Open Access Article
Open Access ArticleFluorescence signal-to-noise optimisation for real-time PCR using universal reporter oligonucleotides†
Michael
Lehnert
 a,
Elena
Kipf
ab,
Franziska
Schlenker
ab,
Nadine
Borst
b,
Roland
Zengerle
abc and
Felix
von Stetten
*ab
a,
Elena
Kipf
ab,
Franziska
Schlenker
ab,
Nadine
Borst
b,
Roland
Zengerle
abc and
Felix
von Stetten
*ab
aLaboratory for MEMS Applications, IMTEK – Department of Microsystems Engineering, University of Freiburg, Georges-Koehler-Allee 103, 79110 Freiburg, Germany
bHahn-Schickard–Institut für Mikroanalysesysteme, Georges-Koehler-Allee 103, 79110 Freiburg, Germany. E-mail: Felix.von.Stetten@Hahn-Schickard.de
cBIOSS – Centre for Biological Signalling Studies, University of Freiburg, Schaenzlestr. 1, 79104 Freiburg, Germany
First published on 26th June 2018
Abstract
In this study we optimised the fluorescence signal generation of contact quenched universal reporter oligonucleotides. These are used as secondary probes in real-time Mediator Probe PCR to detect the sequence-specific cleavage of label-free primary mediator probes. Since the fluorescence signal generation of a universal reporter is not influenced by the target DNA sequence, optimisation of the fluorescence signal-to-noise ratio will improve the performance of all Mediator Probe PCRs that are based on this type of universal reporter. To determine the critical factors influencing signal-to-noise optimisation, we systematically analysed four parameters. These parameters were type of fluorophore, type of quencher molecule, intramolecular orientation of both residuals, and the number of quencher labels. In total, more than 30 different fluorogenic universal reporter structures were analysed, covering the whole fluorescence spectrum from green to crimson. From our results, we deduced a novel set of guidelines for signal-to-noise optimisation in the design of contact quenched, fluorogenic universal reporter oligonucleotides. We confirmed these guidelines in a different thermocycler, and by designing a second set of universal reporters, which were used for multiplex real-time PCR quantification of acute lymphoblastic leukaemia marker sequences. This optimised biplex Mediator Probe PCR showed an improved performance under clinical conditions, with a 10 times higher resolution regarding the limit of quantification. In addition to Mediator Probe PCR, these guidelines may also prove useful in signal-to-noise optimisation of other fluorescence-based assays where contact quenched oligonucleotides or secondary reporter molecules are used.
Introduction
Selective fluorescence signal generation is a key technique in molecular biology. In particular, fluorogenic oligonucleotides are essential in nucleic acid analysis. They are widely applied in real-time PCR, which is the standard method for sequence-specific DNA detection and quantification.1 Furthermore, fluorogenic oligonucleotides are also used in new, upcoming technologies like digital PCR and monochrome multiplexing.2,3 Conventional real-time PCR uses sequence-specific fluorogenic DNA probes which are labelled with a fluorophore and a quencher molecule. There are different kinds of fluorogenic probes and different mechanisms of fluorescent signal generation.4–6 Most initially form a random coil structure, which enables fluorescence energy transfer from the fluorophore to the quencher residual (e.g. hydrolysis probes). In the presence of the target DNA sequence, fluorescence signal generation occurs when the probe is cleaved during polymerase primer extension, separating fluorophore and quencher to a maximum distance.6,7 Other DNA probes adopt a distinct conformation in the initial stage (e.g. molecular beacons). This conformation forces fluorophore and quencher very close to each other, which leads to a very strong fluorescence quenching known as contact quenching.6,8,9 This results in high signal-to-noise ratios when the oligonucleotide unfolds during the process of target DNA detection. Even though signals are generated in different ways, fluorogenic DNA probes generally have two characteristics in common: (1) a high fluorescence signal increase is important to obtain a good real-time PCR performance.10,11 (2) Since a fluorogenic DNA probe binds complementarily to its target site, the probe sequence, and thus its fluorogenic properties, are affected by the DNA sequence. This can be caused by, for example, G-quenching or greater separation of fluorophore and quencher due to an increased probe length. Therefore, the target DNA sequence influences the fluorescence signal generation, which makes probe design less flexible and more laborious.12To avoid the influence of the target DNA sequence, different approaches exist which use sequence-independent reporter molecules for fluorescence signal generation.13 One of these methods is Mediator Probe PCR. Here, label-free, sequence-specific mediator probes are used in combination with fluorogenic universal reporter oligonucleotides (Fig. 1, top). Inside the molecular structure of a universal reporter, the fluorophore and quencher are very close to each other.10,14 The fluorescence signal is therefore initially suppressed by means of contact quenching.6,8,15,16 During PCR, the mediator probe is cleaved and its mediator sequence is released. This mediator sequence binds to the complementary hybridisation site of a distinct universal reporter molecule, where it functions as a primer for polymerase extension. During this process, fluorophore and quencher are separated, leading to a high fluorescence signal increase. Because DNA detection via mediator probes and signal generation using universal reporters are two different steps, it is possible to optimise both processes independently of each other (Fig. 1, bottom).10 General guidelines for the design of label-free mediator probes have already been published.17 Using these guidelines, Mediator Probe PCR has achieved a performance comparable to state-of-the-art real-time PCRs, and often achieves an even better detection limit.18 However, no guidelines for universal reporter fluorescence signal optimisation are available yet, and therefore Mediator Probe PCR is still below its potential capacity. Once optimised, these universal reporters will improve the performance of all real-time PCRs in which they are used. This will be reflected by higher fluorescence signal increases, resulting in better R2-values as well as improved quantification and detection limits.
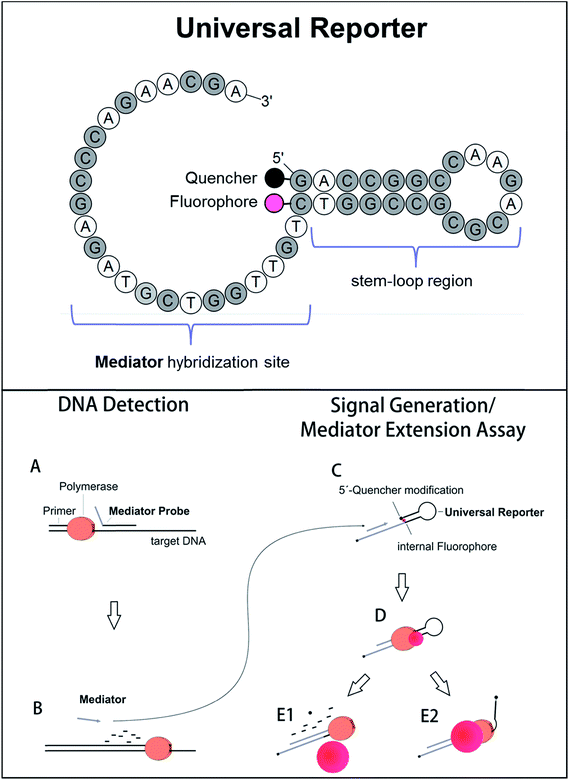 | ||
| Fig. 1 Top: Basic structure of the fluorogenic universal reporter oligonucleotide used in Mediator Probe PCR. Since a universal reporter is target sequence independent, it can be used for tracing DNA-detection events via fluorescence signal generation in many PCRs. In addition, universal reporters may also be applied in additional nucleic acid amplification and detection technologies making use of secondary probes. The universal reporter structure was modelled with OligoPAD (GNWI mbH, Germany). A universal reporter consists of a target-independent mediator hybridisation sequence, a fluorophore–quencher pair, and a stem–loop region which minimises the distance between the fluorophore and quencher residuals and promotes contact quenching.6,8 The quencher is coupled to the 5′-end of the oligonucleotide while the fluorophore is coupled to the base of the nucleotide complementary to the 5′-end. In the case of green fluorophores, the bottom end of the stem-loop region consisted of A and T. In all other cases, a GC-clamp was used. The 3′-end of a universal reporter is blocked using a C3 blocking group. Bottom: Reaction mechanism of the Mediator Probe PCR.10,14 DNA detection via the mediator probe (steps A and B) and signal generation at a universal reporter (steps C–E) are two decoupled processes and can therefore be optimised independently from each other. The mediator extension assay consists of steps C–E, and can be used to analyse the fluorogenic properties of a particular universal reporter by adding a respective mediator sequence directly to the reaction mixture. | ||
Fluorescence signal generation during real-time PCR can be improved by increasing the maximum fluorescence signal in the dequenched state, and by decreasing the fluorescence in the initial state. When successful, both approaches will result in a higher signal-to-noise ratio (SNR), which is the quotient of the maximum fluorescence signal of a positive sample in real-time PCR and the quenched signal of the respective no template control (NTC).10 In the last few years, many studies have been published analysing different factors influencing the SNR, mostly focusing on optimisation of the quenching processes.8,11,15,19,20 However, optimisation of the SNR by decreasing the initial fluorescence signal in the ground state with improved fluorescence quenching, and simultaneously increasing the maximum fluorescence signal has not yet been systematically studied and published for contact quenched oligonucleotides such as universal reporters.
In this study, we analysed the relevant factors impacting on the fluorescence signal generation of contact quenched oligonucleotides in order to improve universal reporter SNRs. Due to highly systematic experimental procedures, we were able to work out the dominant factors which must be considered regarding the design of contact quenched oligonucleotides. These factors are described as guidelines which are presented as results in this paper.
Materials and methods
Oligonucleotides
All oligonucleotides were custom synthesised (biomers.net GmbH, Germany). The secondary structure of each universal reporter was modelled and visualised using the software OligoPAD (Version 0.3.0.2, GNWI mbH, Germany). All universal reporter molecules analysed in this study were derived from a set of five universal reporter sequences (see ESI†) which were recently published.18 Even though these five universal reporters have different DNA sequences, all the molecules have a similar secondary structure, which is presented in Fig. 1. The exact configuration and coupling strategy of fluorophore and quencher depended on the experiment conducted and are described in detail in the relevant sections of this paper.In this study, the fluorophore and quencher residues were located at the clamp of the universal reporter stem–loop region. Each molecule was coupled to one nucleotide of a complementary base pair. In most cases, the 5′-end of the universal reporter was modified with the quencher and the fluorophore was coupled to the base of the complementary nucleotide. Due to the high binding strength of the stem–loop structure and the strong interaction of the potential fluorophore–quencher pair, both residues were brought into close contact to promote contact quenching.6,19
Mediator extension assay and fluorescence measurements
The fluorogenic performance of a universal reporter was characterised by a mediator extension assay (Fig. 1, bottom, C–E).10 In this assay, the universal reporter is activated directly by a mediator in the reaction mix. Five different DNA sequences were used as mediators, each complementary to the hybridisation sequence of a certain universal reporter. Each mediator had a length of 19–20 bases to achieve a binding energy of approximately −16 kcal mol−1, which had been calculated with OligoPAD. Mediators had a concentration of 150 nM and universal reporters of 100 nM. In negative controls, the mediator was replaced by PCR grade water (Qiagen). For mastermix, HotStar TaqPlus Mastermix (Qiagen) was used, with 5 mM MgCl2 and 1× BSA (NEB) added. During universal reporter fluorescence signal optimisation, all experiments were carried out in a RotorgeneQ (RGQ; Qiagen) thermocycler. Once optimisation was finished, additional runs were carried out in a StepOnePlus thermocycler (ABI) to test the robustness of our guidelines. Mediator extension was studied under real-time PCR conditions to consider side effects such as thermobleaching of fluorophores. The thermocycling protocols given in Tables 1 and 2 were used.A fluorescence readout was taken after each annealing and extension step at 60 °C. In each experimental set-up, all samples were tested as triplicates. Since fluorophores are sensitive to different error sources such as degradation due to repetitive freezing and thawing or photobleaching, all universal reporter molecules were aliquoted and stored at −20 °C, protected from light, until use. During experimental set-up, reaction mixes were pipetted on ice and afterwards protected from light. To prevent minor systematic errors due to inter-run deviations, all universal reporters which belonged to one experimental set-up were processed within a single run. For larger set-ups, an inter-run control was added (Table 1).
| Step | Repeats | Substep | T [°C] | t [s] |
|---|---|---|---|---|
| Hot start | 1 | — | 95 | 300 |
| Mediator extension | 45 | Denaturation | 94 | 5 |
| Annealing and extension | 60 | 30 |
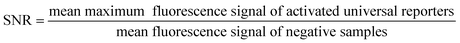 | (1) |
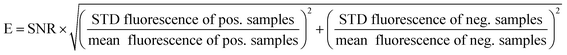 | (2) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 copies of buffy coat DNA. The total amount of the sample DNA per reaction was 500 ng. From this dilution series, R2 and the limit of quantification were determined in order to compare real-time PCR performances. The thermocycling protocol of the SOP is given in Table 2. A fluorescence readout was taken after each annealing and extension step at 60 °C.
000 copies of buffy coat DNA. The total amount of the sample DNA per reaction was 500 ng. From this dilution series, R2 and the limit of quantification were determined in order to compare real-time PCR performances. The thermocycling protocol of the SOP is given in Table 2. A fluorescence readout was taken after each annealing and extension step at 60 °C.
| Step | Repeats | Substep | T [°C] | t [s] |
|---|---|---|---|---|
| Hot start | 1 | 95 | 120 | |
| Cycling | 50 | Denaturation | 94 | 5 |
| Annealing and extension | 60 | 30 |
Results and discussion
Analysing the influence of quencher type, orientation and quantity
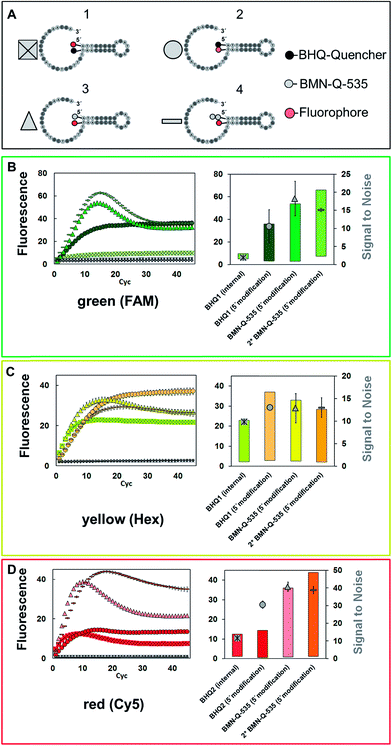
First, the influence of the quencher residues and positions on the SNR was analysed. Four different types of universal reporter were designed, each with a unique quencher configuration (Fig. 2A1–A4). In addition, to minimise the risk of observing effects which are limited to a certain fluorescence spectrum, each universal reporter type was synthesised with three different fluorophores, namely FAM, HEX and Cy5 (Fig. 2B–D). All four universal reporter types are depicted in Fig. 2(A). The results of the mediator extension are shown in Fig. 2B–D (coloured symbols), together with the respective negative controls (Fig. 2B–D, grey symbols). All positive samples showed a significant fluorescence signal increase, while no increase was observed in any NTC. This stresses the high specificity of mediator triggered universal reporter activation. At each wavelength, all NTC baselines were found within the same fluorescence signal range, indicating a similar quenching. The Cy5 labelled universal reporters showed the highest quenching, especially when the fluorophore was coupled internally to the base and a 5′-BHQ2 modification was used. In general, it can be observed that 5′-quencher modifications always lead to higher SNRs compared to an internal quencher modification. Furthermore, the SNRs of universal reporters with a 5′-BMN-Q-535 quencher modification were always among the highest of all universal reporters. Therefore, this quencher can be regarded as the basic 5′-modification for all fluorescence spectra. No significant differences were observed between the SNRs of all other universal reporter variations, where an internal fluorophore and 5′-quencher modification were used. In particular, universal reporters modified with one or two BMN-Q-535 quenchers showed almost identical SNRs due to similar fluorescence quenching and increase. In contrast to double quenched hydrolysis probes, no higher quenching efficiencies were observed for double quenched universal reporters when used in real-time PCR detection.3Analysis of fluorophore positioning considering different quencher types
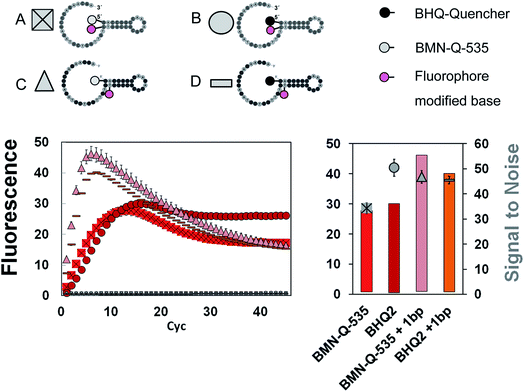
Besides the SNR, differences can also be found regarding the shape of the signal curves (Fig. 2). All universal reporters labelled with a BHQ at their 5′-terminus showed irreversible signal generation. Here, a sigmoidal fluorescence increase was observed, which remained stable in a plateau. In contrast, all universal reporters with a BMN-Q-535 quencher showed a drop in the fluorescence signal after reaching their maximum. To analyse this mechanism of irreversible signal generation in more detail, two additional universal reporter types were designed. These were again labelled with BHQ2 or BMN-Q-535 at the 5′-end, but had an internal Cy5-modification which was not directly opposite the quencher residual (Fig. 3, structures C & D). The results are presented in Fig. 3 (bottom). As can be seen, only when a Cy5 residual was coupled directly opposite the 5′-BHQ2 quencher, an irreversible fluorescence signal generation was observed (Fig. 3, structure B). When Cy5 was not coupled opposite the BHQ2-quencher or another quencher was used, all universal reporters showed a significant slope in the signal curves (Fig. 3, structures A, C and D). These results show that the polymerase has no preference for cleaving BHQ2 residues from a 5′-terminus, which leads to irreversible fluorescence signal generation. Moreover, the type and position of the quencher and fluorophore residuals directly influence this process.6 This indicates that there is an interaction between both residuals (e.g. a hydrogen bond) leading to a conformation promoting irreversible signal generation. This may be due to an enhanced opening of the stem–loop structure or because the respective labels are cleaved off. Another possibility is that the interaction between the two residuals prevents a refolding of the universal reporter's stem. It further can be noticed that there are differences in the height and increase of the fluorescence signal. Universal reporters with a fluorophore modification not directly opposite the quencher position (Fig. 3, structures C and D) reached a higher maximum fluorescence signal and showed a stronger fluorescence increase than those where the fluorophore was placed directly opposite the quencher residual (Fig. 3, structures A and B). However, only a minor influence on the SNR value was found. In addition, we were not able to see any link between the shape of the signal curve and the respective real-time PCR performance. Nevertheless, for applications where an endpoint signal is analysed, such as digital PCR, these effects should be considered.Influence of the selected fluorophore and quencher residues on the signal generation
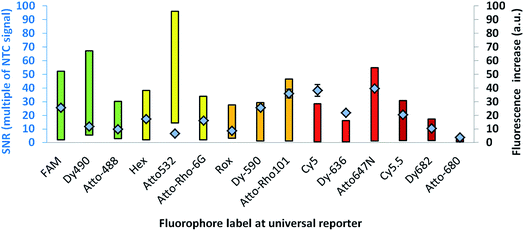
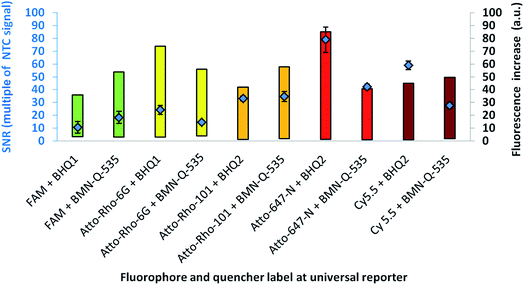
For further optimisation of the SNR, universal reporters with a basic structure according to Fig. 1 were used. Fluorophores were chosen with respect to the 5 fluorescence channels of the RGQ. Each channel allows distinct excitation and detection in a particular fluorescence spectrum. As quencher, BMN-Q-535 was used for all universal reporters. The results of the mediator extension assay are presented in Fig. 4. All tested universal reporters showed a significant fluorescence signal increase after mediator extension, as well as a significant change in the SNR. It can be seen that the selection of the fluorophore has a major impact on the SNR. Differences can be found between universal reporters which have their emission maxima in different fluorescence spectra of the RGQ, but also for universal reporters whose fluorophores share the same spectral range. Nevertheless, at least one universal reporter in each fluorescence spectrum showed a SNR of approximately ten or higher, which can be associated with a good multiplex real-time performance. As can be seen in Fig. 4, the maximal fluorescence signal and/or the initial fluorescence quenching significantly influenced the SNRs of the tested universal reporters. In the case of the orange and crimson fluorescence spectra, universal reporters with the highest signal increase also showed the highest SNRs. As one expects, the highest fluorescence signal increase was observed for universal reporters whose fluorophores had adsorption maxima closest to the excitation wavelength of the RGQ, and had emission maxima within the detection window of the RGQ's broad band filters. However, as can be seen for the green and yellow fluorescence spectra of the RGQ, a high fluorescence signal increase alone does not automatically imply optimum universal reporter SNRs and therefore real-time PCR performance. In these spectral ranges, the best SNRs are found with universal reporters modified with FAM (green) and Atto-Rho-6G or HEX (yellow). In both cases, the highest SNRs were due to a strong fluorescence quenching of the universal reporter in its initial, non-activated stage, which counterbalanced a lower fluorescence signal.For further analysis, the universal reporter with the highest SNR from each fluorescence spectrum was selected and tested with an alternative quencher (BHQ1 or BHQ2). In the case of the yellow fluorescence spectrum, no significant difference was observed between universal reporters modified with Atto-Rho-6G or Hex (Fig. 4). Therefore, Atto-Rho-6G was chosen since the combination Hex + BHQ1 had already been tested in this study (Fig. 2). The results are shown in Fig. 5. The BHQ label led to an additional increase in the SNRs for universal reporters which carried Atto-647-N (red) and Cy5.5 (crimson) as a fluorophore. Even though both universal reporters already showed high SNRs of 42.1 ± 2.1 (Atto-647-N) and 27.6 ± 2.5 (Cy5.5) when they had a BMN-Q-535 quencher, in both cases the SNR was approximately doubled when a BHQ2-label was used instead. Interestingly, the reasons for these significant higher SNRs were different. While the combination Atto-647-N + BHQ2 led to a higher maximum fluorescence signal, the combination Cy5.5 + BHQ2 led to a stronger fluorescence quenching.
These results reflect the high complexity of fluorescence quenching and signal generation processes for contact quenched molecules. In contrast to FRET, which enables energy transfer over a distance of up to approximately 10 nm, contact quenching occurs only at very low distances of approximately 1–2 nm. For this, a weak, non-covalent interaction between fluorophore and quencher like a hydrogen bond is required.16 Furthermore, for contact quenched molecules, quenching efficiency cannot be completely described by the spectral overlap between donor and acceptor.6,20 Therefore, predicting of these interactions is rather complex, since many factors (including quantum mechanics) must be taken into account.23 Additionally, the structures of commercially available fluorophores are not all published, which further hinders predictions.
From the results presented in Fig. 5, the fluorophore–quencher combination leading to the highest SNRs for each spectral range was selected for further testing. Using this set of five differently labelled universal reporters, the reproducibility of the SNRs was tested for intra-device variation in the RGQ with three separate runs (Table 3).
| Fluorescence channel of RGQ | RGQ λmax excitation (nm) | RGQ λmax detection (nm) | Internal fluorophore label | 5′-Quencher modification | SNR Run1 | SNR Run2 | SNR Run3 | CV (%) |
|---|---|---|---|---|---|---|---|---|
| Green | 470 | 510 | FAM | BMN-Q-535 | 26.4 ± 2.4 | 24.5 ± 1.5 | 21.3 ± 2.6 | 19.23 |
| Yellow | 530 | 555 | Atto-Rho-6-G | BHQ-1 | 10.7 ± 1.7 | 13.0 ± 1.0 | 14.0 ± 1.3 | 23.12 |
| Orange | 585 | 610 | Atto-Rho-101 | BMN-Q-535 | 16.7 ± 1.3 | 18.4 ± 2.3 | 19.3 ± 2.7 | 22.07 |
| Red | 625 | 660 | Atto-647-N | BHQ2 | 60.1 ± 7.0 | 76.1 ± 1.9 | 71.0 ± 3.8 | 16.75 |
| Crimson | 680 | 710 | Cy5.5 | BHQ2 | 38.2 ± 6.0 | 37.9 ± 2.7 | 41.3 ± 6.3 | 23.77 |
As can be seen in Table 3, all the universal reporters showed a good reproducibility regarding their SNRs. No significant deviation could be observed between all three runs. In comparison to Fig. 5, slightly lower SNRs were observed for universal reporters in the fluorescence spectra yellow and orange. These variations may be due to minor errors sources as described in the materials and methods section. Since these variations were only observed for these two universal reporters, Atto-Rho-6G and Atto-Rho-101 seem to be especially sensitive to such effects. Nevertheless, the SNRs still were around 10 or higher, which is associated with good multiplex real-time PCR performance.
Guidelines for signal-to-noise optimisation of fluorogenic universal reporters
From the results presented above we were able to define guidelines for optimising the SNRs of fluorogenic universal reporter oligonucleotides. These guidelines can be regarded as an update to those presented before.10(1) A universal reporter with the basic structure presented in Fig. 1 (top) should be used. The quencher must be coupled to the 5′-end of the universal reporter, and the fluorophore to the base directly opposite the quencher residual. To ensure that the right nucleotide is selected for the fluorophore modification, we recommend calculating the secondary structure of the universal reporter first using appropriate bioinformatics software tools. With green fluorophores such as FAM, AT-sequences must be used in the stem–loop structure instead of a GC-clamp to prevent G-quenching. In addition, the 3′-end must be blocked with a C3-spacer.
(2) Perform the mediator extension assay and use high SNRs as target values for the universal reporter optimisation. If a universal reporter reaches a SNR of ten or higher, it can generally be considered for multiplex real-time PCR applications. For some applications, a higher fluorescence signal may be useful. In this case, SNRs of approximately 15–30 should be reached.
(3) For universal reporter optimisation, different fluorophores should be tested first, in some cases even within the same spectral range. While testing different fluorophores, the quencher must be kept constant.
(4) When the fluorophore has been fixed but the SNR is still too low, testing different quenchers may lead to an additional increase. In this case, different quencher modifications should be tested at the 5′-end of the universal reporter while the internal fluorophore modification is kept constant.
Guideline test in second thermocycler
To test the robustness of the described guidelines, additional experiments were carried out in a second real-time thermocycler (SOP). In contrast to the RGQ, the SOP uses one single wavelength of 470 nm for excitation of all fluorophores, and has detection filters for the wavelengths 520 nm, 550 nm, 580 nm and 610 nm. According to guidelines 1, 2 and 3, three different universal reporters were designed, each with a different fluorescence label. Since the wavelength of maximum light absorption of most fluorophores is approximately 20–30 nm below their wavelength of maximum fluorescence emission, using a single wavelength thermocycler for multiplex applications is suboptimal and more challenging. Therefore, Dy494-XL was selected as one of the fluorophores due to its large Stokes shift of 122 nm. The SNRs of these three universal reporters were first compared using the mediator extension assay. The results are given in Table 4.| Name of SOP fluorescence channel | SOP λmax excitation (nm) | SOP λmax detection (nm) | Internal fluorophore label | 5′-Quencher | SNR |
|---|---|---|---|---|---|
| Blue | 470 | 520 | FAM | BMN-Q-535 | 11.56 ± 3.6 |
| Green | 470 | 550 | HEX | BMN-Q-535 | 7.1 ± 0.5 |
| Red | 470 | 610 | Dy494-XL | BMN-Q-535 | 7.4 ± 0.48 |
Student's t-tests were carried out to compare the respective SNR test series of universal reporters labelled with the different fluorophores and to check for significant differences. The results are given in Table 5.
| Fluorophore of universal reporter used in SNR test series A | Fluorophore of universal reporter used in SNR test series B | p-Value calculated by Student's t-test |
|---|---|---|
| Dy494-XL | HEX | 0.42211508 |
| FAM | Dy494-XL | 0.016006779 |
| FAM | HEX | 0.02349825 |
As can be seen in Table 5, significant differences in the SNRs can be found between universal reporters labelled with FAM and HEX (p = 0.023), and FAM and Dy494XL (p = 0.016). In contrast, no significant differences were found between the SNRs of universal reporters labelled with HEX and Dy494-XL (p = 0.42).
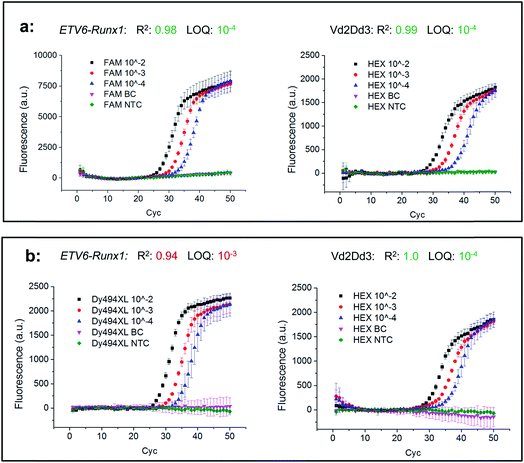
In the next step, two combinations of these universal reports were applied to compare their real-time PCR performances under relevant clinical conditions. For this purpose, an FAM labelled universal reporter was used to detect the gene fusion ETV6-Runx1 in the first biplex Mediator Probe PCR, while in the second Dy494XL was selected. As an inter-reaction control, both biplex Mediator-Probe PCRs used the same HEX labelled universal reporter for Vd2Dd3 detection. The real-time PCR performance was characterised according to the official guidelines of the European MRD Council (Euro-MRD-Group) for ALL monitoring. The results are presented in Fig. 6. Using the Hex labelled universal reporters, both real-time PCRs showed comparable performance characteristics reflected by R2 values of 0.99 and 1.0, an LOD of 10−4 and almost identical fluorescence signal increases. In contrast, major differences can be found in the real-time PCR performances of the FAM and Dy494XL labelled universal reporters. The Dy494XL label gave a comparable fluorescence signal increase to the Hex label, but showed high deviations in the fluorescence signal curves for lower target DNA copy numbers. Because of this, an R2 of 0.94 and an LOQ of 10−3 were achieved, which do not conform to the official guidelines for MRD quantification of ALL targets.22 Using the FAM labelled universal reporter, a significantly higher fluorescence signal increase was achieved in comparison to all other universal reporters. This increase led to smaller deviations in the fluorescence signal curves of all DNA concentrations. Therefore, an R2 of 0.98 and an LOQ of 10−4 were achieved, which do conform to the official MRD guidelines. These results are also reflected by the Student's t-test (Table 5), where the FAM labelled universal reporter showed significantly higher SNRs compared to the Dy494-XL or HEX labelled universal reporters. This does not mean that those fluorophores which led to lower SNRs in our SOP experiments should generally be avoided in Mediator Probe real-time PCRs. Rather, it stresses that the performance of a universal reporter with a distinct fluorophore label may change when applied in a thermocycler with distinct optical properties. Therefore, according to our guidelines, the selection of the fluorophore is of major importance for the universal reporter SNR, and so significantly influences the performance of a Mediator Probe real-time PCR.
Conclusion
In order to derive guidelines for the signal-to-noise optimisation of universal fluorogenic reporter oligonucleotides, we analysed and compared four factors: (1) type of fluorophore (2) type of quencher, (3) position and orientation of fluorophore and quencher residues (4) number of quencher molecules. Even though all tested universal reporters had a similar biochemical structure, with a dual fluorophore–quencher-label with minimum separation, significant differences in the SNRs were still observed. Once the position of fluorophore and quencher had been fixed, we were able to show that the selection of a fluorophore has a major impact on the SNR. Even within the same spectral range, different fluorophores lead to significant variations in the SNRs. The type of quencher molecule also influenced the SNR of a universal reporter, but the observed impact was lower compared to the impact of the selected fluorophore. In contrast, the number of quencher molecules coupled to one universal reporter had no observed effect on the SNR. This can be explained by the different quenching mechanisms of hydrolysis probes and contact quenched universal reporters.From these results we were able to derive guidelines which should be used for SNR optimisation of universal reporter oligonucleotides in Mediator Probe PCRs. These guidelines can be regarded as an update and an addition to those previously presented for Mediator Probe PCR.10,17 These SNR optimisation guidelines will help the community to set up new Mediator Probe PCR systems, or to deduce such systems from other established real-time PCR detection formats such as 5′ nuclease PCRs. We also demonstrated the improvement, with respect to the limit of quantification, of existing Mediator Probe PCRs. By testing different fluorophores according to guideline 3, we were able to improve our limit of quantification by a factor of 10. Since no sophisticated functions like melting curve analysis or curve fitting are needed in our optimisation process, these guidelines are compatible with all real-time PCR thermocyclers. Furthermore, SNR analysis, rather than real-time PCRs, can be conducted during the process of fluorescence signal optimisation, and once one universal reporter has been optimised it can be used in the design of all subsequent real-time PCR assays, independent of the target sequence. This will save time, costs and sample material.
It can be assumed that these SNR guidelines are not restricted to Mediator Probe PCRs alone. They may also be applied to increase the SNRs of other contact quenched molecules (e.g. molecular beacons, scorpion primers), fluorogenic biomolecules (e.g. dual-labelled antibodies) or to improve signal generation in other technologies which make use of primary probes and secondary fluorogenic reporters.4,13,24
Conflicts of interest
No conflict of interest to be declared.Acknowledgements
This study was founded by the German Federal Ministry of Education and Research and the German Federal Ministry of Economic Affairs and Energy as part of the projects IRMA-4-ALL (FKZ 01 EK1508B/INDIMED program) and MP PCR Desire (FKZ KF2162035CR4/ZIM program). We thank PD. Dr Cornelia Eckert from the department of paediatrics, division of oncology and hematology, Charité, Berlin, for providing laboratory resources. We further thank Dr Simon Wadle for scientific discussions.References
- I. M. Mackay, Clin. Microbiol. Infect., 2004, 10, 190–212 CrossRef PubMed.
- (a) L. Dong, Y. Meng, Z. Sui, J. Wang, L. Wu and B. Fu, Sci. Rep., 2015, 5, 13174 CrossRef PubMed; (b) F. Schuler, M. Trotter, M. Geltman, F. Schwemmer, S. Wadle, E. Domínguez-Garrido, M. López, C. Cervera-Acedo, P. Santibáñez, F. von Stetten, R. Zengerle and N. Paust, Lab Chip, 2016, 16, 208–216 RSC; (c) B. Vogelstein and K. W. Kinzler, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 9236–9241 CrossRef PubMed.
- F. Schuler, M. Trotter, R. Zengerle and F. von Stetten, Anal. Chem., 2016, 88, 2590–2595 CrossRef PubMed.
- J. M. Ruijter, P. Lorenz, J. M. Tuomi, M. Hecker and M. J. B. van den Hoff, Microchim. Acta, 2014, 181, 1689–1696 CrossRef PubMed.
- (a) Fluorescent Energy Transfer Nucleic Acid Probes. Designs and Protocols, ed. V. V. Didenko, Humana Press Inc, Totowa, NJ, 2006, vol. 335 Search PubMed; (b) A. Solinas, Nucleic Acids Res., 2001, 29, 96e–96 CrossRef.
- S. A. E. Marras, Methods Mol. Biol., 2006, 335, 3–16 Search PubMed.
- P. M. Holland, R. D. Abramsin, R. Watson and D. H. Gelfand, Proc. Natl. Acad. Sci. U. S. A., 1991, 88, 7276–7280 CrossRef.
- S. A. E. Marras, Nucleic Acids Res., 2002, 30, 122e–122 CrossRef.
- (a) S. Tyagi and F. R. Kramer, Nat. Biotechnol., 1994, 14, 303–308 CrossRef PubMed; (b) X. Dai, W. Yang, E. Firlar, S. A. E. Marras and M. Libera, Soft Matter, 2012, 8, 3067 RSC.
- S. Wadle, S. Rubenwolf, M. Lehnert, B. Faltin, M. Weidmann, F. Hufert, R. Zengerle and F. von Stetten, Methods Mol. Biol., 2014, 1160, 55–73 CrossRef PubMed.
- V. M. Farzan, I. O. Aparin, O. A. Veselova, A. T. Podkolzin, G. A. Shipulin, V. A. Korshun and T. S. Zatsepin, Anal. Methods, 2016, 8, 5826–5831 RSC.
- I. Nazarenko, R. Pires, B. Lowe, M. Obaidy and A. Rashtchain, Nucleic Acids Res., 2002, 30, 2089–2195 CrossRef PubMed.
- B. Faltin, R. Zengerle and F. von Stetten, Clin. Chem., 2013, 59, 1567–1582 CrossRef PubMed.
- B. Faltin, S. Wadle, G. Roth, R. Zengerle and F. von Stetten, Clin. Chem., 2012, 58, 1546–1556 CrossRef PubMed.
- M. K. Johansson, H. Fidder, D. Dick and R. M. Cook, J. Am. Chem. Soc., 2002, 124, 6950–6956 CrossRef PubMed.
- S. Bernacchi and Y. Mély, Nucleic Acids Res., 2001, 29 Search PubMed.
- S. Wadle, M. Lehnert, S. Rubenwolf, R. Zengerle and F. von Stetten, Biomol. Detect. Quantif., 2016, 7, 1–8 CrossRef PubMed.
- S. Wadle, M. Lehnert, F. Schuler, R. Köppel, A. Serr, R. Zengerle and F. von Stetten, BioTechniques, 2016, 61, 123–128 CrossRef PubMed.
- B. G. Moreira, Y. You, M. A. Behlke and R. Owczarzy, Biochem. Biophys. Res. Commun., 2005, 327, 473–484 CrossRef PubMed.
- M. K. Johansson, Methods Mol. Biol., 2006, 335, 17–29 Search PubMed.
- D. Bhojwani, D. Pei, J. T. Sandlund, S. Jeha, R. C. Ribeiro, J. E. Rubnitz, S. C. Raimondi, S. Shurtleff, M. Onciu, C. Cheng, E. Coustan-Smith, W. P. Bowman, S. C. Howard, M. L. Metzger, H. Inaba, W. Leung, W. E. Evans, D. Campana, M. V. Relling and C.-H. Pui, Leukemia, 2012, 26, 265–270 CrossRef PubMed.
- V. H. J. van der Velden, G. Cazzaniga, A. Schrauder, J. Hancock, P. Bader, E. R. Panzer-Grumayer, T. Flohr, R. Sutton, H. Cave, H. O. Madsen, J. M. Cayuela, J. Trka, C. Eckert, L. Foroni, U. Zur Stadt, K. Beldjord, T. Raff, C. E. van der Schoot and J. J. M. van Dongen, Leukemia, 2007, 21, 604–611 CrossRef PubMed.
- (a) L. E. Ratcliff, S. Mohr, G. Huhs, T. Deutsch, M. Masella and L. Genovese, WIREs Comput. Mol. Sci., 2017, 7, e1290 CrossRef; (b) A. Mondal and S. Datta, Proteins, 2017, 85, 1046–1055 CrossRef PubMed.
- Y. Li, P. C. Liu, Y. Shen, M. D. Snavely and K. Hiraga, J. Biomol. Screening, 2015, 20, 869–875 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ay00812d |
| This journal is © The Royal Society of Chemistry 2018 |