimately coupled in all cell types: cells grow in volume before and during replication of the genetic information, and completion of the replication process initiates a complex cascade of highly evolved processes that ultimately leads to cell division.5,6 Because the processes of information copying and division are never truly perfect, living organisms evolve and slowly gain new functions. In fact, the machinery that regulates replication of the genetic information and cell division was itself formed and developed by evolution. However, exactly how these fundamental features of life could have emerged in mixtures of non-living molecules remains one of the biggest mysteries in modern science and is an active area of research in chemistry.To facilitate research into the chemical origins of life, the defining features of modern cells are often divided into hallmarks, such as growth, division, information processing, and compartmentalization.1 In recent years, many chemical systems have been reported in which one or more of these hallmarks have been reconstituted, as steps on the way to creating a living, viable protocell (a primitive cell).5 It is not known what the initial chemical systems or protocells precisely looked like, but they must have originated from the abiotic molecules that were likely present on early Earth,6–8 and they must have been able to undergo a primitive form of open-ended evolution in order to increase in complexity and gain new functions.4 This implies that a chemical ancestor of modern life should have been capable of autonomous proliferation and thus mimic the cell cycle of growth-replication-division described above. Moreover, the mechanisms for growth, replication and division should have been reliable enough to permit evolution.4 Natural variation between the protocells, which could originate from random mutations, must be heritable, that is, they must be transmitted to the next generation. Natural selection by the environment, for instance through competition for resources, will then ensure that the fittest protocell has the highest survival probability. In other words, if these mutations are beneficial for the system and the environment it is currently in, they will survive and be transmitted to the next generation. If not, a new protocell carrying a mutation will, in the worst case, die. This selection process, in which a population gradually changes because certain mutations are passed on and others not, is what is known as Darwinian evolution.
Compartmentalization is commonly thought of as one of the most central hallmarks of life. A compartment serves as a container to keep all ingredients of a living system together and it helps to define the boundaries of a living system, thereby providing it with an “identity”. The compartment and its contents together form a protocell, which is separated from the outside world (the environment) by a membrane or an interface. In theory, compartmentalization is not required for Darwinian evolution: self-replicating chemical systems, consisting of RNA,9,10 peptides,11,12 amyloid fibers13 or supramolecular stacks of macrocyclic rings14,15 are widely investigated. However, the reaction networks underlying these self-replicating systems face three major challenges: dilution, parasitism and compatibility. Compartmentalization by the right type of compartments could help overcome all three challenges, and is therefore, arguably, an essential element of even the earliest life-like chemical systems. Here, we demonstrate that coacervates, which are droplets formed by liquid–liquid phase separation (LLPS), have the characteristics to enable chemical systems capable of self-replication to overcome dilution, parasitism and problems with compatibility between reactions. At the same time, coacervates are well suited to undergo controlled growth and division, thereby opening the way for evolution of protocells.
The first problem of dilution involves the concentration of replicators and their building blocks that are required for the replication reaction to proceed. Model replicators may function well when prepared at relatively high concentrations in a test tube, but in the primordial soup the replicators and molecules required to form them were most likely so rare and dilute, that replication reactions were prohibitively slow. Without a means to concentrate them, degradation would happen faster than replication.4,16 To overcome this, the system needs a way to concentrate the molecules involved in replication. In modern cells, essential molecules such as proteins, ribosomes and RNA are compartmentalized by the outer cell membrane, and in some cases also in smaller subcompartments within the cell, leading to effective local concentrations that can exceed the global concentration by several orders of magnitude.17,18 Inspired by membraneless organelles that form subcompartments in many types of living cells, coacervates have been proposed as compartments that can help overcome dilution of replicators and their building blocks, as they have been found to concentrate nucleic acids, nucleotides, peptides and many types of small molecules.19,20 Here, we will discuss the relevance of this property of coacervates for growth, replication and division.
The second problem is the formation of parasites during reproduction. Parasitism occurs when a mutation that is in principle disadvantageous for the fitness of the population, because it has a decreased complexity or functionality, is replicated faster than the “healthy” species. Parasites can replicate faster by profiting from functionalities of other species, while having a reduced genome size. As they compete for resources for the replication reaction with the healthy species, parasites can overtake a population of replicators in numbers, and if nothing is done to contain the parasites, a population infected by parasites will ultimately become extinct. To overcome this, compartments have been shown to play an important role, either by a stochastic corrector model, or through transient compartmentalization. Here, we will highlight the role of compartments in overcoming parasitism during replication, and discuss the advantages of coacervates as compartments in particular.
Finally, the third problem that protocells may face is the compatibility of chemical reactions. Growth, replication and division are ultimately governed by chemical reactions, such as phosphorylation reactions, template-directed polymerization, and hydrolysis, that operate under specific conditions. In many cases, these conditions are not compatible, as products of one reaction can inhibit another reaction (toxicity), or they undergo side reactions with another reagent (orthogonality). In modern cells, this issue is often circumvented by creating intracellular compartments, such as lysosomes or biomolecular condensates, which separate biomolecules from each other, thereby making it possible that reactions occur simultaneously or can be selectively turned on or off. On early Earth, compartments could have played a similar role. It has recently been shown that coacervates can spontaneously form multiphase droplets, with up to three coexisting phases, which can all localize a distinct set of molecules, while allowing other molecules to be shared between each compartment.21,22 Here, we will focus on the chemical reactions underlying growth, replication and division, and highlight situations in which incompatibility of some processes is expected. We will discuss how compartmentalization by coacervates can be exploited to circumvent these situations.
All three challenges can thus be overcome by introducing compartments, and thereby creating a protocell. As discussed, these protocells should be able to grow, facilitate the replication of information they contain, and divide, in order to proliferate and, ultimately, evolve. Coacervates are a promising class of protocellular compartments that are capable of concentrating reagents, hosting chemical reactions, and protecting sequestered molecules from the outside environment.23,24 These properties make it possible for coacervates to grow, facilitate replication, and divide, with the ultimate aim to undergo open-ended evolution.
In this feature article, we will highlight recent advances in coacervate protocell research aimed at achieving growth, replication of information and division in coacervates, in such a way that the protocell cycle (Fig. 1) is sufficiently robust and reliable to eventually enable their primitive evolution. We will first give a brief introduction of coacervates and the physicochemical properties that make them promising as protocell candidates. We also discuss some alternative types of compartments, of which liposomes are the most important, because strategies, such as the division of liposomes could serve as a blueprint for the investigation scope of realizing division of coacervates in the near future. Then, we discuss the three key steps depicted in the protocell cycle in Fig. 1: growth, replication of information and division. For each step, we will discuss the main requirements, the chemical strategies recently reported to achieve it, and advantages and limitations. We end with a brief discussion of how the systems could be adapted and integrated to show that they are able to evolve, and their opportunities and bottlenecks.
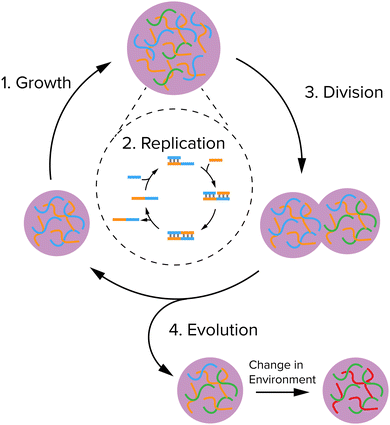 | ||
| Fig. 1 A minimal protocell cycle that can give rise to evolution: a system should be able to grow, replicate its information, divide, and be susceptible to selection by the environment. | ||
Cell-like compartmentalization
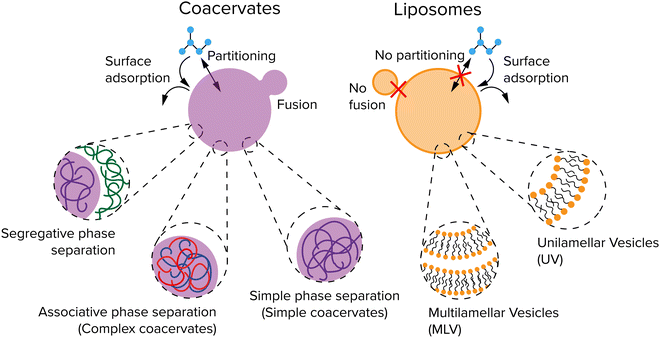
Compartments used for protocells or artificial organelles are commonly divided in two main classes: membrane-enclosed compartments and membraneless compartments. Both types of compartments are present in modern cells. Examples of membrane-enclosed compartments include mitochondria, the nucleus, the Golgi apparatus, lysosomes and transport vesicles in the cell.25 Moreover, the cell cytoplasm is also enclosed by the plasma membrane, which forms an outer boundary of the entire cell.Besides membrane-bound compartments, many cells contain a plethora of membraneless compartments, which are often referred to as biomolecular condensates or membraneless organelles. Well-known examples include nucleoli, Cajal bodies, nuclear speckles, P-granules and processing bodies.26 Membraneless organelles have been proposed to form through LLPS, a process that occurs spontaneously above a certain threshold concentration. Inspired by these biomolecular condensates, liquid-like droplets that form through phase separation are also investigated in protocell research. These droplets are usually called coacervates, a term that originates from colloid science and that was adopted by Oparin and used in a protocell context,24 but that denotes the same state of matter as most biomolecular condensates.
Here, we will mainly focus on coacervates as membraneless protocells, as the lack of a membrane offers some important advantages for growth, replication and division. However, coacervates also have some drawbacks, and other, membrane-enclosed protocells (vesicles) could offer an advantage, for example in effectively separating groups of replicators. In addition, we can take inspiration from strategies developed, for instance, to induce division of vesicles. Therefore, we will also include some examples from research into growth, replication and division in liposomes where appropriate in this article. For a detailed account of the properties and advantages of different protocell types, we refer the reader to several recent reviews.7,8,24,27–29 Interestingly, it is becoming increasingly clear that coacervates can interact strongly with membranes, and that this interaction may have a functional role in modern cells.30–33 As a consequence, it may be plausible that hybrid protocells with a coacervate core and a lipid-based membrane formed at some point during the emergence of cells.
Coacervates are droplets or macroscopically separated phases that form through a type of LLPS and that are more dense than their surrounding solution because they are enriched in one or more solute species.34,35 In general, three types of LLPS are distinguished: segregative, associative, and simple phase separation (Fig. 2). Segregative phase separation involves two soluble polymers or other macromolecules in a common solvent that do not mix due to repulsive interactions, which results in their phase separation in two phases with the same solvent that are both enriched in one of the polymers. The prototypical example of segregative phase separation is the demixing of aqueous solutions of polyethylene glycol (PEG) and dextran. Because both phases are enriched in a polymer, they are not considered to be coacervates. Associative and simple phase separation do result in the formation of a coacervate phase that is selectively enriched in (macro)molecules and denser than the surrounding solution.
Associative phase separation involves two soluble molecules that phase separate into one phase (a complex coacervate) due to attractive interactions between them. The best-known examples of associative phase separation involve oppositely charged polymers or small molecules, such as polylysine (pLL) and ATP. Simple phase separation involves one type of solute, which self-associates via intermolecular attractive interactions, leading to phase separation into a simple coacervate. Simple coacervation is different from precipitation of poorly soluble proteins or polymers in the liquid nature of the condensed phase: a simple coacervate is a strongly hydrated, dense liquid, and sometimes gel-like phase. It typically contains a significant amount of solvent (as much as 90%), and is therefore in a different state from the pure compound. The interactions that underlie the formation of coacervate protocells are charge–charge, dipole–dipole, cation–π interaction, π–π stacking, hydrophobic effect and ligand-bridging.34
The membraneless nature of coacervates makes it possible for a range of small molecules to easily exchange between the coacervate droplets and the dilute phase. The inside of a coacervates can be slightly more hydrophobic than the dilute phase, depending on the composition, which makes it a preferred environment for hydrophobic solutes. Simple and complex coacervates can be formed from a wide array of molecule types, ranging from long polymers, like complex coacervates formed by polycations and polyanions,21 but also small molecules, like simple coacervates formed by a short peptide conjugate bearing a minimal sticker-spacer-sticker motif,20,36 and complex coacervates formed by metabolites and short oligopeptides.37 For protocells, the coacervates composed of the smallest and simplest molecules possible are most interesting, but all types of coacervates can be relevant to understanding the general principles for growth, replication and division.
Liposomes are spherical vesicles that contain at least one lipid bilayer and therefore resemble the lipid membrane of a cell.38 They are generally classified depending on the number of lipid bilayers (Fig. 2): multilamellar vesicles (MLV) consist of multiple lipid bilayers in an onion-like arrangement, while unilamellar vesicles (UV) consist of only one lipid bilayer. UVs are further classified into three types depending on their size: giant unilamellar vesicles (GUV, >1 μm), large unilamellar vesicles (LUV, 100 nm–1 μm), and small unilamellar vesicles (SUV, 10–100 nm). The lumen and outside of a liposome both contain water but are separated by a hydrophobic layer formed by the hydrophobic tails in the lipid bilayer which typically gives liposomes a low permeability. The cell handles this by pore-forming proteins incorporated into the membrane. Studies have shown that small molecules and even ions can actually cross this membrane without the help of membrane proteins,39 but the osmotic pressure differences created by ions for example can also lead to a flux of water causing a change in size,40 or create pores in the membrane to equilibrate the osmotic pressure.41 Finally, fatty acid vesicles have a similar bilayer structure as UVs, but they are typically much more permeable owing to their lower hydrophobicity.40 Fatty acid vesicles have attracted significant interest as protocell models, as their components have been found on meteorites.42 However, they are only stable in a relatively small pH window.43 Phospholipid based liposomes are significantly more stable than their fatty acid counterparts, but prebiotic synthesis of phospholipids remains largely elusive.44
Protocell growth
An important feature of all cell cycles and a hallmark of living systems is their ability to grow. Growth is often linked with division, as cells grow by a combination of metabolic activity and osmotic regulation before they divide in order to maintain a stable size.45 For protocells, this process is not intimately linked to division yet, and it translates to an increase in the contents and thereby volume of the compartment. If self-replicating molecules are present inside the protocell, influx of replicator building blocks and formation of new replicators will be directly linked to growth. Ideally, protocell division should then be initiated at a certain protocell size or replicator concentration. In this section, we will first focus on growth as a separate aspect. By studying growth in protocell models with varying complexity, such as coacervates with small metabolites or macromolecules like RNA, or in the presence of fatty acids and phospholipids, physical and chemical principles that govern this process could be identified, thus helping us understand how growth of the first generation of cells could have emerged.46Passive and active growth
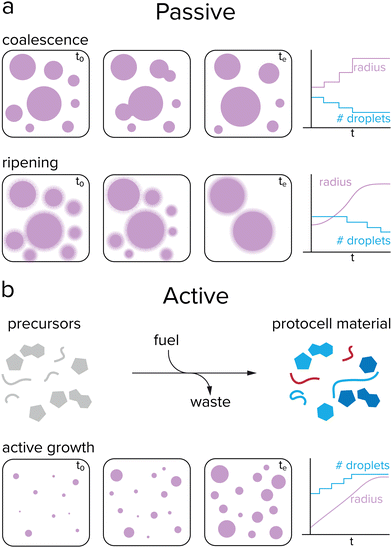
Growth of protocells can be defined as an increase in average size or volume over time. Such an increase in volume could occur either passively or actively. While very primitive cells may have relied on passive mechanisms, active growth is required for sustained growth and proliferation. Passive growth occurs naturally and is driven by a system's convergence towards thermodynamic equilibrium, while active growth requires a reaction that keeps the systems away from equilibrium.47 Phase separated droplets, such as coacervates, coalesce over time to form larger droplets, which decreases the total surface energy of the system (Fig. 3a). Additionally, these droplets can undergo Ostwald ripening, a process during which large droplets grow at the expense of smaller droplets. Small droplets have a relatively high internal pressure compared to large droplets, resulting in an enhanced surface concentration and a flow of droplet material from small droplets towards large droplets.48The previously mentioned processes of coalescence and Ostwald ripening occur spontaneously and are driven by a minimization of the system's free energy. In contrast, (proto)cellular growth is an active process that requires energy to be sustained. Moreover, in the previously mentioned cases of passive growth, the total number of protocells decreases as the average size increases, which is not desired for protocells that need to survive and replicate. For active growth, where single protocells can grow without the need of sacrificing other protocells, the system needs to be maintained away from equilibrium, which can be achieved by an external energy input, such as a chemical reaction or concentration gradient (Fig. 3b).47 This would enable coacervate protocells to grow without a decrease in the total number of compartments.49,50 However, the question remains how controlled growth can be achieved in coacervate protocells. Are chemical reactions required for active growth, or can other processes be used? Does active growth need to compete with passive growth or can both work in synergy, and should growth be controlled in some way?
Chemistry leading to growth
Living systems need energy to sustain themselves and grow. Protocells have this energy available in the form of chemical reactions and gradients. When comparing reactions in coacervates to other protocell systems like liposomes, the presence of the membrane has a significant effect. Coacervates are open systems and allow for free exchange of reactants between the compartment and its surroundings. Liposomes contain a physical barrier that separates their content from the environment, which is desired when containing genetic information like RNA and DNA, but could hinder chemical reactions, as fuel runs out and waste builds up. Chemical reactions that produce protocell material inside or in the presence of coacervates and liposomes are the prime candidates for active growth.49Reactions that produce coacervate components usually involve functionalization reactions that add an interaction motif to a molecule or elongation reactions that increase the number of interaction motifs, such as phosphorylation and (RNA) polymerization.51 As these reactions increase the number of interaction sites of a molecule, such as the number of charged or aromatic groups, they typically increase the tendency for phase separation. The first report of coacervate formation driven by such a reaction was by Aumiller and Keating.52 In this system, peptide/RNA coacervates could be formed by dephosphorylation of phosphoserine residues of an arginine-rich peptide by lambda protein phosphatase (Fig. 4a). Removal of the negatively charged phosphate groups unmasks the positive charge of arginine, which enhances the charge–charge interactions between RRASLRRASL and RNA, and induces coacervation. A similar phosphorylation-based system developed by our group uses pyruvate kinase to convert ADP and phosphoenolpyruvate (PEP) to ATP, which can form coacervates with pLL (Fig. 4b). A second enzyme hexokinase was used to convert ATP with glucose back to ADP to dissolve the droplets, and by varying the PEP/glucose ratio at the start of the reaction the onset of coacervation could be controlled.53
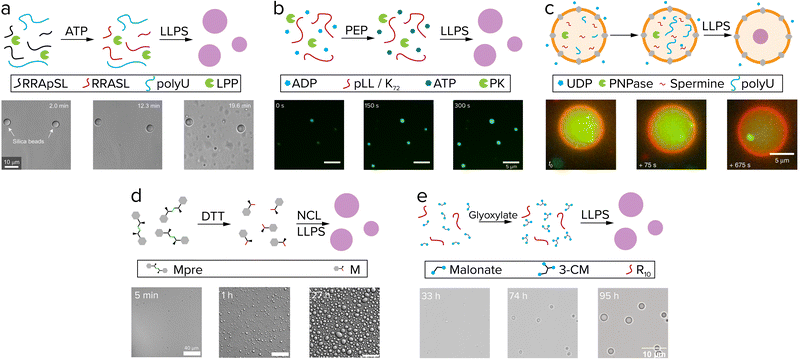 | ||
| Fig. 4 Active growth in coacervates driven by reactions. (a) Phosphorylated (RRASpL)2 is dephosphorylated to (RRASL)2 by lambda protein phosphatase and phase separates with polyU. Adapted with permission from ref. 52. Copyright (2016) Springer Nature Limited. (b) ADP is converted to ATP by pyruvate kinase after addition of phosphoenolpyruvate (PEP) and phase separates with pLL. Adapted from ref. 53 and 58. (c) UDP diffuses through α-hemolysin pores (grey) and is used by PNPase to elongate polyU, after which it phase separates with spermine. Adapted from ref. 55. (d) Disulfide building blocks (Mpre) are reduced by DTT to form amino acid thioester monomers that oligomerize by native chemical ligation (NCL) and subsequently phase separate. Adapted from ref. 59. (e) Coacervate droplets comprising oligoarginine and 3-carboxymalate form and grow upon reaction of malonate with glyoxylate. Adapted from ref. 37. | ||
Although both these systems fulfill the requirements of a reaction that keeps producing droplet material, growth of individual droplet protocells was not investigated. A reason for this is the need to follow individual dispersed droplets over time by microscopy. In recent years, many groups have reported the use of passivated glass surfaces, for example with polyvinyl alcohol (PVA) and PEG, to image coacervate droplets without them wetting the glass.51 Another platform to track single coacervate droplets is by encapsulating them in isolated environments such as water-in-oil droplets54 or liposomes.55 These systems offer high spatial control over coacervates, though one drawback is the limited flow of reagents. Deshpande et al. added α-hemolysin pores to their coacervate-in-liposome platform to allow diffusion of UDP into the liposomes, following conversion by PNPase to polyU, which then forms coacervates with spermine (Fig. 4c).55 The formation of coacervates is taking place exclusively inside liposomes, as both PNPase and coacervate material cannot leave the liposomes. However, when analyzing the droplet size over time, it became clear that after nucleation, coacervate growth is still dominated by fusion events: after the reaction was initiated, the number of coacervates decreased steadily. When studying the same reaction in an ‘open’ system, Spoelstra et al. found that active coacervates adopt nonspherical shapes, and that the degree to which coacervates are driven away from their spherical equilibrium shape is correlated to both enzyme and substrate concentrations.56 Recently, Donau et al. showed that the number and average volume of droplets increased in peptide coacervates formed by a chemical reaction that removes two negative charges, which is indicative of active coacervate formation.57 However, tracking individual coacervate droplets in order to elucidate the mechanisms and limitations underlying active growth and to assess the competition with passive processes, remained elusive.
Growth of single coacervate protocells directly correlated to chemical conversion was reported for the first time by our group.58 Over the course of the reaction, where ATP is synthesized from ADP by pyruvate kinase, radial growth rates of single droplets could be analyzed and connected to the ATP-forming reaction (Fig. 4b). The coacervate growth is limited by diffusion of substrates and proteins into the droplet, and growth rates could be controlled by adjusting their concentrations. The protocells grow in size without dilution of their contents and without decrease of the population size. Moreover, passive events like coalescence and Ostwald ripening could be differentiated from active growth, which is valuable when analyzing the growth as a result of the reaction. This system is the first step to an actively growing protocell, although it still consists of complex components such as proteins and enzymes.
The next steps in active growth of coacervate protocells should focus on designs with smaller molecules, such as peptides and sugars instead of enzymes. Matsuo et al. reported on a system in which coacervates are generated by native chemical ligation (NCL) of peptide building blocks. Dipeptide building blocks with a disulfide linker are reduced with DTT and subsequently oligomerize by NCL to form di-, tri- and tetra-peptides (Fig. 4).59 The hydrophobic interactions that drive phase separation are likely similar to the sticker-spacer coacervates developed in our group,20 however additional associative interactions between the oligopeptide and benzyl mercaptan are crucial to induce LLPS in this system. Although growth of individual coacervates was not shown here, the generation of relatively hydrophobic coacervates controlled by a nonenzymatic reaction is promising, as these droplets could serve as microreactors for chemical reactions that are not efficient in water.20
The most recent work of our group showed how an aldol addition reaction that converts a dicarboxylic acid to a tricarboxylic acid is able to fuel actively growing coacervates (Fig. 4e).37 Similar to the ATP-based system, droplets grow by chemical conversion of fuel to droplet material, however, in this system no large proteins were used. Both these systems show how droplet material producing reactions can lead to active growth in more prebiotically plausible coacervate protocells, though further improvements like control over size dispersity of the population are desirable next steps.
The importance of controlled protocell growth
After having achieved growth of single coacervate protocells, a next challenge would be to have a better control over protocell growth rates. Inducing growth by a reaction already offers some control over the rate, as the concentrations of substrates or catalysts can be tuned, although droplet sizes can vary substantially depending on the preparation, as seen by large standard deviations of the average droplet radii.37,58 A higher level of control can be achieved by introducing an additional reaction that breaks down protocell material.49 If this reaction is localized within the protocell, large droplets may be expected to degrade faster than smaller ones. By having both the anabolic (droplet forming) and catabolic (droplet degrading) reaction taking place at the same time and tuning reaction rates, a so-called steady state of droplet size can be achieved (Fig. 5a).50 Moreover, theoretical studies have predicted that a reaction cycle like this could induce droplet division as a result of a shape instability for certain combinations of interfacial tension, reaction rates and diffusivities.49,60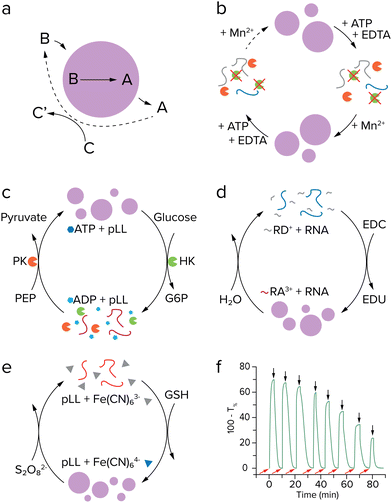 | ||
| Fig. 5 Coacervate formation and dissolution cycles. (a) Active droplet model described by Zwicker et al. Stable droplets and division were predicted for such a system. Adapted with permission from ref. 49. Copyright (2017) Springer Nature Limited. (b) (De)phosphorylation cycle of (RRASL)2 by lambda protein phosphatase (green) and protein kinase A (orange). Adapted from ref. 52. (c) Enzymatic control over LLPS by ATP formation and hydrolysis, with both enzymes operating under the same conditions. Adapted from ref. 53. (d) Coacervation by EDC-fueled anhydride formation of peptide RD+ to form RA3+, which phase separates with RNA (blue). Adapted from ref. 57. (e) Redox controlled coacervation by ferricyanide reduction with pLL (red). (f) Turbidity measurement of chemically controlled coacervate formation and dissolution. Adapted from ref. 64. | ||
For a practical implementation of reaction networks that form and eventually dissolve coacervates, we can take another look at the examples discussed above of actively growing coacervates, as they can in principle combine reactions that produce and break down droplet material. The system developed by Aumiller and Keating presents both a droplet forming and dissolving reaction. However, the reaction cycle cannot run continuously because protein kinase A does not function in presence of Mn2+, an essential cofactor for the second enzyme lambda protein phosphatase (Fig. 5b).52 The enzymatic network developed by our group uses two enzymes which are compatible, allowing for subsequent cycles of coacervate formation and dissolution under the same conditions.53 After formation of coacervates by pyruvate kinase, hexokinase can dissolve the coacervates by converting ATP to ADP with glucose as substrate (Fig. 5c). It was found that after six cycles, the system lost its ability to generate new coacervates, as a result of the build-up of waste products from the reactions. This issue could possibly be solved by introducing a flow in the system that feeds new reactants and subsequently removes waste,61 or introduction of additional enzymes that convert waste to new substrate. However, both of these options introduce additional experimental challenges, as flow of reactants requires some sort of immobilization of coacervate droplets, and additional enzymes would greatly complicate the network.
The reaction driven coacervate generation reported by Donau et al. is a reaction cycle that does not require enzymes.57 The negatively charged C-terminal aspartate in an arginine-rich peptide precursor is converted to an anhydride by EDC, removing two negative charges from the peptide (Fig. 5d). The anhydride is able to phase separate with RNA and form coacervate droplets, similar to the system by Aumiller and Keating.52 A distinctive characteristic of this system is that the anhydride is slowly hydrolyzed in water and forms the precursor again, completing a reaction cycle and also dissolving the droplets. The hydrolysis of these anhydrides was intensively studied by the Boekhoven lab in other fuel-driven assemblies before its application to coacervate growth.62,63 This research showcases some of the additional control that can be gained over a system by introducing a reaction that breaks down droplet material. By varying the initial concentration of EDC fuel, the behavior of the formed droplets ranges from metastable to dynamic, where dynamic droplets form vacuoles and fragment into smaller satellite droplets that eventually decay completely. Because the backward reaction is initiated by water, which is present in large amounts inside the complex coacervate droplets, control of the dissolution reaction is limited. Recently, our lab developed redox-active coacervates consisting of pLL or oligoarginine with the redox pair ferri- and ferrocyanide (Fig. 5e).64 Coacervates can be generated when ferricyanide with three negative charges is reduced to ferrocyanide with four negative charges, and dissolved again when ferrocyanide is oxidized. This cycle can be repeated more times than the pyruvate-hexokinase system and does not need enzymes to function, showing more signs of prebiotic plausibility (Fig. 5f). Moreover, the oxidation reaction that dissolves the coacervates could be driven by an oxidizing agent that is concentrated in the coacervates. The extent to which this could result in a stable size of active coacervates remains to be seen. These advances in constructing coacervate generating and dissolving networks could enable further steps in developing a protocell division cycle, as discussed later.
Replication of information
In order to create a protocell that can undergo Darwinian evolution, the protocell needs to be able to store hereditary information, in addition to growth and division. In biological systems this information is plain to see, as the DNA sequence relates to properties of the organism such as anatomy, size and colour. This information gives the species a unique “identity”, distinguishing it from similar species and creating phenotypic differences on which natural selection can act. In simple prebiotic systems, however, this storage of information is less obvious, but the molecular structure or specific sequence of subunits of a molecule can be seen as a form of information.4In order to maintain the identity of the species across generations, the hereditary information has to be transmitted from the parent to the offspring, which requires its replication followed by distribution among the offspring. In modern-day organisms, the hereditary information that is stored in the DNA is passed on to future generations by a highly coordinated replication process involving numerous enzymes. It is unlikely that such advanced biomolecules were present during the emergence and early development of life. Therefore, scientists have shown great interest in the development of self-replicating systems that replicate without enzymatic aid. Such systems are based on autocatalysis: they can selectively catalyze their own formation through specific recognition between a catalytic template product and the reactant building blocks. Because of this selectivity, information in the form of molecular structure or subunit sequence can be transferred from the original molecule to its copy.65,66 Additionally, a self-replicating system contains all the necessary information for replication. It can thus autonomously catalyse its own copying, without a need for external (enzyme) catalysts that the system does not encode for.4,67
Chemistry leading to replication
The self-replicating systems that have been developed to date can be divided into two groups according to the mechanism of replication: template-directed ligation and template-directed polymerization (Fig. 6). In template-directed replication (Fig. 6a), a template T catalyses its own formation from two building blocks, A and B, that are complementary to the template.4,66,68A and B first react via an uncatalyzed bimolecular reaction to generate template T. A new pair of building blocks A and B then binds to the template via non-covalent interactions, bringing them in close proximity and in the right orientation for bond formation. In other words, their effective molarity is increased by the template. This leads to acceleration of the reaction between A and B, forming a new copy of the template molecule within the product duplex [T·T]. Dissociation of this complex yields two identical T molecules. In most template-directed self-replicating systems, however, this dissociation is slow, as for entropic reasons the template duplex is usually more stable than the termolecular [A·B·T] complex.68 This is called product inhibition, and is an important limiting factor in many self-replicating systems.4,65,66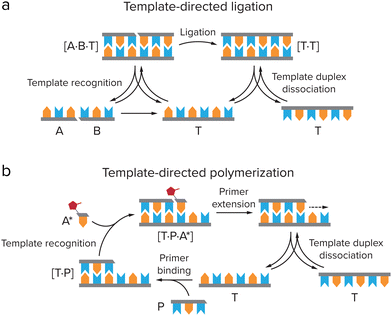 | ||
| Fig. 6 Two types of mechanisms for self-replication. (a) Template directed ligation, in which a template T catalyses its own formation from ligation of two building blocks, A and B, that are complementary to the template. Adapted from ref. 4. (b) Template-directed polymerization, in which T is copied by binding of a primer P, which is elongated by activated monomers A* that are complementary to the template. | ||
Several systems have been developed based on template-directed ligation, including purely synthetic, peptide-based and nucleic acid replicators.65–69 Here, we will focus on peptide and nucleic acid replicators, as they bear the largest similarity to extant life, and have the strongest similarity to the most common ingredients of coacervates. Peptide replicators are typically based on secondary structure formation in which the template and building blocks coordinate to each other, instead of on direct recognition between the template and building block sequences. In 1996, Ghadiri and co-workers developed a self-replicating system based on the leucine-zipper motif, where recognition between the α-helical peptide template and its building blocks is based on hydrophobic interactions, which drive the formation of a coiled-coil structure in which the hydrophobic groups are shielded.11 The building blocks are then ligated through NCL, which yields a copy of the original template peptide. A similar peptide replicator system has been developed by Ashkenasy and co-workers, which is based on anti-parallel β-sheet formation.12,13,70 Short peptides with alternating hydrophobic and hydrophilic residues can form β-sheets in water. Two short peptide fragments can coordinate to these β-sheets by hydrogen bonding, which catalyses their coupling via NCL. The newly formed template does not dissociate, but instead stacks on the β-sheet to form fibrillar structures which catalyse replication. This way, product inhibition is avoided. The self-replicating system based on stacking macrocyclic rings that was developed by the Otto group avoids product inhibition in a similar way.14,15 They use a dynamic combinatorial library of building blocks consisting of an aromatic core that is functionalized with two thiol groups and an amphiphilic peptide chain. Through oxidation, these building blocks can reversibly form macrocyclic rings of different sizes, which subsequently stack due to self-assembly of the peptide chains into β-sheets. Information in terms of macrocycle size can then be transferred from one generation of stacks to the next.
The first nucleic acid-based replicator was developed by Von Kiedrowski in 1986, and made use of a hexadeoxynucleotide template, to which two complementary trinucleotide building blocks could coordinate via Watson–Crick–Franklin base pairing interactions.9 Ligation was achieved through carbodiimide activation, resulting in formation of a phosporamidate bond.71,72 In this system, product inhibition was a significant limiting factor, which could only be overcome by tethering the template to a solid support, dissociating the product template by heating after replication, and tethering the newly formed template before entering the next replication cycle.73 The replicating R3C ligase ribozyme system that was developed by Joyce is not limited by product inhibition.10,74 In this system, two variants of the ribozyme can bind each other's building blocks and promote their ligation to form the other ribozyme variant, in a cross-catalytic manner. When several cross-catalytic ribozymes and their building blocks were combined, this system was able to select and amplify the most efficient ribozymes, thus exhibiting a rudimentary form of evolution. In addition, the R3C ligase replicator was shown to encode functional information. Modification of the stem loop region resulted in encoding of aptamer binding functionality, creating an aptazyme replicator that only functions in the presence of a specific ligand.75 Introduction of functions beyond the ability to replicate is an essential step in evolution of self-replicating systems, as it allows for a partial decoupling of genotype and phenotype, which significantly increases the variation within the replicator population, as one genotype can result in a range of different phenotypes.4,69,76
Replication through template-directed ligation of building blocks is distinctly different from the replication of genetic information in extant biology. In an attempt to bridge this gap, template-directed polymerization was developed (Fig. 6b), in which a primer P is elongated on a template sequence T by activated monomers A*. Following pioneering work by Orgel,77,78 the Szostak lab developed a prebiotic replication mechanism capable of template-directed addition of mononucleotide building blocks.79,80 In their system, a copy of the template is formed by primer extension on the nucleic acid template, using imidazole-activated nucleotides that form an imidazolium-bridged dinucleotide intermediate, which then binds to the template. Subsequent attack of the 3′-hydroxyl of the primer displaces an activated nucleotide as the leaving group and results in extension of the primer by one nucleotide. The reaction is catalysed by Mg2+ or Fe2+,81 and can be made more efficient by the use of 2-methylimidazole- or 2-aminoimidazole-activated nucleotides.78,82 Although the system is still hindered by the low rate of extension, due to strand reannealing and hydrolysis of activated monomers, and by low copying fidelities,69,83 it can replicate both DNA and RNA segments of up to 25 nucleotides long, which is long enough to encode for simple active ribozymes and/or aptamers.84,85
Dealing with parasites in replication
In order for these self-replicating systems to undergo evolution towards greater complexity, they need to overcome product inhibition and be amplified (close to) exponentially, but also be able to mutate and show dynamic kinetic stability, as was reviewed by Duim and Otto.4 Another important hurdle that self-replicating systems have to overcome eventually to proliferate and gain new functions is to deal with the emergence of parasites during the replication process. As the most elementary selection of replicators is based on replication efficiency, shorter sequences that replicate more quickly will be selected over slower replicating longer sequences. Replicators that are simpler in construction can therefore come to dominate the reaction mixture, as was made painfully clear by Spiegelman's experiments on the Qβ replicase, an RNA-dependent RNA polymerase from the Qβ bacteriophage that specifically replicates its own genome.86,87 By creating a selection pressure for faster replicators, due to 75 rounds of serial transfer of the Qβ replicase into fresh mixtures of nucleoside triphosphates (NTPs) after ever shorter incubation times, a replicator was created that had a replication rate 15 times as high as that of the original Qβ RNA, while the size was reduced by 83%. Although these shortened RNAs were still a valid template for the replicase, they were no longer infectious and no longer encoded any genes.88 The evolution experiment had thus created parasites for the original Qβ replicase. Even though this biological system is more complex than the simple self-replicators we discuss here, it provides a clear example of how Darwinian evolution of a replicator does not necessarily result in complexification and spontaneous emergence of new function.Compartmentalization is an important step in overcoming the formation of parasites. Theoretical models such as the stochastic corrector model and transient compartmentalization model predict that through compartmentalization, cooperative groups of replicators that all aid a common metabolism can be protected from parasites overtaking the population.89,90 Because parasites that might be formed in the replication process do not contribute to the metabolism while they do use up resources, groups of replicators that contain parasites have a less efficient metabolism, leading to a lower growth rate or even extinction. Groups of replicators that do not contain parasites will replicate more efficiently and can therefore survive, while the local groups that are infested with parasites vanish.67 This selection on group level can be realised by (transient) compartmentalization of the replicator groups (Fig. 7). When the compartments divide or when they are dissolved and reformed, e.g. through coacervate compartment formation and dissolution,49,53 replicators are randomly grouped in compartments, leading to variation between the compartments on which natural selection can act.89–91 The Yomo and Griffiths group managed to show that this theoretical prediction also holds in practice.92,93 When revisiting the Spiegelman experiment in water-in-oil droplets that were manually divided or dissolved and reformed, they observed that it is possible to avoid parasite take-over in the Qβ replicase system. In addition to overcoming parasitism, compartmentalization of replicators can help to concentrate the replicator and enhance the rate of reactions that might be prohibitively slow in dilute solution.20,23,54
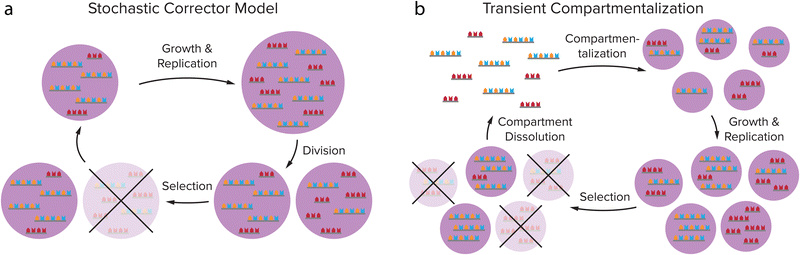 | ||
| Fig. 7 Theoretical models predict that compartmentalization can protect cooperative groups of replicators from parasites overtaking the population through selection on group level. (a) Stochastic Corrector Model: compartments containing both replicators and parasites grow and the replicators and parasites are replicated, after which the compartment divides, leading to a random distribution of replicators and parasites in the daughter cells. This gives variation between the compartments on which natural selection can act. (b) Transient compartmentalization model: in a pool of replicators and parasites, compartments are formed that take up random groups of sequences, leading to variation between the compartments. The fittest compartments will grow and replicate most efficiently, and are selected. Adapted with permission from ref. 91. Copyright (2020) Elsevier. | ||
Achieving replication in coacervates
In order to create self-replicating systems that evolve towards greater complexity, compartmentalization and division of the compartment are thus important steps. Therefore recently, the functioning of self-replicating systems, related to the ones discussed above, has started to be investigated inside coacervate (and liposome) protocells. In order for self-replicating systems to function inside coacervate protocells, each step of the replication process has to remain functional: selective template recognition by the building blocks, ligation/polymerization and template duplex dissociation (Fig. 8). The ligation/polymerization reaction is likely the easiest step to function inside coacervate protocells, as it was recently shown by several groups that reaction rates can be significantly enhanced inside coacervates if the reagents are enriched inside the droplets (see above).20,37 Recently, Fraccia and Martin also showed that ligation of DNA segments functionalized with a 3′ terminal phosphate group to produce phosphodiester bonds upon activation by EDC, is efficient inside liquid crystalline coacervate droplets, and still functions in isotropic coacervate droplets (Fig. 8b). The DNA ligation also had a direct effect on the coacervate morphology, forming more solid-like or multiphase structures as the reaction progressed.94 In addition to efficiently supporting DNA ligation, our group has also shown that coacervates can significantly increase the rate of peptide bond formation through amino thioacid oxidation by ferricyanide, which forms coacervates with pLys(Me)3.64 Furthermore, coacervates were shown to enhance the activity of ribozymes, opening the door towards enhancement of ribozyme self-replicating systems.95–97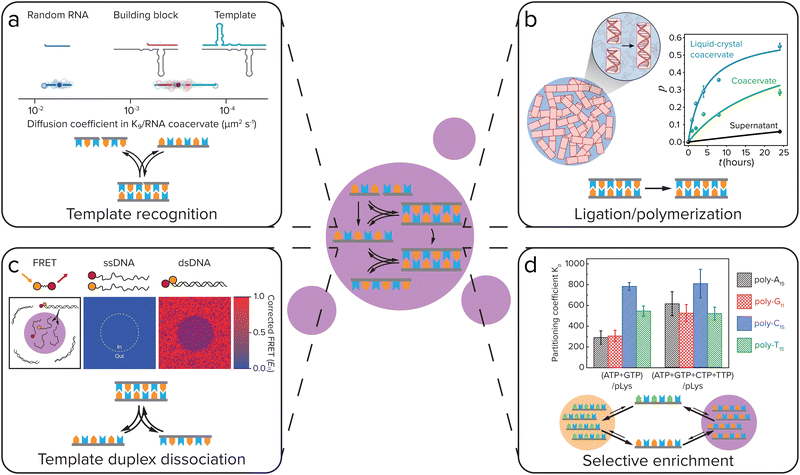 | ||
| Fig. 8 For self-replicating systems to function inside coacervate protocells, each step of the replication process has to remain functional: selective template recognition by the building blocks, ligation/polymerization and template duplex dissociation. Additionally, coacervates can give selective enrichment of certain replicator sequences. (a) Template recognition between building blocks and the template. Iglesias-Artola et al. showed by Fluorescence Recovery After Photobleaching (FRAP) that the R3C ligase replicator template and building block have similar diffusion coefficients in coacervates made from K9 and the replicator RNA. Adapted from ref. 100. (b) Efficient ligation/polymerization. Fraccia and Martin observed efficient ligation of dsDNA inside both liquid crystalline and isotropic coacervate droplets. Adapted from ref. 94. (c) Dissociation of the template duplex to release the copied template. Nott et al. observed melting of 12-mer DNA duplexes inside Ddx4 coacervates using Förster Resonance Energy Transfer (FRET). Adapted with permission from ref. 98. Copyright (2016) Springer Nature Limited. (d) Selective enrichment of replicator sequences inside coacervates. Lu et al. showed that in coacervates of polylysine with different nucleoside triphosphates (NTPs), polynucleic acid sequences that can form Watson–Crick–Franklin base pairs with the NTP are selectively enriched, while partitioning is less selective in coacervates made with all four NTPs. Adapted from ref. 101. | ||
Getting both efficient template recognition and template duplex dissociation in coacervates will, however, likely be a larger challenge. Coacervates have been reported to melt DNA double helices, due to competitive interaction of the coacervate material with the DNA, disrupting DNA base-pairing (Fig. 8c),98 although this depends on the specific coacervate material.99 Compartmentalization of DNA replicators into coacervates can therefore likely help to overcome product inhibition by weakening the template duplex. However, since weakening of template–template interactions will inevitably also weaken template-building block interactions, this means template recognition will be less efficient. The outcome of this delicate balance will be highly dependent on the nature of the coacervate. Poudyal et al. investigated the template-directed polymerization reaction developed by the Szostak lab inside coacervates.96 They observed a strong dependence on the charge density and interaction strength of the polycation that is used to form coacervates. Polyallylamine, oligoarginine and to a lesser extent also oligolysine inhibited template-directed polymerization, while the quaternary ammonium polymer polydiallyldimethylammonium chloride (PDAC) that has weaker ion-pairing interactions and a lower charge density was able to support the reaction, and even allowed for a more efficient reaction at low Mg2+ concentrations than in buffer. They further showed that RNA diffusion is significantly faster in PDAC/RNA coacervates than in the coacervates that inhibited template-directed polymerization, indicating that coacervates used to compartmentalize self-replicating systems should be dilute enough to not disrupt template-recognition interactions too much.
Iglesias-Artola et al. elaborated on this by investigating the R3C ligase replicator that was developed by the Joyce lab inside coacervates.100 When comparing oligoarginine and oligolysine homopeptides with charge-interspaced (RGG)4 and (KGG)4 heteropeptides, they too observed a strong dependence on the charge density, RNA diffusion rates and replicator–peptide interaction strength for the functioning of template-directed replication inside coacervates. Replication only took place in the (RGG)4 and (KGG)4 coacervates, while it was strongly inhibited in oligolysine and oligoarginine coacervates. Even though in both oligolysine and heteropeptide coacervates strong template-building block complexes were formed (Fig. 8a), the difference in reactivity was explained by ITC experiments that showed a strong binding of oligolysine and oligoarginine to the RNA, which leads to competitive replacement of the Mg2+, that is required for (template-directed) ribozyme ligation, from the coacervates. In (RGG)4 and (KGG)4 droplets, however, enough Mg2+ remains present for the ribozyme ligation to function. Although in both of these papers, the self-replicating systems do not yet function as well as in buffer, these results show that self-replicating systems can function inside coacervate protocells, but that their efficiency is highly dependent on a delicate balance of interaction strength between the coacervate materials and the replicator.
In addition to having an effect on ligation/polymerization rates and template-building block recognition and dissociation, the coacervate environment can also impose selectivity on the replicator sequences that are taken up and replicated. Our group has previously shown that in coacervates formed from pLL with NTPs, certain polynucleic acid sequences can be selectively enriched depending on the NTPs that form the coacervates, with polyC and polyT partitioning strongly into coacervates made with ATP and GTP, while in coacervates made with all four NTPs partitioning was less selective (Fig. 8d).101
Such a difference in uptake of different RNA sequences was also observed by Wollny et al., who analysed the partitioning of the different RNA sequences in total RNA from human induced pluripotent stem cells by single-cell RNA sequencing technologies.102 They too observed that for certain RNA sequences, the enrichment inside coacervates depended on the nature of the coacervate material. RNAs with high sequence complementarity within or across RNA sequences were significantly enriched in single coacervates. A similar observation was made by Aumiller et al., who saw that polyA15 partitions around 50× more strongly into polyU/spermine coacervates than other polynucleotides of the same length.103
Through this bias for uptake of certain RNA sequences, different types of coacervates can localize different RNA replicators to create a population of genetically different protocells, on which natural selection can act. Furthermore, we have observed that not only longer polynucleic acid sequences are selectively enriched in coacervates, but also single nucleotides show differences in partitioning. The extent of partitioning of NTPs depends on the nucleobase, with GTP and ATP – which have a higher base stacking free energy – partitioning significantly more than CTP, UTP and TTP.101,103 Such a difference in partitioning can have important consequences for replication: different partitioning of activated nucleotides can bias which sequences are most efficiently replicated, and implies that it is favourable to colocalize the template and building blocks inside a coacervate. Additionally, such a preference for complementary sequences could give the system a certain resistance towards mutants, which would have a lower complementarity to the building blocks. Taken together, coacervates can impose selectivity on which replicator templates and building blocks are enriched, and can thereby affect the efficiency of replication of different replicator sequences inside the droplets. In this way, a population of genetically different coacervate protocells can be created, which have a different fitness level. Once these droplets can also divide, this would allow for evolution of the coacervate protocells.
Division of protocells
The division of a protocell and its information is a crucial step in the protocell cycle (Fig. 1), and essential for the production of offspring and for evolution. At the same time, division is the most elusive of steps in the coacervate protocell cycle, as it involves overcoming the tendency of coacervate droplets to relax to a spherical shape as a result of surface tension. Moreover, division must be reasonably well controlled to make sure that most or all offspring that is produced contains the basic number of components to be viable. In this section we use division to indicate the splitting of a single protocellular compartment in two or more daughter compartments, which is sometimes also called fission, splitting or fragmentation.The same challenges surrounding the bottom-up engineering of a division mechanism play a role in research aimed at building a synthetic cell, and it is helpful to take inspiration from the strategies that have been proposed there.104 The mechanisms to induce protocell or synthetic cell division can be divided into externally and internally driven division mechanisms. In current-day cells the division and thereby the multiplication of a cell occurs internally: it is controlled by the cell's internal state.105 Internally driven and controlled division is also the ultimate goal for division of protocells. However, it is possible that division was initially controlled and driven by external factors, and therefore, we will discuss the possibilities of both types in this section. In addition, we chose to discuss both division of coacervates and liposomes, as it is possible that a hybrid protocell with a coacervate core and a lipid or fatty acid-based membrane was present or even required at the stage of protocell division.31,106,107
Externally driven division
External factors that can drive cell division include osmotic pressure,108 light,108,109 temperature,110 and shear force,111 and these have all been used to induce division of liposomes. Dreher et al. have designed a GUV that has a defined division plane by segregation of lipids in the membrane (Fig. 9a).108 When the GUV is divided at the interface between the two regions, it splits in precisely two GUVs. The actual division is driven by a change in osmotic pressure, in this case due to evaporation of water on the outside, which leads to an osmotic flux of water out of the GUV and an increase of the GUV surface-to-volume ratio. The GUV deforms and eventually divides at the designated interface into two smaller GUVs. The same group also showed that light induced local division of GUVs is possible.108 Upon irradiation with UV-light their caged fluorescein is uncaged, which causes an osmolarity change and division is observed as a result of the removal of water. The advantage of using light is that division only takes place in the region that was irradiated, giving full spatiotemporal control over which GUVs are divided. In both examples, the division is asymmetric in the sense that the segregated lipids in the membrane of the parent vesicle are unequally divided over the daughter vesicles. Additionally, this division cannot be repeated. To generate new parent GUVs, single-phased vesicles were added, which fused with one of the daughter vesicles.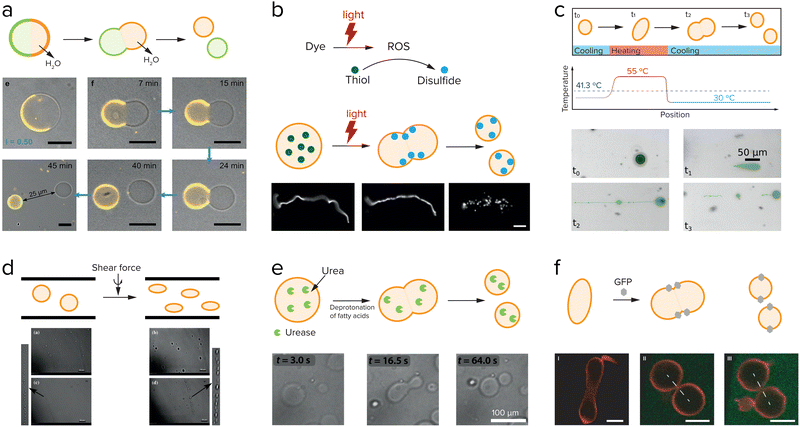 | ||
| Fig. 9 Research aimed at division of liposomes focusses both on externally driven division (a–d), and internally driven division (e and f). (a) GUVs with a segregated lipid bilayer divide upon increasing the outside osmotic pressure either by an enzymatic reaction or by a light-driven reaction. Adapted from ref. 108. (b) Division of fatty acid vesicles containing either a fluorescent dye or hydroxypyrene upon irradiation with light due to the formation of reactive oxygen species (ROS) that react with thiols to form disulfides, which interact with and deform the membrane. Adapted from ref. 109. (c) Division of UVs by heating and cooling. Adapted from ref. 110. (d) Division of GUVs in shear flow. Adapted from ref. 111 with permission from the Royal Society of Chemistry. (e) GUVs can be deformed and divided by the use of an enzyme, urease, which causes an increase in the inner pH and deprotonation of the fatty acids. Adapted from ref. 112 and 113. (f) Division of GUVs by membrane-anchored GFP proteins which induce a membrane curvature and in certain cases, division. Adapted from ref. 114. | ||
Another example of light-induced division is shown by the group of Zhu et al.109 They showed that upon irradiation of a fatty acid sample containing either fluorescent dyes or hydroxypyrene, a reactive oxygen species is formed (Fig. 9b). This reactive oxygen species can oxidize thiols that were present inside the vesicles into disulfides, which then interacted with the membrane to cause a change in surface tension and subsequently division. Temperature changes have also been shown to induce division of liposomes. On early Earth the temperature changes might have been higher than in the current environment, which could have influenced the early division of protocells. Kudella et al. showed that upon heating a vesicle above their main melting point a deformation of the shape of the vesicle occurs (Fig. 9c).110 Upon cooling down again the vesicles divided. An interesting variation on this temperature-induced division of vesicles is the temperature-induced phase transition between a vesicle solution and an oil droplet phase.112,113
By heating and cooling, a vesicle population can be dissolved and reformed again, resulting in redistribution of the vesicle content, and a potential increase in population size if the nucleation occurs quickly. Such a mechanism could be used to achieve transient compartmentalization, as proposed in the previous section, to prevent parasites from taking hold. Finally, shear force is the most frequently used to drive liposome division (Fig. 9d).111,114
The same external factors that have been used to drive division of liposomes can in principle also be used to divide coacervate protocells. In general, coacervates are sensitive to many different environmental factors, including salt concentration, pH and temperature, which can lead to an unwanted dissolution instead of division. Close to the critical point of a coacervate dispersion, large size and shape fluctuations are expected that could lead to fragmentation or division of coacervates, although it can be difficult to avoid crossing the phase boundary. Donau et al. showed that active coacervate droplets that have run out of fuel and slowly become instable indeed fragment into several small daughter droplets moments before they completely dissolve (Fig. 10c).57 Lu et al. showed that temperature can also be used to dissolve coacervates composed of NTPs or RNA and cationic peptides.101 In analogy to examples with liposomes, temperature cycling could be used to destabilize coacervates enough that they fragment. Alternatively, this strategy could be used to realize transient compartmentalization required for robust self-replication.
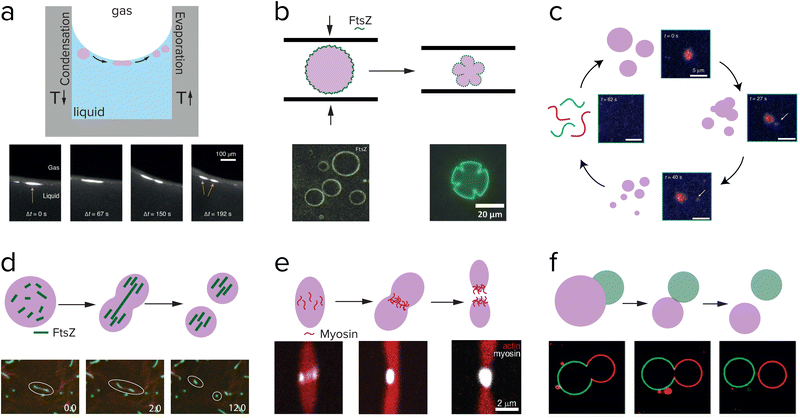 | ||
| Fig. 10 Research aimed at division of coacervates also explores externally driven division (a–f), and internally driven division (d and e). (a) Division of complex polymeric coacervates is observed in rock pores with a temperature gradient. Adapted from ref. 121. (b) pLL/GTP coacervates coated with FtsZ are pressed in between two glass plates, and deformed into shapes that resemble dividing cells. Adapted with permission from ref. 120. Copyright (2018) John Wiley & Sons. (c) Peptide/polyU coacervates in a chemically fuelled reaction cycle reach an unstable state and fragment just before dissolving. Adapted from ref. 58. (d) RNA/GFP-K72 coacervates are divided into fragments by FtsZ filament growth. Adapted from ref. 122. (e) Myosin proteins self-organize into a ring in the middle of actin coacervates which eventually results in division. Adapted from ref. 123. (f) Asymmetric division of a pair of PEG/dextran droplets in a liposome, which bud off due to an osmotic pressure difference. Adapted from ref. 124. | ||
One of the most straightforward methods to break up a coacervate protocell into smaller daughter droplets is the application of shear forces. Early work on the formation of coacervate droplets in a microfluidic device illustrates that this is indeed possible.115,116 Recently, Matsuo and Kurihara used extrusion to fragment a population of peptide-containing coacervate droplets.59 Apart from shear forces, compression may have resulted in deformation and potential destabilization of coacervates, although this requires a stabilized coacervate interface to avoid wetting. Fanalista et al. demonstrated that upon squeezing a pLL/GTP coacervate solution between two slides, the coacervate droplets became strongly deformed into flower-like shapes to the extent that fragmentation becomes possible (Fig. 10b).117 The coacervates were coated with FtsZ filaments, which formed a rigid surface layer that stabilized the coacervates. Interestingly, FtsZ is one of the key proteins involved in cell division in bacteria where it forms a ring around the dividing plane. Prebiotic analogues of this protein could have played a role in guiding the division of coacervate protocells. Finally, Ianeselli et al. showed how rock pores could have facilitated coacervate division.118 Porous rocks are believed to have been abundantly present on early Earth, and are a good candidate to aid in the emergence and evolution of life. At the interface of gas bubbles inside heated rock pores, a combination of capillary flows, perturbative fluxes after precipitation of water and Marangoni flows results in accumulation of coacervate droplets and shear-induced division (Fig. 10a).
Internally driven division
The ultimate goal of protocell division is to drive division by a reaction or process occurring inside the protocell, which can also be controlled internally. However, what a minimal internally driven division could have looked like is still a mystery. Many researchers are focusing on the division of liposomes with the use of either an enzyme or membrane-bound proteins, for implementation in a synthetic cell. An enzyme that is compartmentalized in liposomes can catalyze a reaction that affects the surface charge or structure of the membrane components. Miele et al. used urease to convert externally supplied urea into ammonia, which resulted in a deprotonation of the oleic acids located in the membrane.119 Upon deprotonation, the membrane started deforming and a mild shear force could then complete the division of the liposomes (Fig. 9e). In their follow-up work they show they have control over this reaction and can reverse the initial deformation.120 Not only enzymes can influence the membrane of liposomes, but also membrane-bound proteins can have an effect. Steinkühler et al. showed that low densities of a membrane-bound protein can induce a curvature and this can generate strains on the membrane that lead to division (Fig. 9f).121A complete cycle of growth, replication and internally driven division of coacervates is to the best of our knowledge not realized yet. Several studies show promising results on deformation or even fission of coacervates driven by the assembly of protein filaments inside coacervates. Te Brinke et al. used FtsZ in coacervates composed of RNA and a disordered elastin-like protein (Fig. 10d).122 FtsZ filaments form inside these coacervates in the presence of GTP, and because of the local crowded environment they form bundles that start deform the coacervate phase into an elongated droplet. Because the influx of the GTP fuel and FtsZ monomers is higher at the tips of the elongated droplet, the filaments grow more easily near the ends and become thinner and more fragile in the middle. Eventually, this leads to a break-up of the filaments and the surrounding coacervate phase. This process could be repeated several times, before the coacervate droplets had become too small to divide. Another active protein that can be used to divide coacervates, is the molecular motor protein myosin.123 Myosin filaments were found to assemble at the midplane of a droplet of liquid crystalline actin, and could deform and divide the droplet into two daughter droplets (Fig. 10e).
A type of asymmetric division that has also been studied in coacervate droplets is budding.124,125 In the case of Andes-Koback and Keating the asymmetric division was governed by an aqueous interface between a PEG-rich phase and a dextran-rich phase inside the vesicles, and domains of a liquid ordered and liquid disordered lipid phase in the membrane.124 After budding, which is driven by an osmotic pressure disbalance, two chemically distinct daughter protocells are generated, which could provide a route towards differentiation in protocell populations (Fig. 10f). Similar budding could be realized with coacervates. Our group and others have recently shown that multiphase coacervates can be formed from mixtures of polycations and polyanions.21,22 For some systems, the coexisting coacervates are partially wetted, and tuning of the interfacial tensions could result in a transition to a non-wetting state and budding.
Finally, an interesting and completely different mechanism of internally controlled division was shown in theory by Zwicker et al. They theoretically investigated the size and shape of active droplets, in which a fuel-driven chemical reaction produces molecules that can condense as droplets, while a second reaction taking place inside the droplets degrades these molecules.49 Not only can such a reaction network produce a population of droplets with a stable size, they can also undergo division as a result of a shape instability if a constant energy supply is present. The authors suggest that this energy supply could be a chemical fuel, but could also originate from tides or temperature gradients.
Taken together, coacervates can be divided by a variety of external and internally driven mechanisms, although all mechanisms appear to require a mild shear force for the final abscission. Internally driven mechanisms for division could be linked to replicator copying, for example by inducing multiphase separation above a certain concentration or length of replicators. Once division has been completed, small variations in the composition and size of the offspring protocells can give rise to evolutionary advantages for some coacervates.
Towards evolution
Compartmentalization can help primitive life-like systems increase local concentrations to survive in dilute conditions, enable reactions that are not compatible with each other or with the surrounding phase, and overcome parasitism. These are essential to achieve a sustainable cell cycle that includes growth, replication and division. In order for these protocells to undergo a primitive form of evolution, they must also acquire mutations, which should influence their survivability by selection pressure from the environment. Here, we explore the possibility that the protocellular compartments themselves could also provide selectivity for certain reactions. Can protocellular compartments favor a certain reaction and thereby guide evolution? A first step to answer this question is to investigate if coacervates or vesicles can influence chemical reactions. Our group and others have shown that coacervates can increase the rate of a reaction that hardly works in a dilute solution.20,126–128 The reaction rate is increased due to a combination of increased local concentrations of reactants inside the coacervates and a lowering of the effective energy barrier for the reaction caused by the different physicochemical environment.The next step is to see if this physicochemical environment could induce selectivity for certain reactants or reactions. It has been reported by many research groups that coacervates can show selective partitioning of guest molecules, which in turn can influence the rate of chemical reactions.95 Interestingly, our group recently reported that the oxidative aminoacylation of thioacids, which yields a peptide bond, occurs more readily with some amino acids than with others, despite their small size and comparable nucleophilicities.64 We found up to tenfold higher yield of certain dipeptide products over the alternative dipeptide, despite starting with equimolar concentrations of the two amino acid building blocks. This preference was explained by a stronger interaction of the less reactive species with the coacervate matrix. It appears therefore that strong partitioning and high reactivity may be inversely related, as strong interactions of solute species with the coacervate matrix can yield high local concentrations, but also stabilize the reactants.50,129 When the apparent transition state energies remain unchanged, this suppresses their reactivity.
For liposomes there are a few examples that indicate that these compartments can show selectivity for certain reactions. Cornell et al. show that amino acids can increase the stability of fatty acid vesicles against magnesium and sodium chloride.130 This stabilization requires a high concentration of amino acids present in the vesicles, but it also creates a positive feedback loop. The bound amino acids stabilize the fatty acid vesicle, which results in more binding of amino acids and thereby further stabilization of the vesicle. This positive feedback loop could have been a scenario in which the amino acids become concentrated near a membrane and possibly form short peptides which could have an even larger stabilizing effect. Adamala and Szostak have created a similar positive feedback loop, but combined it with selectivity.131 In their system, a catalyst is present inside the fatty acid vesicle that can form dipeptides. These dipeptides will partition into the membrane and enhance the uptake of fatty acids and thereby vesicle growth. The fatty acid vesicles can grow by consuming vesicles that do not contain dipeptides in their membrane, thereby introducing a form of competition that could be the onset of primitive evolution.
A further step towards true evolution of these protocellular compartments would be to apply the substrate preference and reaction selectivity to a replication reaction, as explained before. Ultimately, such replicators should have evolved from replicating a certain chemical sequence to replicating a sequence containing crucial information that encodes a function, such as catalysis of another (metabolic) reaction.132 By selecting for certain substrates or reactions, compartments could enhance the emergence of mutations in the sequence, and thereby the modulation of the function. This initial shift could be the precursor for the uncoupling of genotype and phenotype, in which encoded information (genotype) results in a function (phenotype) that is subject to selection pressure.67
One of the ultimate goals of research into coacervates and other types of protocells is to combine growth, replication and division into a single cell cycle that can repeat itself perpetually, in order to create an evolving protocell. In this Feature Article, we have analyzed the progress that has been made to date to achieve this goal. We have discussed how the free exchange of material between membraneless coacervate protocells and their surroundings can be an advantage over membrane-bound protocells such as liposomes and fatty acid vesicles. This free exchange of material can be used to construct phase separating systems based on chemical or enzymatic reactions that generate coacervate components. Careful tuning of the reaction rates would allow for controlled, active growth of coacervate protocells and keeps the system away from equilibrium. In addition to being able to grow, the protocell should be able to store (simple forms of) genetic information. A way to achieve this is by uptake of self-replicating systems. Functioning of these systems inside coacervates depends on a delicate balance of interaction strength between the coacervate material and the replicator, but their compartmentalization would help to solve problems like dilution and parasitism and could impose selectivity on the sequences that are formed. In order for coacervate protocells to undergo evolution, the droplets should also be able to divide. We have discussed externally and internally driven division of both liposome-based and coacervate protocells and showed how coacervates can be divided by several mechanisms, such as shear force or shape instabilities resulting from reaction cycles. Once true controlled growth of droplets, efficient replication of longer (encoding) sequences of genetic information and controlled division into equal daughter cells are realized, such a system can undergo evolution, provided the replication or division is not perfect and there is a natural selection of certain phenotypes. The encoded information will not only be required to let the system grow and divide, but also for other functions. A rudimentary form of evolution will make this primitive self-sustained protocell adapt to changes in its environment, and gradually gain new functions and increase in complexity until it can be truly called alive.
Author contributions
All authors contributed equally to the conceptualization, writing and reviewing of this manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the European Research Council (ERC) under grant agreement No. 851963.Notes and references
- D. E. Koshland, Jr., Science, 2002, 295, 2215–2216 CrossRef PubMed.
- J. W. Szostak, D. P. Bartel and P. L. Luisi, Nature, 2001, 409, 387–390 CrossRef CAS PubMed.
- L. Cronin and S. I. Walker, Science, 2016, 352, 1174–1175 CrossRef CAS PubMed.
- H. Duim and S. Otto, Beilstein J. Org. Chem., 2017, 13, 1189–1203 CrossRef CAS PubMed.
- N. A. Yewdall, A. F. Mason and J. C. M. van Hest, Interface Focus, 2018, 8, 20180023 CrossRef.
- A. I. Oparin, Origins Life, 1976, 7, 3–8 CrossRef CAS PubMed.
- P. A. Monnard and P. Walde, Life, 2015, 5, 1239–1263 CrossRef CAS PubMed.
- I. Gözen, E. S. Köksal, I. Põldsalu, L. Xue, K. Spustova, E. Pedrueza-Villalmanzo, R. Ryskulov, F. Meng and A. Jesorka, Small, 2022, 18, 2106624 CrossRef PubMed.
- G. von Kiedrowski, Angew. Chem., Int. Ed. Engl., 1986, 25, 932–935 CrossRef.
- T. A. Lincoln and G. F. Joyce, Science, 2009, 323, 1229–1232 CrossRef CAS PubMed.
- D. H. Lee, J. R. Granja, J. A. Martinez, K. Severin and M. R. Ghadiri, Nature, 1996, 382, 525–528 CrossRef CAS.
- B. Rubinov, N. Wagner, H. Rapaport and G. Ashkenasy, Angew. Chem., Int. Ed., 2009, 48, 6683–6686 CrossRef CAS PubMed.
- B. Rubinov, N. Wagner, M. Matmor, O. Regev, N. Ashkenasy and G. Ashkenasy, ACS Nano, 2012, 6, 7893–7901 CrossRef CAS PubMed.
- J. M. Carnall, C. A. Waudby, A. M. Belenguer, M. C. Stuart, J. J. Peyralans and S. Otto, Science, 2010, 327, 1502–1506 CrossRef CAS PubMed.
- M. Colomb-Delsuc, E. Mattia, J. W. Sadownik and S. Otto, Nat. Commun., 2015, 6, 7427 CrossRef CAS PubMed.
- A. Pross, Pure Appl. Chem., 2005, 77, 1905–1921 CrossRef CAS.
- W. Peeples and M. K. Rosen, Nat. Chem. Biol., 2021, 17, 693–702 CrossRef CAS PubMed.
- A. Klosin, F. Oltsch, T. Harmon, A. Honigmann, F. Jülicher, A. A. Hyman and C. Zechner, Science, 2020, 367, 464–468 CrossRef CAS PubMed.
- S. Koga, D. S. Williams, A. W. Perriman and S. Mann, Nat. Chem., 2011, 3, 720–724 CrossRef CAS PubMed.
- M. Abbas, W. P. Lipiński, K. K. Nakashima, W. T. S. Huck and E. Spruijt, Nat. Chem., 2021, 13, 1046–1054 CrossRef CAS PubMed.
- G. A. Mountain and C. D. Keating, Biomacromolecules, 2020, 21, 630–640 CrossRef CAS.
- T. Lu and E. Spruijt, J. Am. Chem. Soc., 2020, 142, 2905–2914 CrossRef CAS PubMed.
- K. K. Nakashima, M. A. Vibhute and E. Spruijt, Front. Mol. Biosci., 2019, 6, 21 CrossRef CAS PubMed.
- M. H. I. van Haren, K. K. Nakashima and E. Spruijt, J. Syst. Chem., 2020, 8, 107–120 Search PubMed.
- B. Alberts, D. Bray, K. Hopkin, A. D. Johnson, J. Lewis, M. Raff, K. Roberts and P. Walter, Essential cell biology, Garland Science, 2015 Search PubMed.
- A. Aguilera-Gomez and C. Rabouille, Dev. Biol., 2017, 428, 310–317 CrossRef CAS PubMed.
- O. D. Toparlak and S. S. Mansy, Exp. Biol. Med., 2019, 244, 304–313 CrossRef CAS PubMed.
- M. Imai, Y. Sakuma, M. Kurisu and P. Walde, Soft Matter, 2022, 18, 4823–4849 RSC.
- M. Li, X. Huang, T. Y. Tang and S. Mann, Curr. Opin. Chem. Biol., 2014, 22, 1–11 CrossRef CAS PubMed.
- M. G. F. Last, S. Deshpande and C. Dekker, ACS Nano, 2020, 14, 4487–4498 CrossRef CAS PubMed.
- F. Pir Cakmak, A. T. Grigas and C. D. Keating, Langmuir, 2019, 35, 7830–7840 CrossRef CAS PubMed.
- H. Kusumaatmaja, A. I. May and R. L. Knorr, J. Cell Biol., 2021, 220, e202103175 CrossRef CAS PubMed.
- T. Lu, S. Liese, L. Schoenmakers, C. Weber, H. Suzuki, W. Huck and E. Spruijt, J. Am. Chem. Soc., 2022, 144, 13451–13455 CrossRef CAS PubMed.
- M. Abbas, W. P. Lipiński, J. Wang and E. Spruijt, Chem. Soc. Rev., 2021, 50, 3690–3705 RSC.
- J. van der Gucht, E. Spruijt, M. Lemmers and M. A. Cohen Stuart, J. Colloid Interface Sci., 2011, 361, 407–422 CrossRef CAS.
- M. Abbas, J. O. Law, S. N. Grellscheid, W. T. S. Huck and E. Spruijt, Adv. Mater., 2022, 34, e2202913 CrossRef.
- I. B. A. Smokers, M. H. I. van Haren, T. Lu and E. Spruijt, ChemSystemsChem, 2022, 4, e202200004 CAS.
- A. Akbarzadeh, R. Rezaei-Sadabady, S. Davaran, S. W. Joo, N. Zarghami, Y. Hanifehpour, M. Samiei, M. Kouhi and K. Nejati-Koshki, Nanoscale Res. Lett., 2013, 8, 102 CrossRef PubMed.
- H. Sugiyama, T. Osaki, S. Takeuchi and T. Toyota, Commun. Chem., 2020, 3, 1–10 CrossRef.
- R. Bittman and L. Blau, Biochim. Biophys. Acta, 1986, 863, 115–120 CrossRef CAS.
- W. C. Su, D. L. Gettel, M. Chabanon, P. Rangamani and A. N. Parikh, J. Am. Chem. Soc., 2018, 140, 691–699 CrossRef CAS PubMed.
- H. Naraoka, A. Shimoyama and K. Harada, Origins Life Evol. Biospheres, 1999, 29, 187–201 CrossRef CAS PubMed.
- W. R. Hargreaves and D. W. Deamer, Biochemistry, 1978, 17, 3759–3768 CrossRef CAS PubMed.
- C. Gibard, S. Bhowmik, M. Karki, E. K. Kim and R. Krishnamurthy, Nat. Chem., 2018, 10, 212–217 CrossRef CAS PubMed.
- M. Björklund, Biochim. Biophys. Acta, Mol. Cell Res., 2019, 1866, 409–417 CrossRef PubMed.
- I. A. Chen, Science, 2006, 314, 1558–1559 CrossRef CAS.
- C. A. Weber, D. Zwicker, F. Jülicher and C. F. Lee, Rep. Prog. Phys., 2019, 82, 064601 CrossRef CAS.
- W. Ostwald, Z. Phys. Chem., 1897, 22, 289–330 CrossRef CAS.
- D. Zwicker, R. Seyboldt, C. A. Weber, A. A. Hyman and F. Jülicher, Nat. Phys., 2017, 13, 408–413 Search PubMed.
- J. Kirschbaum and D. Zwicker, J. R. Soc., Interface, 2021, 18, 20210255 CrossRef CAS PubMed.
- K. K. Nakashima, A. A. M. André and E. Spruijt, Methods Enzymol., 2021, 646, 353–389 CAS.
- W. M. Aumiller, Jr. and C. D. Keating, Nat. Chem., 2016, 8, 129–137 CrossRef PubMed.
- K. K. Nakashima, J. F. Baaij and E. Spruijt, Soft Matter, 2018, 14, 361–367 RSC.
- E. Sokolova, E. Spruijt, M. M. Hansen, E. Dubuc, J. Groen, V. Chokkalingam, A. Piruska, H. A. Heus and W. T. Huck, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 11692–11697 CrossRef CAS PubMed.
- S. Deshpande, F. Brandenburg, A. Lau, M. G. F. Last, W. K. Spoelstra, L. Reese, S. Wunnava, M. Dogterom and C. Dekker, Nat. Commun., 2019, 10, 1800 CrossRef PubMed.
- W. K. Spoelstra, E. O. van der Sluis, M. Dogterom and L. Reese, Langmuir, 2020, 36, 1956–1964 CrossRef CAS.
- C. Donau, F. Späth, M. Sosson, B. A. K. Kriebisch, F. Schnitter, M. Tena-Solsona, H. S. Kang, E. Salibi, M. Sattler, H. Mutschler and J. Boekhoven, Nat. Commun., 2020, 11, 5167 CrossRef CAS PubMed.
- K. K. Nakashima, M. H. I. van Haren, A. A. M. André, I. Robu and E. Spruijt, Nat. Commun., 2021, 12, 3819 CrossRef CAS.
- M. Matsuo and K. Kurihara, Nat. Commun., 2021, 12, 5487 CrossRef CAS PubMed.
- R. Seyboldt and F. Jülicher, New J. Phys., 2018, 20, 105010 CrossRef.
- S. N. Semenov, A. S. Wong, R. M. van der Made, S. G. Postma, J. Groen, H. W. van Roekel, T. F. de Greef and W. T. Huck, Nat. Chem., 2015, 7, 160–165 CrossRef CAS PubMed.
- M. Tena-Solsona, B. Rieß, R. K. Grötsch, F. C. Löhrer, C. Wanzke, B. Käsdorf, A. R. Bausch, P. Müller-Buschbaum, O. Lieleg and J. Boekhoven, Nat. Commun., 2017, 8, 15895 CrossRef CAS PubMed.
- M. Tena-Solsona, C. Wanzke, B. Riess, A. R. Bausch and J. Boekhoven, Nat. Commun., 2018, 9, 2044 CrossRef PubMed.
- J. Wang, M. Abbas, J. Wang and E. Spruijt, ChemRxiv, 2021 DOI:10.26434/chemrxiv-2021-1zt9k.
- G. Clixby and L. Twyman, Org. Biomol. Chem., 2016, 14, 4170–4184 RSC.
- A. Robertson, A. J. Sinclair and D. Philp, Chem. Soc. Rev., 2000, 29, 141–152 RSC.
- P. Adamski, M. Eleveld, A. Sood, Á. Kun, A. Szilágyi, T. Czárán, E. Szathmáry and S. Otto, Nat. Rev. Chem., 2020, 4, 386–403 CrossRef.
- V. Patzke and G. von Kiedrowski, ARKIVOC, 2007, 5, 293–310 Search PubMed.
- K. Le Vay, L. I. Weise, K. Libicher, J. Mascarenhas and H. Mutschler, Adv. Biosyst., 2019, 3, e1800313 Search PubMed.
- V. Bourbo, M. Matmor, E. Shtelman, B. Rubinov, N. Ashkenasy and G. Ashkenasy, Origins Life Evol. Biospheres, 2011, 41, 563–567 Search PubMed.
- G. von Kiedrowski, B. Wlotzka, J. Helbing, M. Matzen and S. Jordan, Angew. Chem., Int. Ed. Engl., 1991, 30, 423–426 CrossRef.
- W. S. Zielinski and L. E. Orgel, Nature, 1987, 327, 346–347 CrossRef CAS PubMed.
- A. Luther, R. Brandsch and G. von Kiedrowski, Nature, 1998, 396, 245–248 CrossRef CAS PubMed.
- D. E. Kim and G. F. Joyce, Chem. Biol., 2004, 11, 1505–1512 CrossRef CAS PubMed.
- B. J. Lam and G. F. Joyce, Nat. Biotechnol., 2009, 27, 288–292 CrossRef CAS PubMed.
- K. Ruiz-Mirazo, J. Umerez and A. Moreno, Biol Philos, 2008, 23, 67–85 CrossRef.
- R. Lohrmann, P. K. Bridson and L. E. Orgel, Science, 1980, 208, 1464–1465 CrossRef CAS PubMed.
- T. Inoue and L. E. Orgel, J. Mol. Biol., 1982, 162, 201–217 CrossRef CAS.
- T. Walton and J. W. Szostak, J. Am. Chem. Soc., 2016, 138, 11996–12002 CrossRef CAS PubMed.
- T. Walton and J. W. Szostak, Biochemistry, 2017, 56, 5739–5747 CrossRef CAS PubMed.
- L. Jin, A. E. Engelhart, W. Zhang, K. Adamala and J. W. Szostak, J. Am. Chem. Soc., 2018, 140, 15016–15021 CrossRef PubMed.
- L. Li, N. Prywes, C. P. Tam, D. K. O'Flaherty, V. S. Lelyveld, E. C. Izgu, A. Pal and J. W. Szostak, J. Am. Chem. Soc., 2017, 139, 1810–1813 CrossRef CAS PubMed.
- M. Sosson and C. Richert, Beilstein J. Org. Chem., 2018, 14, 603–617 CrossRef CAS.
- N. Prywes, J. C. Blain, F. Del Frate and J. W. Szostak, eLife, 2016, 5, e17756 CrossRef PubMed.
- D. K. O'Flaherty, L. Zhou and J. W. Szostak, J. Am. Chem. Soc., 2019, 141, 10481–10488 CrossRef PubMed.
- I. Haruna, K. Nozu, Y. Ohtaka and S. Spiegelman, Proc. Natl. Acad. Sci. U. S. A., 1963, 50, 905–911 CrossRef CAS PubMed.
- I. Haruna and S. Spiegelman, Proc. Natl. Acad. Sci. U. S. A., 1965, 54, 579–587 CrossRef CAS.
- D. R. Mills, R. L. Peterson and S. Spiegelman, Proc. Natl. Acad. Sci. U. S. A., 1967, 58, 217–224 CrossRef CAS PubMed.
- G. Laurent, L. Peliti and D. Lacoste, Life, 2019, 9, 78 CrossRef CAS PubMed.
- E. Szathmáry and L. Demeter, J. Theor. Biol., 1987, 128, 463–486 CrossRef.
- A. Blokhuis, P. Nghe, L. Peliti and D. Lacoste, J. Theor. Biol., 2020, 487, 110110 CrossRef CAS PubMed.
- S. Matsumura, Á. Kun, M. Ryckelynck, F. Coldren, A. Szilágyi, F. Jossinet, C. Rick, P. Nghe, E. Szathmáry and A. D. Griffiths, Science, 2016, 354, 1293–1296 CrossRef CAS PubMed.
- H. Urabe, N. Ichihashi, T. Matsuura, K. Hosoda, Y. Kazuta, H. Kita and T. Yomo, Biochemistry, 2010, 49, 1809–1813 CrossRef CAS PubMed.
- T. P. Fraccia and N. Martin, ChemRxiv, 2021 DOI:10.26434/chemrxiv-2021-zg20r.
- B. Drobot, J. M. Iglesias-Artola, K. Le Vay, V. Mayr, M. Kar, M. Kreysing, H. Mutschler and T. D. Tang, Nat. Commun., 2018, 9, 3643 CrossRef PubMed.
- R. R. Poudyal, R. M. Guth-Metzler, A. J. Veenis, E. A. Frankel, C. D. Keating and P. C. Bevilacqua, Nat. Commun., 2019, 10, 1–13 CrossRef PubMed.
- R. R. Poudyal, C. D. Keating and P. C. Bevilacqua, ACS Chem. Biol., 2019, 14, 1243–1248 CrossRef CAS PubMed.
- T. J. Nott, T. D. Craggs and A. J. Baldwin, Nat. Chem., 2016, 8, 569–575 CrossRef CAS PubMed.
- Y. Chen, M. Yuan, Y. Zhang, S. Liu, X. Yang, K. Wang and J. Liu, Chem. Sci., 2020, 11, 8617–8625 RSC.
- J. M. Iglesias-Artola, B. Drobot, M. Kar, A. W. Fritsch, H. Mutschler, T. Y. Dora Tang and M. Kreysing, Nat. Chem., 2022, 14, 407–416 CrossRef CAS.
- T. Lu, K. K. Nakashima and E. Spruijt, J. Phys. Chem. B, 2021, 125, 3080–3091 CrossRef CAS PubMed.
- D. Wollny, B. Vernot, J. Wang, M. Hondele, A. Safrastyan, F. Aron, J. Micheel, Z. He, A. Hyman, K. Weis, J. G. Camp, T. D. Tang and B. Treutlein, Nat. Commun., 2022, 13, 2626 CrossRef CAS PubMed.
- W. M. Aumiller, Jr., F. Pir Cakmak, B. W. Davis and C. D. Keating, Langmuir, 2016, 32, 10042–10053 CrossRef PubMed.
- L. Olivi, M. Berger, R. N. P. Creyghton, N. De Franceschi, C. Dekker, B. M. Mulder, N. J. Claassens, P. R. Ten Wolde and J. van der Oost, Nat. Commun., 2021, 12, 4531 CrossRef CAS PubMed.
- K. Schafer, Vet. Pathol., 1998, 35, 461–478 CrossRef CAS.
- T. Y. Dora Tang, C. Rohaida Che Hak, A. J. Thompson, M. K. Kuimova, D. S. Williams, A. W. Perriman and S. Mann, Nat. Chem., 2014, 6, 527–533 CrossRef CAS PubMed.
- Y. Zhang, Y. Chen, X. Yang, X. He, M. Li, S. Liu, K. Wang, J. Liu and S. Mann, J. Am. Chem. Soc., 2021, 143, 2866–2874 CrossRef CAS PubMed.
- Y. Dreher, K. Jahnke, E. Bobkova, J. P. Spatz and K. Göpfrich, Angew. Chem., Int. Ed., 2021, 60, 10661–10669 CrossRef CAS PubMed.
- T. F. Zhu, K. Adamala, N. Zhang and J. W. Szostak, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 9828–9832 CrossRef CAS PubMed.
- P. W. Kudella, K. Preißinger, M. Morasch, C. F. Dirscherl, D. Braun, A. Wixforth and C. Westerhausen, Sci. Rep., 2019, 9, 18808 CrossRef CAS PubMed.
- A. Pal and D. V. Khakhar, Soft Matter, 2019, 15, 1979–1987 RSC.
- R. Rubio-Sánchez, D. K. O'Flaherty, A. Wang, F. Coscia, G. Petris, L. Di Michele, P. Cicuta and C. Bonfio, J. Am. Chem. Soc., 2021, 143, 16589–16598 CrossRef.
- L. Zhou, Y. Fan, Z. Liu, L. Chen, E. Spruijt and Y. Wang, CCS Chem., 2021, 3, 358–366 CrossRef CAS.
- T. F. Zhu and J. W. Szostak, J. Am. Chem. Soc., 2009, 131, 5705–5713 CrossRef CAS PubMed.
- D. van Swaay, T. Y. Tang, S. Mann and A. de Mello, Angew. Chem., Int. Ed., 2015, 54, 8398–8401 CrossRef CAS PubMed.
- H. Cheung Shum, J. Varnell and D. A. Weitz, Biomicrofluidics, 2012, 6, 12808–128089 CrossRef PubMed.
- F. Fanalista, S. Deshpande, A. Lau, G. Pawlik and C. Dekker, Adv. Biosyst., 2018, 2, 1800136 CrossRef.
- A. Ianeselli, D. Tetiker, J. Stein, A. Kühnlein, C. B. Mast, D. Braun and T. Y. Dora Tang, Nat. Chem., 2022, 14, 32–39 CrossRef CAS PubMed.
- Y. Miele, Z. Medveczky, G. Holló, B. Tegze, I. Derényi, Z. Hórvölgyi, E. Altamura, I. Lagzi and F. Rossi, Chem. Sci., 2020, 11, 3228–3235 RSC.
- Y. Miele, G. Holló, I. Lagzi and F. Rossi, Life, 2021, 11, 634 CrossRef PubMed.
- J. Steinkühler, R. L. Knorr, Z. Zhao, T. Bhatia, S. M. Bartelt, S. Wegner, R. Dimova and R. Lipowsky, Nat. Commun., 2020, 11, 1–11 Search PubMed.
- E. Te Brinke, J. Groen, A. Herrmann, H. A. Heus, G. Rivas, E. Spruijt and W. T. S. Huck, Nat. Nanotechnol., 2018, 13, 849–855 CrossRef CAS PubMed.
- K. L. Weirich, K. Dasbiswas, T. A. Witten, S. Vaikuntanathan and M. L. Gardel, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 11125–11130 CrossRef CAS.
- M. Andes-Koback and C. D. Keating, J. Am. Chem. Soc., 2011, 133, 9545–9555 CrossRef CAS.
- Y. Song, T. C. Michaels, Q. Ma, Z. Liu, H. Yuan, S. Takayama, T. P. Knowles and H. C. Shum, Nat. Commun., 2018, 9, 1–7 CrossRef PubMed.
- M. I. Jacobs, E. R. Jira and C. M. Schroeder, Langmuir, 2021, 37, 14323–14335 CrossRef CAS PubMed.
- W. A. Wee, H. Sugiyama and S. Park, iScience, 2021, 24, 103455 CrossRef CAS PubMed.
- W. Mu, Z. Ji, M. Zhou, J. Wu, Y. Lin and Y. Qiao, Sci. Adv., 2021, 7, eabf9000 CrossRef CAS.
- D. Zwicker, arXiv, 2022, preprint, arXiv:2202.13646 DOI:10.48550/arXiv:2202.13646.
- C. E. Cornell, R. A. Black, M. Xue, H. E. Litz, A. Ramsay, M. Gordon, A. Mileant, Z. R. Cohen, J. A. Williams, K. K. Lee, G. P. Drobny and S. L. Keller, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 17239–17244 CrossRef CAS PubMed.
- K. Adamala and J. W. Szostak, Nat. Chem., 2013, 5, 495–501 CrossRef CAS PubMed.
- J. Ottelé, A. S. Hussain, C. Mayer and S. Otto, Nat. Catal., 2020, 3, 547–553 CrossRef.
Footnote |
| † These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2022 |