Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Dual purpose fibre – SERS pH sensing and bacterial analysis†
Holly
Fleming
ab,
Sarah
McAughtrie
ab,
Bethany
Mills
 b,
Michael G.
Tanner
b,
Michael G.
Tanner
 bc,
Angus
Marks
a and
Colin J.
Campbell
bc,
Angus
Marks
a and
Colin J.
Campbell
 *a
*a
aEaStCHEM, School of Chemistry, University of Edinburgh, Edinburgh, EH9 3FJ, UK. E-mail: colin.campbell@ed.ac.uk
bCentre for Inflammation Research, Queen's Medical Research Institute, University of Edinburgh, Edinburgh, EH16 4TJ, UK
cScottish Universities Physics Alliance (SUPA), Institute of Photonics and Quantum Science (IPAQS), Heriot-Watt University, Edinburgh EH14 4AS, UK
First published on 5th October 2018
Abstract
The exploitation of fibre based Raman probes has been challenged by often complicated fabrication procedures and difficulties in reproducibility. Here, we have demonstrated a simple and cost-effective approach for sensing pH through an optical fibre, by employing a wax patterned filter paper-based substrate for surface enhanced Raman spectroscopy (SERS). Through this method, high reproducibility between fibres was achieved. In addition to sensing pH, it was possible to extract fluid samples containing P. aeruginosa for further analysis. This dual purpose fibre is bronchoscope deployable, and is able to gather information about both the host and pathogen, which may lead to an improved treatment plan in future in vivo applications.
Introduction
With the rising global challenge of antimicrobial resistance, there is an urgent need to reduce unnecessary antimicrobial prescriptions.1 Ventilator-associated pneumonia (VAP) is the most common infection among the critically unwell in intensive care units (ICUs), with Pseudomonas aeruginosa behind many of these infections.2Studies have shown that using the standard methods to identify the infectious agents, often by interpreting non-specific clinical or radiological features combined with culture techniques from sputum samples, was not adequate to diagnose lower tract respiratory infection but required processing of bronchoalveolar lavage fluid (BALF) samples.3 However, even the use of these samples has a number of limitations. Bacterial cultures typically take up to 3 days for results, but specifically, these samples can suffer from a lack of sensitivity due to aspirated fluid being prone to contamination from the upper respiratory tract. Molecular sequencing methods such as polymerase chain reactions (PCR) can be overly sensitive, potentially leading to overtreatment of patients. Combined with poor sampling techniques, there is a significant impact on the treatment and management of patients.4–7
The correct identification of pathogens causing respiratory infections and the monitoring of their antimicrobial sensitivity is of great importance. In addition to identification, knowing the physiological environment of the site of interest can also hold great value. pH is tightly regulated within cells and their microenvironments, with any deviations from the homeostasis indicating disease processes. Within the lung, an acidic pH can encourage the growth of bacteria, reduce the efficiency of endogenous cationic antimicrobial peptides and inactivate some antibiotics – all factors that contribute to antimicrobial resistance and worsening patient outcome.8,9
The need for bed-side based monitoring has driven research efforts with a focus on robust and rapidly responding optical sensor devices. Fibre-based Raman spectroscopy is becoming a popular method for in vivo investigations due to its being a non-destructive and minimally invasive technique. Furthermore, it exhibits high spatial resolution and chemical sensitivity, important factors for in situ monitoring. However, as Raman scattering is an inherently weak phenomenon, its use for dynamic physiological sensing can be somewhat limited.
Surface enhanced Raman spectroscopy (SERS), where a reporter molecule is adsorbed on to the surface of a metal nanoparticle (NP), can increase the signal intensity by several orders of magnitude over conventional Raman scattering. SERS has been used in many applications, from intracellular physiological sensing, drug delivery sensing, explosive detection and many more.10,11
SERS has been demonstrated through fibre previously, however, it has been challenged by difficulties in reproducibility from fibre to fibre, as well as generating SERS signal intensities great enough to overcome the intrinsic silica fibre background. As a result, much of the focus in this area has been on background suppression through complex fibre designs and correction methods.12–14
Paper based substrates for SERS sensing and detection are gaining traction as point-of-care systems due to their low cost and flexible nature.15–17 One of the difficulties is directing the deposition of NP, due to the inherent wicking ability of filter paper which is both quick and uncontrolled. The use of patterning with a hydrophobic ink can assist with some of these issues, by defining a specified SERS sensing region.18,19
Here, we demonstrate a facile and cost-effective SERS substrate capable of ratiometrically measuring pH using SERS, whilst retrieving a biological fluid sample through combining paper and fibre-based systems. The advantages of a wax patterned paper sensing substrate are two-fold, it allows simple controlling of particle deposition and also facilitates an easy way to retrieve biological fluid samples. Our method has been designed to be small enough to be bronchoscope deployable with the ability to reach alveolar regions within the lung, allowing for site specific information to be gathered and so gaining information about both host and pathogen. This approach has also overcome many of the outlined challenges by achieving an easily repeatable fabrication process and generating high signal to noise ratios.
Results and discussion
Paper SERS substrate design
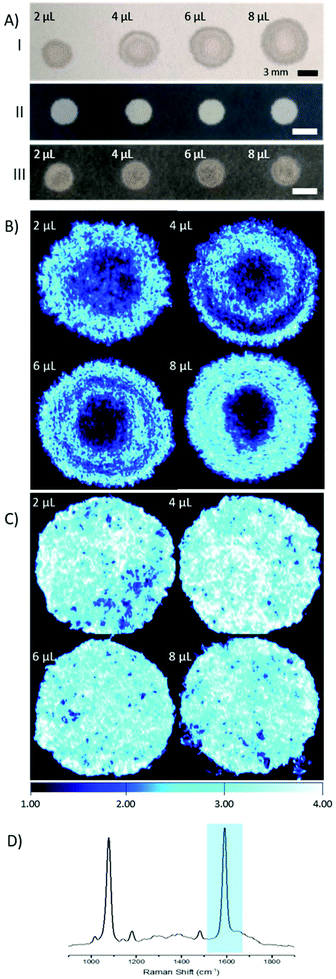
The main goal of this research was to provide a simple SERS substrate to combine with a fibre-based approach to sensing, in particular pH. For this purpose, filter paper was chosen as the preferred substrate due to its wide availability and porous nature.4-Mercaptobenzoic acid functionalised AuNPs (4-MBA; 150 nm, 3.6 × 108 particles per μL) were pipetted on to filter paper substrates. Repeated depositions of a low volume of AuNPs (2 μL droplets) were applied, to a final volume of 2–8 μL, with drying stages in between, to avoid a loss of AuNPs through soaking and washing through the paper. The size of the spots that contain AuNPs was defined by the volume of the solution, however there is much variance in shape and the spread, or wicking, of the AuNPs within the spots (Fig. 1A, I).
In an effort to overcome the influence of non-uniform capillary wicking, and therefore the low reproducibility of using a filter paper-based SERS substrate, a hydrophobic wax surround was printed onto the filter paper. Solid wax printing is a simple, inexpensive, and quick method, amenable to mass production. Patterning paper with a wax surround allows control of the wicking action of aqueous solutions, confining the dispersion to the hydrophilic areas.18–20 By limiting the solution containing the AuNPs to a defined area, there is a reduction in variability in AuNP concentration at any point within the hydrophilic area. A substantial difference between applying AuNPs to filter paper with and without a wax barrier can be clearly seen by eye (Fig. 1A). The image shows both an increasing spot size, as well as an uneven spread of AuNPs across the filter paper where a wax barrier was not used. The unevenness is further revealed by SERS mapping (Fig. 1B). The patterned paper allows AuNPs to be deposited on the filter paper in a controlled manner, filling the entire hydrophilic area evenly (Fig. 1C). SEM images from paper substrates with a printed wax barrier show a high density of AuNPs on the cellulose fibres, which may help in the formation of SERS “hot-spots”, further enhancing the signal intensity (Fig. S1†).
Fibre sensing
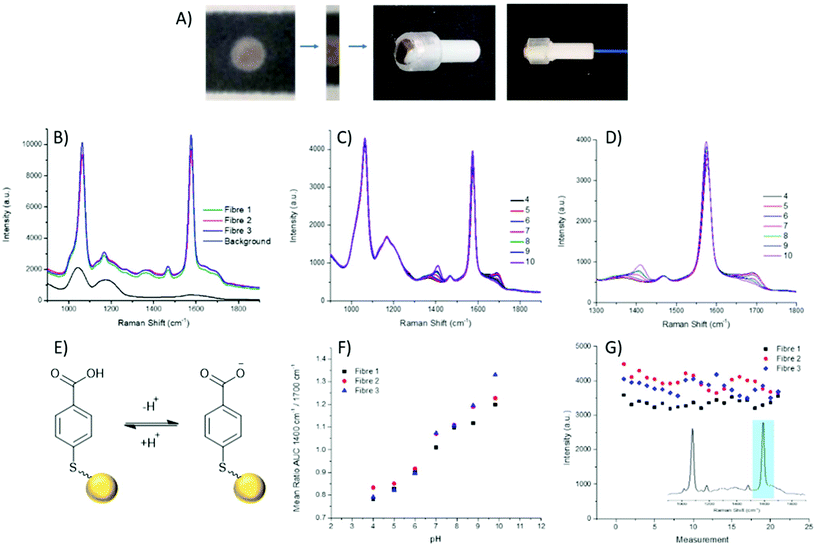
The fabrication of the packaged ferrule end is a simple process (Fig. 2A), whereby a 2 mm wide strip of the wax printed AuNP paper was placed, facing upwards, on a ring-shaped section of rubber. The ferrule was pushed through the ring with the paper flush across the top surface. A commercially available 200 μm single core (NA 0.39), multi-mode fibre was then threaded through the ferrule until abutted against the paper, and secured in place at the base of the ferrule.
Through combining the fibre with the paper-based SERS substrate, a controlled particle deposition is achieved, providing confidence that the same signal intensity can be reached in any location where the fibre tip is placed on the AuNP paper. This extends to being able to reliably reproduce a strong signal when moving between fibres. Three separate fibres with packaged distal ends were prepared and their spectra recorded in air. It can be seen that across the three fibres reproducibility of the paper-based system is demonstrated by strong signals of similar intensities (Fig. 2B). Moreover, the strong signal intensity generated by the paper-based SERS sensor easily overcomes the intrinsic fibre background using relatively low laser power and integration time (1 mW and 1 s respectively). Importantly, the fibre background does not impose significantly on the pH sensitive peaks at 1380 cm−1 and 1700 cm−1.
Aqueous pH buffers were prepared from pH 4–10 and verified with an electrochemical pH meter (Mettler-Toledo). The packaged distal end of the fibre was submerged in buffer and the spectrum recorded after 10 s. Between readings the AuNP-paper substrate was rinsed in dH2O and blotted dry. Each fibre had three replicate calibrations where pHs were recorded in a random order. Three fibres in total were measured. The spectra were analysed by first normalising the spectra to the magnitude of a reference peak (1070 cm−1), followed by evaluating the area under the curve (AUC) within a ±25 cm−1 window of the peaks at 1380 cm−1 and 1700 cm−1. Plotting pH against the AUC ratio, all fibres demonstrated consistent variation within the physiological range (Fig. 2F), again indicating the suitability of the paper SERS substrate for fibre sensing in future in vivo applications.
The loss of nanoparticles over time from the distal end of the fibre would be problematic, not only due to loss of signal, but also because loss of AuNPs is undesirable for use in in vivo applications. With the fibres which have been dip-coated with nanoparticles, typically, a porous sol–gel layer is used to protect the distal end. However, this coating can also suffer from variations in both coating depth and the porosity of the sol–gel layer, which may affect the speed at which measurements can be acquired. The paper SERS substrate showed no significant signal loss from over the course of the pH measurements (approximately 60 min per fibre). The intensity of the 1587 cm−1 peak was plotted over time, with the slight oscillations being attributed to the drying and wetting of the SERS samples. The average intensities between the first and last sets of pH measurements differ by less than 250 counts, less than 10% of overall signal intensity (Fig. 2G).
Extraction and culture of P. aeruginosa
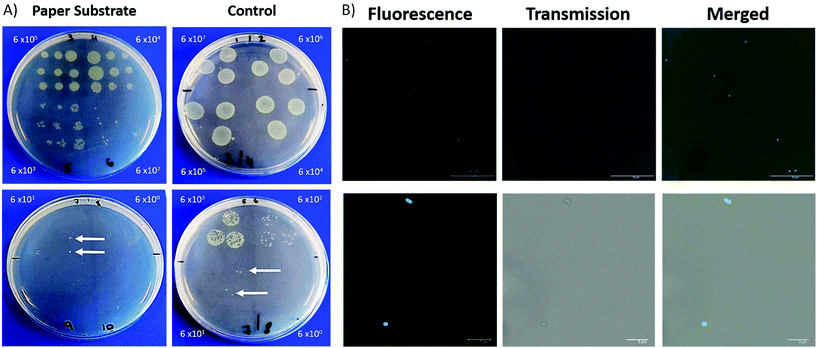
There has been a considerable increase in the number of infections due to antimicrobial resistant bacteria with lower respiratory tract infections responsible for the second highest burden of disease globally.25–27 Within ICUs, the development of pneumonia is associated with high mortality rates.28 The investigation of respiratory disease can involve biopsies, an invasive procedure, and the collection of BALF, which can become contaminated by bacteria found in the upper respiratory tract. The ability to sample fluid at specific sites can alleviate issues related to contamination. This fibre has been designed keeping in mind that it should be deployable through a conventional bronchoscope. In this way, it can be extended and retracted at specific region of interest, therefore minimising the contact between the distal end of the probe and upper respiratory tract.The porous nature of the filter paper not only provides a suitable substrate for the deposition of nanoparticles but also lends itself to retrieving a sample of fluid. We demonstrate that the filter paper is capable of sampling liquid containing bacteria which can then be cultured and counted. A clinical isolate of P. aeruginosa 3284 was cultured and prepared for this study.
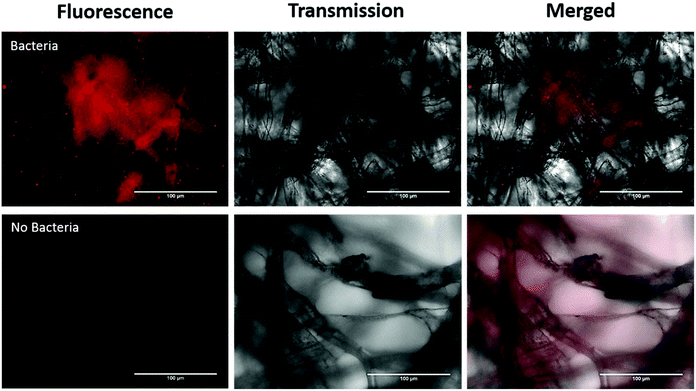
In addition, we investigated imaging bacteria directly on the paper itself (Fig. 4). Due to the autofluorescence of the paper (Fig. S2†), a far-red dye was used. The bacteria were labelled with a Syto60 (5 μM), a fluorescent nuclear stain, before introducing the AuNP-paper strips into the solution containing 6 × 105 CFUs per mL. While this strategy suppressed much of the autofluorescence seen in the green channel, there is still some homogeneous signal observed in the Cy5 fluorescence channel. However, despite the background signal, the labelled bacteria can clearly be detected by widefield imaging, indicating that augmenting collected samples with an appropriate far-red bacteria-specific stain could enable in vivo bacterial detection on fibre without any need for a processing step. This could pave the way for simultaneous measurements for bacteria and pH through a single fibre.
Experimental section
Materials and reagents
Gold nanoparticles in citrate buffer (AuNPs, 150 nm, put concentration), poly-L-lysine (30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–70
000–70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 MW), 4-mercaptobenzoic acid (4-MBA) were all purchased from Sigma-Aldrich. Filter paper (Whatman, grade 114) was purchased from Scientific Laboratory Supplies Ltd. Lysogeny Broth was purchased from Thermofisher. Both the optical fibre (200 μm core diameter, NA 0.39) and fibre ferrules were purchased from Thorlabs.
000 MW), 4-mercaptobenzoic acid (4-MBA) were all purchased from Sigma-Aldrich. Filter paper (Whatman, grade 114) was purchased from Scientific Laboratory Supplies Ltd. Lysogeny Broth was purchased from Thermofisher. Both the optical fibre (200 μm core diameter, NA 0.39) and fibre ferrules were purchased from Thorlabs.
Instrumentation
Solid wax printing was carried out using a Xerox ColourQube8580.SERS maps were collected on a Renishaw In Via system, using a 785 nm laser, 5× objective, at ∼1.5 mW laser power with a 1.4 s integration time.
The fibre-based SERS spectra were collected using a home built optical set up specifically for optical fibres.12 A 785 nm laser line (Thorlabs) was used as the excitation source, coupled to an OceanOpticsPro spectrometer. The output of the laser power was set as 1 mW, and an integration time of 10 s was used.
Images of bacteria extracted from paper fluorescent were taken by confocal laser scanning microscopy (Leica SP8, 488 nm excitation, 63× oil immersion). Images were brightness and contrast enhanced with proprietary software. Bacteria imaged directly on waxed AuNP paper were taken using an EVOS® microscope with GFP and Cy5 light cubes, images were brightness and contrast enhanced using ImageJ software.
Methods
The functionalised nanoparticles were then concentrated and combined. The samples were centrifuged at 5500 rpm for 10 min, and the supernatant (950 μL) was removed without disturbing the pellet. The AuNPs were forced back into suspension in the remaining volume of dH2O (∼50 μL) through sonication and vortexing. The 5 aliquots were combined, centrifuged at 5500 rpm for 10 min, and the appropriate amount of supernatant was removed to give a final volume of 50 μL.
For fabrication of the SERS-active substrate, 2 μL droplets of the 4-MBA functionalised AuNPs were pipetted on to the filter paper disk and allowed to dry at room temperature for an hour. A further 6 μL of AuNPs was dropped on to each disk in 2 μL aliquots with drying in between to a total of 8 μL (2.9 × 109 total particles deposited).
A SERS spectrum was obtained using the packaged distal end of the fibre from 7 separate pH buffers from pH 4–10. SERS spectra were acquired while the fibre tip was fully submerged in each buffer. Each of the 3 fibres were used for 3 replicate measurements between pH 4–10, with the order of the measurements within each replicate being random.
The optical density at 595 nm (OD595) of the resulting culture was measured and adjusted to a value of 1 to give an approximate bacterial concentration of 6 × 108 CFU mL−1. Serial dilutions (6 × 108 to 6 × 100 CFU mL−1) were prepared with sterile PBS.
For each dilution, the packaged ferrule was dipped into the bacteria containing solution and pressed in succession across an LB agar plate. To compare, 3 × 20 μL samples of each dilution was dropped on lysogeny broth (LB) agar plates for CFU analysis. Plates were incubated at 37 °C with 5% CO2 overnight, with CFUs manually counted.
After the paper ferrule had been pressed into LB agar, it was placed in an Eppendorf containing PBS (0.5 mL) and lightly vortexed. The paper and ferrule were removed and the bacteria containing solution labelled with a ubiquicidin-based probe (5 μM), without a washing step. Immediately following the addition of the label, the solutions were placed in a confocal imaging chamber, pre-coated with poly-D-lysine (0.1 mg mL−1) and imaged by confocal laser scanning microscopy.
For direct imaging of bacteria on the paper, bacteria were first stained with Syto60 (5 μM; Thermofisher), as per manufacturer's instructions, and washed twice with PBS by centrifugation at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 1 min. The AuNP paper was dipped into the stained bacterial solution containing 6 × 105 CFUs per mL and imaged using widefield microscopy.
000 rpm for 1 min. The AuNP paper was dipped into the stained bacterial solution containing 6 × 105 CFUs per mL and imaged using widefield microscopy.
Conclusions
In this study, a facile, inexpensive and reproducible paper-based SERS sensor has been integrated with optical fibre technology for use in pH sensing across a physiological relevant range. Using a patterned wax printed stencil to control the wicking boundary of AuNPs, the distribution of particles can be controlled across the paper. In addition, due to the wicking nature of the filter paper, it was possible to extract the bacteria, P. aeruginosa, demonstrating the dual-purpose ability of the paper substrate to acquire physiological and pathogenic information.Conflicts of interest
The authors have no conflicts to declare.Acknowledgements
This work was supported by the Engineering and Physical Sciences Research Council (EPSRC) and Medical Research Council (MRC) under grant number EP/L016559/1 (OPTIMA), and the School of Chemistry, University of Edinburgh; and the EPSRC Interdisciplinary Research Collaboration (EP/K03197X/1).References
- J. O'Neill, Tackling drug-resistant infections globally: final report and recommendations, H M Government/Wellcome Trust, 2016 Search PubMed.
- S. Ramírez-Estrada, B. Borgatta and J. Rello, Infect. Drug Resist., 2016, 9, 7–18 Search PubMed.
- R. P. Baughman, D. A. Keeton, C. Perez and R. W. Wilmott, Am. J. Respir. Crit. Care Med., 1997, 156, 286–291 CrossRef CAS PubMed.
- H. Wang, X. Gu, Y. Weng, T. Xu, Z. Fu, W. Peng and W. Yu, BMC Pulm. Med., 2015, 15, 94 CrossRef PubMed.
- J. R. Lentino and D. A. Lucks, J. Clin. Microbiol., 1987, 25, 758–762 CAS.
- K. C. Carroll, J. Clin. Microbiol., 2002, 40, 3115–3120 CrossRef PubMed.
- A. Zumla, J. A. Al-Tawfiq, V. I. Enne, M. Kidd, C. Drosten, J. Breuer, M. A. Muller, D. Hui, M. Maeurer, M. Bates, P. Mwaba, R. Al-Hakeem, G. Gray, P. Gautret, A. A. Al-Rabeeah, Z. A. Memish and V. Gant, Lancet Infect. Dis., 2014, 14, 1123–1135 CrossRef PubMed.
- A. Dalhoff, S. Schubert and U. Ullmann, Infection, 2005, 33, 36–43 CrossRef CAS PubMed.
- W. F. Walkenhorst, J. W. Klein, P. Vo and W. C. Wimley, Antimicrob. Agents Chemother., 2013, 57, 3312–3320 CrossRef CAS PubMed.
- A. Jaworska, L. E. Jamieson, K. Malek, C. J. Campbell, J. Choo, S. Chlopicki and M. Baranska, Analyst, 2015, 140, 2321–2329 RSC.
- L. E. Jamieson, V. L. Camus, P. O. Bagnaninchi, K. M. Fisher, G. D. Stewart, W. H. Nailon, D. B. McLaren, D. J. Harrison and C. J. Campbell, Nanoscale, 2016, 8, 16710–16718 RSC.
- D. Choudhury, M. G. Tanner, S. McAughtrie, F. Yu, B. Mills, T. R. Choudhary, S. Seth, T. H. Craven, J. M. Stone, I. K. Mati, C. J. Campbell, M. Bradley, C. K. I. Williams, K. Dhaliwal, T. A. Birks and R. R. Thomson, Biomed. Opt. Express, 2017, 8, 243 CrossRef CAS PubMed.
- K. Ehrlich, A. Kufcsák, S. McAughtrie, H. Fleming, N. Krstajic, C. J. Campbell, R. K. Henderson, K. Dhaliwal, R. R. Thomson and M. G. Tanner, Opt. Express, 2017, 25, 30976 CrossRef CAS PubMed.
- J. C. C. Day and N. Stone, Appl. Spectrosc., 2013, 67, 349–354 CrossRef CAS PubMed.
- R. P. Gandhiraman, D. Nordlund, V. Jayan, M. Meyyappan and J. E. Koehne, ACS Appl. Mater. Interfaces, 2014, 6, 22751–22760 CrossRef CAS PubMed.
- E. P. Hoppmann, W. W. Yu and I. M. White, Methods, 2013, 63, 219–224 CrossRef CAS PubMed.
- C. H. Lee, M. E. Hankus, L. Tian, P. M. Pellegrino and S. Singamaneni, Anal. Chem., 2011, 83, 8953–8958 CrossRef CAS PubMed.
- R. Derda, S. K. Y. Tang, A. Laromaine, B. Mosadegh, E. Hong, M. Mwangi, A. Mammoto, D. E. Ingber and G. M. Whitesides, PLoS One, 2011, 6, 5 CrossRef PubMed.
- D. A. Bruzewicz, M. Reches and G. M. Whitesides, Anal. Chem., 2008, 80, 3387–3392 CrossRef CAS PubMed.
- A. W. Martinez, S. T. Phillips, M. J. Butte and G. M. Whitesides, Angew. Chem., Int. Ed., 2007, 46, 1318–1320 CrossRef CAS PubMed.
- P. R. Stoddart and D. J. White, Anal. Bioanal. Chem., 2009, 394, 1761–1774 CrossRef CAS PubMed.
- G. F. S. Andrade, M. Fan and A. G. Brolo, Biosens. Bioelectron., 2010, 25, 2270–2275 CrossRef CAS PubMed.
- S. W. Bishnoi, C. J. Rozell, C. S. Levin, M. K. Gheith, B. R. Johnson, D. H. Johnson and N. J. Halas, Nano Lett., 2006, 6, 1687–1692 CrossRef CAS PubMed.
- F. Wang, R. G. Widejko, Z. Yang, K. T. Nguyen, H. Chen, L. P. Fernando, K. A. Christensen and J. N. Anker, Anal. Chem., 2012, 84, 8013–8019 CrossRef CAS PubMed.
- R. P. Dickson, J. R. Erb-Downward and G. B. Huffnagle, Lancet Respir. Med., 2014, 2, 238–246 CrossRef PubMed.
- A.-P. Magiorakos, C. Suetens, D. L. Monnet, C. Gagliotti and O. E. Heuer, Ant. Res. Infect. Control, 2013, 2, 6 CrossRef PubMed.
- R. Lozano, M. Naghavi, K. Foreman and S. Lim, et al. , Lancet, 2012, 380, 2095–2128 CrossRef.
- J. Chastre and J.-Y. Fagon, Am. J. Respir. Crit. Care Med., 2002, 165, 867–903 CrossRef PubMed.
- J. B. J. Scholte, H. A. van Dessel, C. F. M. Linssen, D. C. J. J. Bergmans, P. H. M. Savelkoul, P. M. H. J. Roekaerts and W. N. K. A. van Mook, J. Clin. Microbiol., 2014, 52, 3597–3604 CrossRef PubMed.
- A. R. Akram, N. Avlonitis, A. Lilienkampf, A. M. Perez-Lopez, N. McDonald, S. V. Chankeshwara, E. Scholefield, C. Haslett, M. Bradley and K. Dhaliwal, Chem. Sci., 2015, 6, 6971–6979 RSC.
- N. Krstajić, B. Mills, I. Murray, A. Marshall, D. Norberg, T. H. Craven, P. Emanuel, T. R. Choudhary, G. O. S. Williams, E. Scholefield, A. R. Akram, A. Davie, N. Hirani, A. Bruce, A. Moore, M. Bradley and K. Dhaliwal, J. Biomed. Opt., 2018, 23, 076005 Search PubMed.
- B. Mills, A. R. Akram, E. Scholefield, M. Bradley and K. Dhaliwal, J. Visualized Exp., 2017, 129, e56284 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8an01322e |
| This journal is © The Royal Society of Chemistry 2018 |