Open Access Article
Open Access ArticleDependence of click-SELEX performance on the nature and average number of modified nucleotides†
Julia
Siegl
a,
Olga
Plückthun
a and
Günter
Mayer
 *ab
*ab
aChemical Biology & Chemical Genetics, Life and Medical Sciences (LIMES) Institute, University of Bonn, Gerhard-Domagk-Str. 1, 53121 Bonn, Germany. E-mail: gmayer@uni-bonn.de
bCenter of Aptamer Research & Development (CARD), University of Bonn, Gerhard-Domagk-Str. 1, 53121 Bonn, Germany
First published on 21st February 2022
Abstract
The click-SELEX procedure enables the identification of nucleobase-modified aptamers in which chemical entities are introduced by a copper(I)-catalysed alkyne-azide ‘click’ reaction. Here we report on the impact of modified nucleobases on PCR conditions and the average amount of modified nucleobases on click-SELEX performance. We demonstrate click-SELEX being strongly dependent on which and on how many modifications are used. However, when using C3-GFP the number of modifications did not impact the overall success of the selection procedure.
Introduction
Molecules that specifically interact with proteins are important tools in biomedical research. Besides antibodies and nanobodies, nucleic acid-based molecules have emerged over the past decades as promising macromolecular ligands, with applications in basic science and synthetic biology, but also as therapeutics and diagnostics.1,2 Chemically functionalized DNA libraries using click chemistry3,4 provide a valuable strategy to generate nucleobase-modified aptamers, so-called clickmers, binding to a variety of target molecules.5 This approach enables the incorporation of virtually any chemical modification, which is available as an azide and compatible with the conditions of copper(I) catalysed alkyne-azide cycloadditions and subsequent enzymatic steps during the click-SELEX procedure.6 Nucleobase-modified DNA aptamers and deoxyribozymes, oligonucleotides with an altered phosphate-sugar backbone and aptamers with extended alphabet, e.g., bearing non-canonical base pairs, are of high interest in biomedical applications and as superior DNA-based catalysts.7–13 Each of these technologies bear intrinsic advantages, limitations and challenges, e.g., compatibility with amplification steps of the SELEX procedure or the requirement of specifically evolved polymerases or newly established sequencing approaches. The click-SELEX procedure overcomes some of these limitations in regard of nucleobase-modifications and compatibility with PCR, as the ethynyl modified nucleobase C5-ethynyl-deoxyuridine can be used during PCR simply by replacing thymidine triphosphates. However, currently, the methodology is limited to one modification per DNA strand but has been shown to be adaptable to multiplex formats, thereby assaying several chemical entities simultaneous.14,15 Here we describe the suitability of different chemical entities for being used in the click-SELEX procedure, the impact of modifications on PCR performance, and the relation of selection output on the statistical number of modifications per DNA strand in the starting library. Our experiments reveal that the PCR performance is more reliable when using less modifications per DNA sequence. Using a model target, the success of the selection experiment, however seems to be less affected and not directly impacted by the average amount of chemical modifications per DNA strand in the starting library.Results
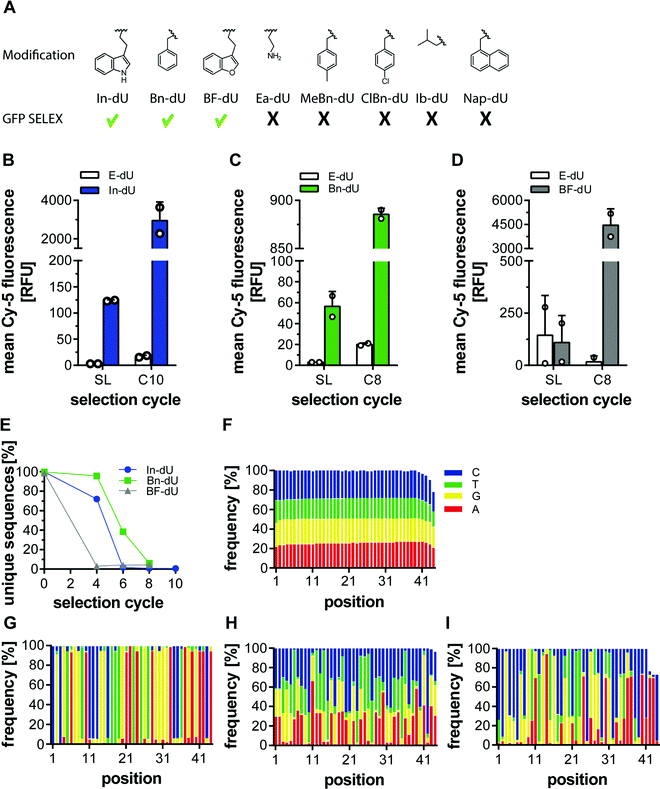
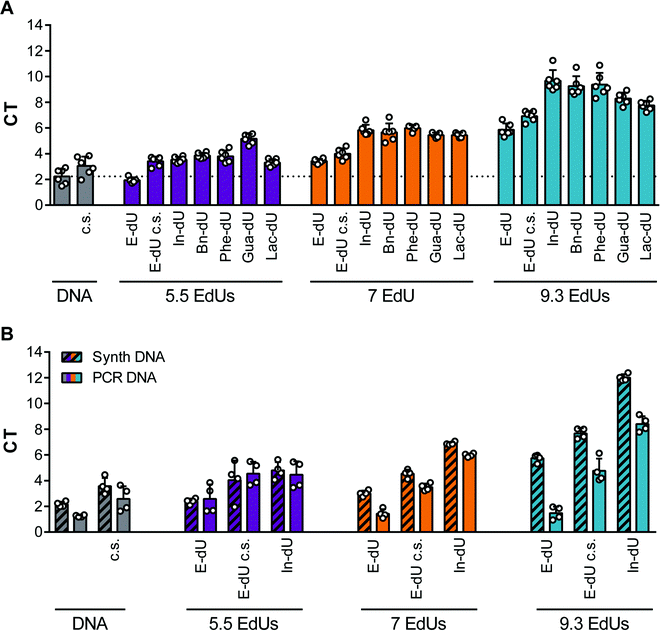
Previously, we employed several azides for CuAAC to modify DNA libraries and subjected these to click-SELEX procedures. Among them, we used indole, benzyl, and other aromatic residues and successfully identified clickmers binding to cycle 3 Green Fluorescent Protein (C3-GFP),5 streptavidin,14 CXCL9,15 and Δ9-tetrahydrocannabinol.16 To more systematically investigate the suitability of individual azide moieties for the click-SELEX procedure, we performed click-SELEX targeting C3-GFP using eight different representative and available azides (Fig. 1A). Among those, we were able to enrich libraries with improved binding capabilities using indole (In-dU, Fig. 1B), benzyl (Bn-dU), Fig. 1C) and benzofuran (BF-dU, Fig. 1D). None of the other entities lead to the enrichment of binding species. Moreover, the selection procedure had to be terminated due to the occurrence of PCR by-products (ESI† Fig. S1) and were thus not further characterized. This finding is in accordance with recently published results from split-combine selection procedures,14 in which also In-dU, Bn-dU, and BF-dU as well as ethylamine (Ea-dU) and 4-chloro benzyl (ClBn-dU) were used, leading to clickmers only when using In-dU, BF-dU and Bn-dU. The enriched libraries revealed click-dependent binding (Fig. 1B–D) and were further analysed by next-generation sequencing (NGS). This analysis revealed a strong reduction of the number of unique sequences, most pronounced by using BF-dU, followed by In-dU and Bn-dU (Fig. 1E). Likewise, the distribution of nucleotides changed strongly from the staring library (Fig. 1F) to the enriched libraries from selection cycles 10 (In-dU, Fig. 1G) and 8 (Bn-dU, and BF-dU, Fig. 1H and I), respectively.We next investigated the impact of the clicked-in modifications on the PCR performance. Therefore, we employed qPCR and compared Ct values obtained from the DNA library (built from canonical nucleotides), the respective EdU-modified library (replacing thymidine by EdU) and library variants modified with different azides. For this analysis we chose In-dU, Bn-dU, phenol-dU (Phe-dU), guanidinium-dU (Gua-dU), and lactose-dU (Lac-dU) as representative entities covering aromatic, polar, aliphatic, and larger residues. Likewise, we varied the average number of EdU residues per DNA strand, having either 5.5, 7.0, or 9.3 EdU (Table 1). The qPCR data reveal that less EdU content results in a PCR performance more similar to the one of the naïve DNA, whereas applying click chemistry conditions only (in the absence of an azide) already impacts PCR yields (Fig. 2A). The introduction of modifications further reduces PCR performance, although less pronounced when using libraries having an average of 5.5. EdU molecules and gradually decreasing when using libraries with 7.0 to 9.3 EdU (Fig. 2A). Interestingly, the chemical nature of the clicked-in modification seems to have less impact on PCR performance. We also applied variations of the PCR conditions, e.g., change of elongation time and temperature, variation of the Mg2+-ion concentration, addition of DMSO, and using different polymerases (ESI† Fig. S2 and S3). None of these variations improved PCR performance significantly. We also analysed the PCR performance of synthetic libraries compared to pre-amplified libraries and different EdU content. As expected, pre-amplified libraries reveal a better performance, as these libraries are already adapted to the amplification procedure and the number of non- or hardly amplifiable sequences are lower (Fig. 2B). Applying click conditions only, In-modifications and different EdU content (Fig. 2B) follows the same trend when using synthetic libraries as observed in the previous experiments (Fig. 2A).
| Nucleotide [%] | ||||
|---|---|---|---|---|
| Library | A | EdU | C | G |
| 9.3 | 8.6 | 9.3 | 8.8 | 15.2 |
| 7 | 12.3 | 7.0 | 12.0 | 10.7 |
| 5.5 | 11.3 | 5.5 | 11.9 | 13.2 |
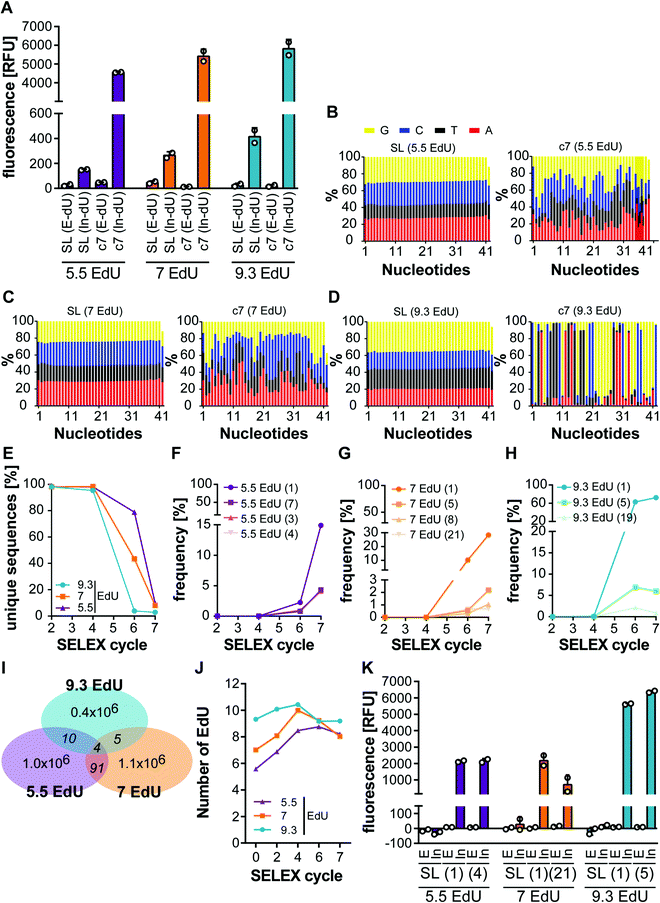
Having shown the impact of the average number of EdUs on PCR performance, we next analysed its impact on enrichment during click-SELEX. Therefore, we performed three click-SELEX experiments targeting C3-GFP using previously reported conditions and DNA libraries with the indicated average number of EdUs modified with In. We analysed the binding properties of enriched compared to the starting libraries by flow cytometry and observed enrichment of binding species independent of the amount of EdU employed after 7 selection cycles (Fig. 3A). Interestingly, the signal intensity of the starting library is increasing with the average number of EdUs (Fig. 3A), which is in line with previous data revealing dependency of interaction properties of the starting library when using different types of click-in entities. We subsequently performed next-generation sequencing (NGS) of the selected libraries. This analysis revealed a distribution of EdU in the starting libraries of 5.5, 7, 9.3 EdUs, respectively (Fig. 3B and Table 1). The libraries of selection cycles 7 in turn, reveal a defined pattern of enrichment of specific nucleotides at different positions of the random region, which is most pronounced in the library having an average number of 9.3 EdU per strand (Fig. 3B and ESI† Fig. S4–S6). In accordance with this observation, the number of unique sequences in each population of enriched libraries declines rapidly in the library having an average number of EdU of 9.3 and gradually to a lesser extent when using a lower amount of EdUs in the starting libraries (Fig. 3C). We chose two representative sequences (Table 2 and ESI† Table S3–S5) from each of the three selections and performed binding experiments to validate that all conditions indeed yield monoclonal clickmers binding to C3-GFP. Besides the most frequent sequence found in the enriched DNA populations from selection cycle 7, we choose additional sequences with similar enrichment profiles (Fig. 3F–H). The overlap of sequences among the three selections was found to be very low, i.e. 4, 5, 10, 91 out of 0.4–1.0 million sequences (Fig. 3I and ESI† Table S6). Interestingly, the EdU content of the library starting from an average number of 5.5 per sequence changed significantly over the course of the selection, whereas the EdU content of the 9.3 and 7.0 libraries remained similar when comparing round 7 DNA with the starting library (Fig. 3J). All sequences were found to bind to C3-GFP and only when modified with In (Fig. 3K). These data are in line with results obtained from click-SELEX procedures targeting C3-GFP. With exception of clickmer 7EdU(1) these clickmers share a sequence motif (CTTTGAATATGTAG) with the previously identified clickmer I10,14 indicating a robust and reproducible enrichment process.
| Name | Random region | EdU content |
|---|---|---|
| 9 EdU (1) | CGTAGCTTTCGATATGTTGCCGGGGGGTAACGAGGCCGGGGC | 9 |
| 9 EdU (5) | CGCGGTGAACAGAACCGGCTTTCGATATGTAGGTGCGAACAC | 8 |
| 9 EdU (19) | CTGTCGGACGAGTTGTAGCCCCGGCTTTGCATATGTAGGGG | 11 |
| 7 EdU (1) | TGGACAGACCATAACCTGTAACCCCTACACCTTCCAGGGGC | 7 |
| 7 EdU (5) | AGCAACTCGGCTTTCAATATGTAGAGATGAAACGGGAACCA | 8 |
| 7 EdU (8) | TACGTGCTGACATCGACGGCTTTGAATATGTAGGAGGACCGG | 10 |
| 7 EdU (21) | TGAGCCTACTTTAACGGCATATCGAACCCCCGGGGAACTGC | 8 |
| 5.5 EdU (1) | CCGGCCTTTCGATATGTAGGACCCGGGGTCATTGCCAAGGAA | 9 |
| 5.5 EdU (3) | CGGAGGACCTACGGAAGCGCCACGGCTTTCGATACGTAGGGG | 6 |
| 5.5 EdU (4) | AGGGCCACGAACTAGCGCGCCTTTCGATATGTAGCACGGTAA | 8 |
| 5.5 EdU (7) | CAGCAACTTTCGATATGTACGGCCGAGGTCTAAGACGGTCGC | 9 |
Discussion
In conclusion, we describe the impact of several parameters on the click-SELEX process and the amplification behaviour of chemical modified libraries used for this procedure. Using the model protein C3-GFP as target molecule in the selection experiment, we found that not all used chemical entities were compatible with the click-SELEX approach. Importantly, when using individual chemical entities, the selection could not be driven to completion and had to stop early on in the procedure. This was mainly due to the onset of by-products,17 which usually counteract on the enrichment of binding species. This limitation can putatively be overcome by changing the library design, e.g., by using less modifications per DNA strand, different primer binding sites or by implementing emulsion or digital droplet PCR. As our study is limited to C3-GFP using other target proteins might reveal a different set of modifications which indeed lead to clickmers. ClBn-dU has been used in a split-combine click-SELEX approach,14 in which 5 different chemical entities were used simultaneously. In this process, by-product formation was not observed, probably due to the presence of other entities, i.e. Bn-dU, In-dU and BF-dU that enable smooth PCR performance and enrichment of binding species. But also, in this approach ClBn-dU dependent clickmers were not enriched. In most of our previous studies, In-dU, BF-dU, and Bn-dU were used successfully in click-SELEX and found leading to target-specific clickmers.PCR performance assessment revealed that the average number of EdU residues per DNA strand as well as the conditions of the click reaction do have an impact, most likely due to minor DNA damage occurring during the click reaction.18,19 Surprisingly, the nature of the clicked-in molecule had less impact on PCR performance than previously expected. Even larger molecules, such as lactose were tolerated very well. This might be due to the imprinting step during PCR, i.e. the replacement of the chemical modified EdU-residue in the DNA by EdU during the first extension cycle of the PCR reaction, which subsequently is used as template for the following PCR cycles. Although it has been shown previously, that fully and modified DNA templates can be amplified by PCR,20,21 a reduction of the number of EdUs per strand was found to have no strong impact on the overall enrichment of binding species, although the occurrence of high copy number sequences, correlating with the number of unique sequences in the selected DNA populations, was found shifted to later selection cycles. Indeed, using an average of 5 EdUs per strand has become a standard in out lab for click-SELEX experiments. The investigated parameters represent important details to finetune and optimize selections using click-modified nucleic acid libraries for the identification of interaction partners, not only for proteins but also for small molecules and other targets.
Author contributions
G. M. conceived the study, discussed results and wrote the manuscript. OP conducted experiments using individual click-in moieties and commented on the manuscript. J. S. conducted qPCR experiments, the selections using different amounts of EdU residues per DNA strand, commented and revised the manuscript.Conflicts of interest
G. M. holds and has filed patents on the click-SELEX procedure.Acknowledgements
This work has been made possible by funds from the German Research Foundation to G. M. (DFG, SFB 1089 and MA3442/7-1) and from the Federal Ministry of Economic Affairs and Energy to G. M. (ZF 4613701CR8).References
- J. C. Rohloff, A. D. Gelinas, T. C. Jarvis, U. A. Ochsner, D. J. Schneider, L. Gold and N. Janjic, Nucleic Acid Ligands With Protein-like Side Chains: Modified Aptamers and Their Use as Diagnostic and Therapeutic Agents, Mol. Ther. – Nucleic Acids, 2014, 3, e201 CrossRef CAS PubMed.
- F. Pfeiffer, M. Rosenthal, J. Siegl, J. Ewers and G. Mayer, Customised nucleic acid libraries for enhanced aptamer selection and performance, Curr. Opin. Biotechnol, 2017, 48, 111–118 CrossRef CAS PubMed.
- P. M. E. Gramlich, S. Warncke, J. Gierlich and T. Carell, Click–Click–Click: Single to Triple Modification of DNA, Angew. Chem., Int. Ed., 2008, 47, 3442–3444 CrossRef CAS PubMed.
- P. M. E. Gramlich, C. T. Wirges, A. Manetto and T. Carell, Postsynthetic DNA Modification through the Copper-Catalyzed Azide–Alkyne Cycloaddition Reaction, Angew. Chem., Int. Ed., 2008, 47, 8350–8358 CrossRef CAS PubMed.
- F. Tolle, G. M. Brändle, D. Matzner and G. Mayer, A Versatile Approach Towards Nucleobase-Modified Aptamers, Angew. Chem., Int. Ed., 2015, 54, 10971–10974 CrossRef CAS PubMed.
- F. Pfeiffer, F. Tolle, M. Rosenthal, G. M. Brändle, J. Ewers and G. Mayer, Identification and characterization of nucleobase-modified aptamers by click-SELEX, Nat. Protoc., 2018, 13, 1153–1180 CrossRef CAS PubMed.
- M. Kimoto, R. Yamashige, K. Matsunaga, S. Yokoyama and I. Hirao, Generation of high-affinity DNA aptamers using an expanded genetic alphabet, Nat. Biotechnol., 2013, 31, 453–457 CrossRef CAS PubMed.
- K. Sefah, Z. Yang, K. M. Bradley, S. Hoshika, E. Jiménez, L. Zhang, G. Zhu, S. Shanker, F. Yu, D. Turek, W. Tan and S. A. Benner, In vitro selection with artificial expanded genetic information systems, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 1449–1454 CrossRef CAS PubMed.
- V. B. Pinheiro, A. I. Taylor, C. Cozens, M. Abramov, M. Renders, S. Zhang, J. C. Chaput, J. Wengel, S.-Y. Peak-Chew, S. H. McLaughlin, P. Herdewijn and P. Holliger, Synthetic Genetic Polymers Capable of Heredity and Evolution, Science, 2012, 336, 341–344 CrossRef CAS PubMed.
- S. Arangundy-Franklin, A. I. Taylor, B. T. Porebski, V. Genna, S. Peak-Chew, A. Vaisman, R. Woodgate, M. Orozco and P. Holliger, A synthetic genetic polymer with an uncharged backbone chemistry based on alkyl phosphonate nucleic acids, Nat. Chem., 2019, 11, 533–542 CrossRef CAS PubMed.
- M. Renders, E. Miller, C. H. Lam and D. M. Perrin, Whole cell-SELEX of aptamers with a tyrosine-like side chain against live bacteria, Org. Biomol. Chem., 2016, 15, 1980–1989 RSC.
- M. Hollenstein, C. J. Hipolito, C. H. Lam and D. M. Perrin, A self-cleaving DNA enzyme modified with amines, guanidines and imidazoles operates independently of divalent metal cations (M2+), Nucleic Acids Res., 2009, 37, 1638–1649 CrossRef CAS PubMed.
- Y.-W. Cheung, P. Röthlisberger, A. E. Mechaly, P. Weber, F. Levi-Acobas, Y. Lo, A. W. C. Wong, A. B. Kinghorn, A. Haouz, G. P. Savage, M. Hollenstein and J. A. Tanner, Evolution of abiotic cubane chemistries in a nucleic acid aptamer allows selective recognition of a malaria biomarker, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 16790–16798 CrossRef CAS PubMed.
- O. Plückthun, J. Siegl, L. L. Bryant and G. Mayer, Dynamic changes in DNA populations revealed by split–combine selection, Chem. Sci., 2020, 11, 9577–9583 RSC.
- J. Siegl, C. Nikolin, N. L. Phung, S. Thoms, C. Blume and G. Mayer, Split-Combine Click-SELEX Reveals Ligands Recognizing the Transplant Rejection Biomarker CXCL9, ACS Chem. Biol., 2022, 17(1), 129–137 CrossRef PubMed.
- M. Rosenthal, F. Pfeiffer and G. Mayer, A Receptor-Guided Design Strategy for Ligand Identification, Angew. Chem., Int. Ed., 2019, 58, 10752–10755 CrossRef CAS PubMed.
- F. Tolle, J. Wilke, J. Wengel and G. Mayer, By-product formation in repetitive PCR amplification of DNA libraries during SELEX, M. Isalan, ed., PloS one, 2014, 9, e114693 Search PubMed.
- M. Slavíčková, M. Janoušková, A. Šimonová, H. Cahová, M. Kambová, H. Šanderová, L. Krásný and M. Hocek, Turning Off Transcription with Bacterial RNA Polymerase through CuAAC Click Reactions of DNA Containing 5-Ethynyluracil, Chem. – Eur. J., 2018, 24, 8311–8314 CrossRef PubMed.
- B. Sauter, L. Schneider, C. Stress and D. Gillingham, An assessment of the mutational load caused by various reactions used in DNA encoded libraries, Bioorg. Med. Chem., 2021, 52, 116508 CrossRef CAS PubMed.
- M. Ondruš, V. Sýkorová, L. Bednárová, R. Pohl and M. Hocek, Enzymatic synthesis of hypermodified DNA polymers for sequence-specific display of four different hydrophobic groups, Nucleic Acids Res., 2020, 48, 11982–11993 CrossRef PubMed.
- S. Jäger, G. Rasched, H. Kornreich-Leshem, M. Engeser, O. Thum and M. Famulok, A versatile toolbox for variable DNA functionalization at high density, J. Am. Chem. Soc., 2005, 127, 15071–15082 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d2cb00012a |
| This journal is © The Royal Society of Chemistry 2022 |