Mercury-mediated cross-resistance to tellurite in Pseudomonas spp. isolated from the Chilean Antarctic territory†
F.
Rodríguez-Rojas
a,
W.
Díaz-Vásquez
ab,
A.
Undabarrena
c,
P.
Muñoz-Díaz
a,
F.
Arenas
a and
C.
Vásquez
*a
aLaboratorio de Microbiología Molecular, Facultad de Química y Biología, Universidad de Santiago de Chile, Alameda 3363, Santiago, Chile. E-mail: claudio.vasquez@usach.cl
bFacultad de Ciencias de la Salud, Universidad San Sebastián, Lota 2465, Santiago, Chile
cLaboratorio de Microbiología Molecular y Biotecnología Ambiental, Facultad de Química, & Centro de Biotecnología Daniel Alkalay Lowitt, Universidad Técnica Federico Santa María, Av. España 1680, Valparaíso, Chile
First published on 11th November 2015
Abstract
Mercury salts and tellurite are among the most toxic compounds for microorganisms on Earth. Bacterial mercury resistance is established mainly via mercury reduction by the mer operon system. However, specific mechanisms underlying tellurite resistance are unknown to date. To identify new mechanisms for tellurite detoxification we demonstrate that mercury resistance mechanisms can trigger cross-protection against tellurite to a group of Pseudomonads isolated from the Chilean Antarctic territory. Sequencing of 16S rRNA of four isolated strains resulted in the identification of three Pseudomonads (ATH-5, ATH-41 and ATH-43) and a Psychrobacter (ATH-62) bacteria species. Phylogenetic analysis showed that ATH strains were related to other species previously isolated from cold aquatic and soil environments. Furthermore, the identified merA genes were related to merA sequences belonging to transposons commonly found in isolated bacteria from mercury contaminated sites. Pseudomonas ATH isolates exhibited increased tellurite resistance only in the presence of mercury, especially ATH-43. Determination of the growth curves, minimal inhibitory concentrations and growth inhibition zones showed different tellurite cross-resistance of the ATH strains and suggested a correlation with the presence of a mer operon. On the other hand, reactive oxygen species levels decreased while the thiol content increased when the isolates were grown in the presence of both toxicants. Finally, qPCR determinations of merA, merC and rpoS transcripts from ATH-43 showed a synergic expression pattern upon combined tellurite and mercury treatments. Altogether, the results suggest that mercury could trigger a cell response that confers mercury and tellurite resistance, and that the underlying mechanism participates in protection against oxidative damage.
Introduction
Certain heavy metals and metalloids are harmful for most microorganisms even at very small concentrations.1–3 Among them, mercury(II) and tellurite exhibit very low minimal inhibitory concentrations (MICs) for E. coli.4,5 Both toxicants generate cell damage mainly by thiol depletion, enzyme inactivation and oxidative stress.6–10 Bacterial mercury resistance lies mainly on the MerA flavoenzyme, which transforms toxic Hg+2 to the volatile and less toxic elemental mercury.11 On the other hand, specific mechanisms for detoxifying tellurite have not been reported so far. However, it has been shown that some metabolic flavoenzymes such as dihydrolipoamide dehydrogenase, thioredoxin reductase and glutathione reductase are able to reduce tellurite to the less-toxic elemental tellurium.12–14 In addition, an enhanced response to oxidative stress can confer tellurite resistance in some bacteria.15Several studies have demonstrated that sites polluted with heavy metals and/or metalloids contain bacterial species that cope with the toxicity of most metals through different molecular mechanisms, either by direct resistance or by conferring cross-resistance to a wide variety of toxicants. In this line, some naturally occurring examples are Pseudomonas putida SP1,16Raoultella planticola17 and Cupriavidus metallidurans CH344,18,19 which are considered as multimetal-resistant bacteria. In this context, it has been evidenced that common resistance mechanisms to heavy metals have been developed during bacterial evolution. Such is the case of the Czc system, which is used by some bacteria for detoxifying cadmium, zinc and cobalt.20 Also, previous studies demonstrated that the ars operon, involved in arsenate and arsenite resistance, increases up to 60-fold the MIC of tellurite for E. coli.21 On the other hand, more recently it was observed that the tellurite resistant determinant trgB, from the trgAB operon, confers copper resistance in Rhodobacter capsulatus by a still undetermined mechanism.22
In the last century and mainly because of the industrial revolution, it became evident the dramatic increase of heavy metal/metalloid-polluted sites across the Earth surface, including pristine places such as Antarctica.23,24 The Antarctic continent is constantly exposed to diverse abiotic stresses such as low temperatures, high salinity, and UV radiation, among others, which can cause oxidative stress in living organisms. Moreover, mercury levels have increased in Antarctica mostly because of the grasshopper effect and mercury depletion events (MDEs).25,26 Hence, the Antarctic continent has become an interesting place for sampling and isolating bacterial strains that are resistant to multiple stresses, including that produced by heavy metals.27 In this context, mercury-resistant bacteria have been isolated from several polar environmental samples.28 Furthermore, our laboratory identified and characterized, for the first time, highly tellurite resistant psychrotolerant bacteria from Antarctic samples.29
The understanding of the molecular mechanisms underlying mercury and tellurite resistance, and the oxidative damage associated with these psychrotolerant bacteria, could allow the design of new bioremediation strategies for polluted places at low temperatures. In this work, psychrotolerant bacteria from the Chilean Antarctic territory showing resistance to mercury and tellurite were analyzed. Some of them are resistant to tellurite only in the presence of mercury, suggesting that this metal could trigger a molecular response that confers cross-resistance to both tellurite and mercury.
Experimental
Bacterial isolation
All samples were collected during the Chilean Antarctic Expedition ECA-48, sponsored by Instituto Antártico Chileno (INACH), 2012. Bacterial strain ATH-5 was isolated from the Collins Glacier (73° 41′ S; 65° 55′ E), ATH-41 and ATH-43 from Greenwich Island (62° 30′ S; 59° 39′ W), and ATH-62 from Deception Island (62° 55′ S; 60° 37′ W), all located in the Antarctic Peninsula. For bacterial enrichment, volumes of 100 μL of each sample were suspended in 1 mL of LB media supplemented with amphotericin B 2.5 μg mL−1 to inhibit fungal proliferation, and incubated for 16 h at room temperature. Then, 60 μL of each culture were spread on LB agar plates containing 40 μM HgCl2 and 40 μM K2TeO3, and incubated at 25 °C for 48 h. Pure isolates were obtained by repeated streaking and preserved in 15% glycerol stocks at −80 °C. To determine the optimal growth temperature, the isolates were grown individually at 4, 25 and 37 °C.16S rRNA sequencing and detection of merA in the Antarctic strains
For 16S rRNA sequencing, colony PCR was carried out using GoTaq®DNA Polymerase Master Mix (Promega) and universal 16S primers 8F7 and 1492R30 at 55 °C (annealing temperature). PCR products were sequenced (Macrogen®) and subjected to nBLAST analysis against bacterial 16S rRNA sequences of the National Center for Biotechnology Information (NCBI) database to determine strain genus. The merA gene was identified in ATH strains by PCR using purified genomic DNA as the template and universal specific primers for the mer operon (Forward P1: GGCTATCCGTCCAGCGTCAA, and Reverse A5: ACCATCGTCAGGTAGGGGACCAA) as described by Felske et al.31 The PCR consisted in 30 cycles that included a denaturation step at 95 °C for 30 s, annealing at 60 °C for 30 s and extension at 72 °C for 2 min. PCR products were sequenced (Macrogen®) and then subjected to nBLAST. The sequenced products allowed determining the start codon of merA sequences to design forward primers for all analyzed merA genes. For designing reverse primers for merA encoding open reading frames, sequences of the first hits of the nBLAST analysis were used. ATH-5, ATH-41 and ATH-43 merA cloning included start codons and excluded stop codons in each case. Primer sequences for ATH-43 were: forward 5′-ATGACCGAACTGAAAATCATTG-3′ and reverse 5′-CCCCCCTGCGCAGCA-3′ (designed from Pseudomonas putida SP1, accession No ADJ53344); for ATH-41: forward 5′-ATGACTGAATTGCAAATCGTTGG-3′ and reverse 5′-GCCGGCGCAGCAGGAAAG-3′ (designed from Xanthomonas campestris, accession No Y17691); for ATH-5: forward 5′-ATGACTACTCTGCAAATCAGC-3′ and reverse the same as for ATH-41. PCR was the same as for P1-A5 primers, except the annealing temperature was 55 °C. All PCR products were sequenced (Macrogen®) to obtain the complete open reading frames of each merA gene.Phylogenetic trees
Almost complete sequences of 16S rRNA genes and merA ORFs (translated sequence) were used to construct phylogenetic trees. To obtain almost complete sequences of each 16S rRNA gene, PCR products previously amplified were additionally sequenced with universal primers 518F and 800R (Macrogen®). Sequence edition, alignment and contigs analysis were carried out using the Vector NTI v10 software package (Invitrogen). Contigs were evaluated by nBLAST against 16S ribosomal RNA sequences of the Bacteria and Archaea databases to establish the closest type strain match, and Nucleotide Collection Databases (NCBI), to search for other phylogenetically related organisms. Construction of phylogenetic trees was based on the V1-V9 region of 16S rRNA gene sequences using the Neighbor Joining Algorithm32 and for the merA tree Maximum-likelihood algorithm, with bootstrap values of 1000 replications33 and MEGA software version 6.0.34 The 16S rRNA and merA sequences of ATH strains were deposited in GenBank under the following accession numbers: KR15900 (16S rRNA_ATH-5), KR15899 (16S rRNA_ATH-41), KR15898 (16S rRNA_ATH-43), KR18929 (16S rRNA_ATH-62), KR818931 (merA_ATH-5), KR827601 (merA_ATH-41), and KR818930 (merA_ATH-43).Antibiotic susceptibility and biochemical characterization
For antibiotic susceptibility analysis, cells were grown to the exponential phase and spread onto Mueller-Hinton agar plates. Antibiotic displays for Gram (−) bacteria (Valtek®) were used and growth inhibition zone diameters were measured and compared with the values from the Clinical and Laboratory Standards Institute (CLSI) to determine susceptibility or resistance to each tested antibiotic.For biochemical characterization, analytical profile indexes (API®) were used according to manufacturer’s instructions.
Growth curves, growth inhibition zones and MIC determinations
Minimal inhibitory concentrations (MICs) of K2TeO3 and HgCl2 were determined in LB media supplemented with increasing concentrations of the respective toxicants. Cultures were started with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 dilutions of saturated cultures. To determine the MIC of K2TeO3 in the presence of HgCl2, all cultures contained HgCl2 at a concentration equivalent to 1/4 of the MIC (determined previously for each strain) and increasing concentrations of K2TeO3. For growth curve construction, cells were inoculated in a LB broth using 1
100 dilutions of saturated cultures. To determine the MIC of K2TeO3 in the presence of HgCl2, all cultures contained HgCl2 at a concentration equivalent to 1/4 of the MIC (determined previously for each strain) and increasing concentrations of K2TeO3. For growth curve construction, cells were inoculated in a LB broth using 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 dilutions of overnight cultures and subjected to the following conditions: 1 MIC of K2TeO3, 1/4 MIC of HgCl2, and both toxicants combined at the same concentrations, at 25 °C for 15 h. Optical density at 600 nm was monitored using a microplate multireader Tecan Infinite® 200 PRO.
100 dilutions of overnight cultures and subjected to the following conditions: 1 MIC of K2TeO3, 1/4 MIC of HgCl2, and both toxicants combined at the same concentrations, at 25 °C for 15 h. Optical density at 600 nm was monitored using a microplate multireader Tecan Infinite® 200 PRO.
To determine growth inhibition zones, ATH cells grown to OD600 ∼ 0.3 were seeded onto LB agar plates supplemented or not with HgCl2 (1/4 MIC for the respective strain) and sterile circular paper discs embedded with 10 μL of K2TeO3 400 mM were placed at the centers of the plates. Plates were incubated at 25 °C for 48 h.
Reactive oxygen species and thiol quantification
Reactive oxygen species (ROS) were determined using the oxidant-sensitive probe 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA, Sigma-Aldrich) essentially as described by Pérez et al.8 Briefly, cultures were grown to OD600 ∼ 0.3 and treated either with 1/4 MIC of HgCl2, 1/2 MIC of K2TeO3 or simultaneously with both toxicants at the same concentration at 25 °C for 30, 60 and 90 min. Measurements were carried out after each treatment by exciting the probe at 490 nm and measuring the fluorescence intensity at 519 nm using a microplate multireader Tecan Infinite® 200 PRO. Fluorescence intensity values were normalized by OD600.The thiol content was quantified by Ellman's method using 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB, Sigma-Aldrich), as previously described by Turner et al.7 Experimental conditions were the same as for ROS determination. TNB−2 absorbance was measured spectrophotometrically at 412 nm (an extinction coefficient of 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 M−1 cm−1). A standard curve of reduced glutathione was constructed for assay calibration. Data values were normalized by protein concentration as determined by the Bradford method.35
600 M−1 cm−1). A standard curve of reduced glutathione was constructed for assay calibration. Data values were normalized by protein concentration as determined by the Bradford method.35
All experiments were performed in triplicate and Student's t-test was applied for statistical analysis when required.
RNA purification and real time PCR
For RNA isolation, ATH-43 was grown to OD600 ∼ 0.3 and treated with mercury, tellurite or both toxicants for 10 min at the same concentrations as for ROS and thiol quantification. Briefly, cells were harvested and RNA was isolated using the Favorprep tissue total RNA purification mini Kit (Favorgene). RNA quantification was carried out by using a Quant-it Ribogreen Kit (Invitrogen), following the manufacturer’s instructions. Quantitative PCR was conducted using a Light Cycler 1.5 thermocycler with the RNA MasterSYBR Green Kit (Roche Applied Science). Real time PCR program involved reverse transcription at 40 °C for 10 min, followed by 45 cycles of denaturation at 95 °C for 30 s; annealing at 55 °C for 30 s, and extension 72 °C, 30 s. Specific primers for merA amplification (forward 5′-GTTCCACCGATTCCAGGACTG-3′; reverse 5′-CAGTACGAACTCGCCGTTGG-3′), merC (forward 5′-TTTCCCGCCCTTGCTAGTCT-3′; reverse 5′-ATGAGCGGGCGACACGAGAT-3′) and rpoS (forward 5′-TAAAGAAGTGCCGGAGTTTGACA-3′; reverse 5′-TTGCTTTCGATCATGCGCTT-3′) from ATH-43 were used. The D-glyceraldehyde-3-phosphate dehydrogenase gene, gapA (forward 5′-CCAGTGTCTTGGTGAGTGGATTG-3′; reverse 5′-CCATTGCCTTCCTGCTTGA-3′), and the 16S rRNA gene (forward 5′-GATTCCAACGGCTAGTTCACATC-3′; reverse 5′-GGTGCAAGCGTTAATCGGAA-3′) were used as reference genes for normalization. All transcript levels were calculated using the ΔΔCt method.36Results
Identification and characterization of mercury and tellurite-resistant bacteria from Antarctica
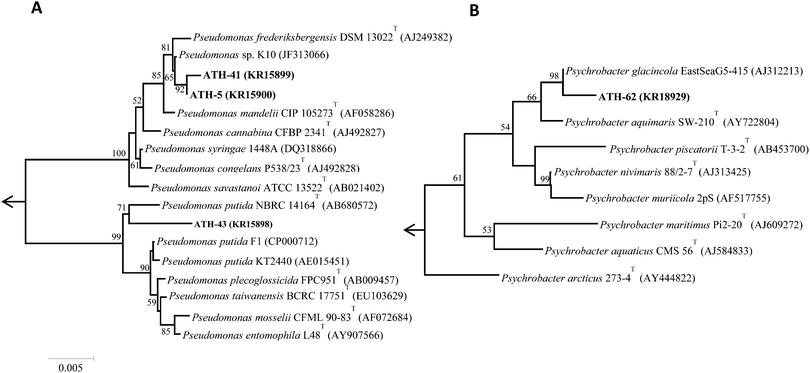
Of nineteen Antarctic samples analyzed, four bacterial isolates named ATH strains (Table 1) exhibiting simultaneous resistance to mercury and tellurite were obtained. While ATH-5 was isolated from a sample collected at Glacier Collins (King George Island), ATH-41 and ATH-43 were from soil samples at the Chilean Prat military base (Greenwich Island). In turn, ATH-62 was obtained from Péndulo Cove, Deception Island, a location that was devastated by a volcanic eruption in 1964. All strains were Gram (−) and grew optimally between 20 and 25 °C, except ATH-43, which grew between 20 and 37 °C. The 16S rRNA sequence data revealed that ATH-5, ATH-41 and ATH-43 belonged to the Pseudomonas genus, whereas ATH-62 was related to the Psycrobacter genus. Further comparisons with 16S sequences from NCBI showed that the closest type strains were Pseudomonas mandelii CIP 105273T for ATH-5 and ATH-41, Pseudomonas putida NBRC 14164T for ATH-43, and the halophilic bacteria Psychrobacter aquimaris SW-210T![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 37 for ATH-62. Phylogenetic trees based on the almost complete 16S rRNA gene sequences showed that ATH-5 and ATH-41 were also related to Pseudomonas frederiksbergensis DSM 13022T
37 for ATH-62. Phylogenetic trees based on the almost complete 16S rRNA gene sequences showed that ATH-5 and ATH-41 were also related to Pseudomonas frederiksbergensis DSM 13022T![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 38 and to Pseudomonas sp. K10, while ATH-43 phylogeny was related to various Pseudomonas putida species (Fig. 1A) and ATH-62 to Psychrobacter glacincola EastSeaG5-415 (Fig. 1B).
38 and to Pseudomonas sp. K10, while ATH-43 phylogeny was related to various Pseudomonas putida species (Fig. 1A) and ATH-62 to Psychrobacter glacincola EastSeaG5-415 (Fig. 1B).
| Strain | Isolation site | Optimal growth temperature (°C) | Closest type strain (% identity) | Most closely related merA (species, % identity) |
|---|---|---|---|---|
| ATH-5 | Collins Glacier | 20–25 | Pseudomonas mandelii CIP 105273T (99.65%) | Tn5044 (Xanthomonas campestris, 98.03%) |
| ATH-41 | Prat base, Greenwich Island | 20–25 | Pseudomonas mandelii CIP 105273T (99.43%) | Tn5044 (Xanthomonas campestris, 86.96%) |
| ATH-43 | Prat base, Greenwich Island | 20–37 | Pseudomonas putida NBRC 14164T (99.77%) | Tn5041 (Pseudomonas putida DLL-E4, 99.76%) |
| ATH-62 | Péndulo cove, Deception Island | 20–25 | Psychrobacter aquimaris SW-210T (99.50%) | ND |
Antibiotic susceptibility tests for Gram (−) bacteria determined that while ATH-5, ATH-41 and ATH-43 were resistant to almost all antibiotics (except for amikacin, gentamicin and ciprofloxacin), ATH-62 was sensitive to all of them (Table S1, ESI†). The biochemical characteristics of ATH-5, ATH-41 and ATH-43 were very similar, unlike ATH-62, which used all the tested carbon sources (Table S2, ESI†).
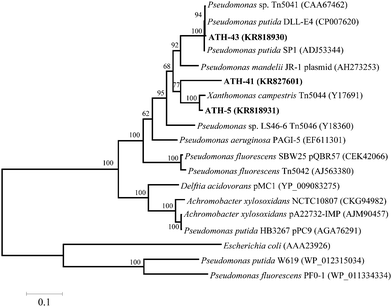
To identify mer operons, universal primers that amplify a specific operon sequence including part of the 3′ end of merP and part of the 5′ end of merA gene were used. The mer operon was detected in ATH-5, ATH-41 and ATH-43 utilizing primers P1 and A5 (see the Experimental section) (Table 1). Even though several sets of primers were utilized as described in ref. 31 the mer operon was not found in ATH-62 (not shown). The P1-A5 sequences determined for ATH-5, ATH-41 and ATH-43 were used to design specific primers to amplify complete merA sequences. BLAST analysis showed that merA from ATH-5 and ATH-41 was related to that of transposon Tn5044 found in Xanthomonas campestris (Table 1). On the other hand, merA from ATH-43 exhibited high identity with merA from Pseudomonas putida DLL-E4 transposon Tn5044 (Table 1). A phylogenetic tree constructed with merA sequences showed that merA genes from ATH isolates were phylogenetically related mainly to sequences found in transposable elements (Tn5041, Tn5044 and Tn5046) from different Pseudomonas species isolated from several environments (Fig. 2).
Mercury enhances tellurite resistance in Pseudomonas spp. isolated from Antarctica
Preliminary experiments carried out in our laboratory showed that the ATH-43 isolate grew in the presence of tellurite up to 40 μM only if the cultures were supplemented with sublethal mercury concentrations (∼40 μM). This observation prompted us to assess if mercury caused the same phenotype in other isolates. While ATH-43 showed the highest and lowest MICs of mercury (HgMIC) and tellurite (TeMIC), respectively, ATH-5 and ATH-41 exhibited almost similar sensitivity to both toxicants. On the other hand, and although exhibiting a rather similar HgMIC regarding the other strains, ATH-62 showed the highest resistance to tellurite (Table 2). To assess if mercury can induce resistance to tellurite in ATH strains, the TeMIC was determined in the presence of mercury. The results showed that it increased 8.3-fold in ATH-43 when compared to the tellurite treatment alone. TeMIC increased 1.5-fold in ATH-5, while it remained unaltered in ATH-41. In contrast, it decreased 2.3-fold for ATH-62 in the presence of mercury (Table 2).| Strain | Hg+2 (μM) | TeO3−2 (μM) | TeO3−2 (μM) w/Hg+2 |
|---|---|---|---|
| ATH-5 | 100 | 80 | 120 |
| ATH-41 | 100 | 120 | 120 |
| ATH-43 | 200 | 24 | 200 |
| ATH-62 | 80 | 700 | 300 |
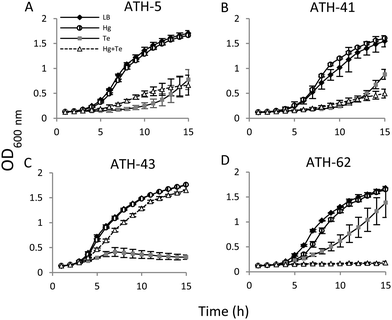
Then the growth curves were constructed under four different conditions: no toxicant amended (controls), mercury, tellurite, and combined mercury/tellurite treatments (Fig. 3). The growth curves of ATH-5 and ATH-41 were similar for all treatments (Fig. 3A and B). They grew as fast as the control in the presence of mercury, but tellurite and mercury/tellurite treatments delayed drastically the bacterial growth, with a slight increase of ATH-5 growth in the presence of mercury/tellurite (Fig. 3A). On the other hand, mercury alone did not alter ATH-43 growth, and as expected, tellurite amendment inhibited growth rapidly (Fig. 3C). Interestingly, the combined mercury/tellurite treatment almost did not affect ATH-43 growth, which was similar to that observed in control and mercury-amended cultures. In contrast to the results observed with the Pseudomonads isolates, ATH-62 showed delayed growth with tellurite and a total growth inhibition in the presence of both toxicants (Fig. 3D).
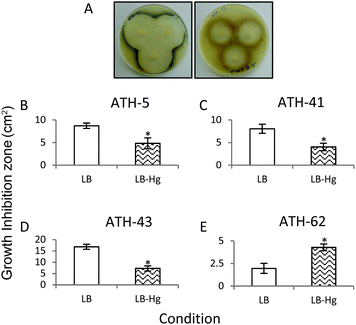
In a third approach to characterize the potentiation of tellurite resistance phenotype by mercury, tellurite growth inhibition zones were determined in the presence and absence of mercury (Fig. 4). As shown before, ATH-43 exhibited the most evident potentiation effect on tellurite resistance. In fact, the growth inhibition zone decreased 2.5 fold in the presence of mercury (Fig. 4A and D). In turn, ATH-5 and ATH-41 exhibited a slight but significant growth decrease in mercury amended medium (Fig. 4B and C). The opposite effect was observed with ATH-62, i.e., the growth inhibition zone increased 2.19 fold in the presence of mercury as compared to controls (Fig. 4E).
Altogether, these results suggest that mercury potentiates tellurite resistance in the Antarctic Pseudomonads isolates, especially ATH-43, but not in Psychrobacter sp. ATH-62.
ROS and the thiol content in Antarctic isolates exposed to mercury and tellurite
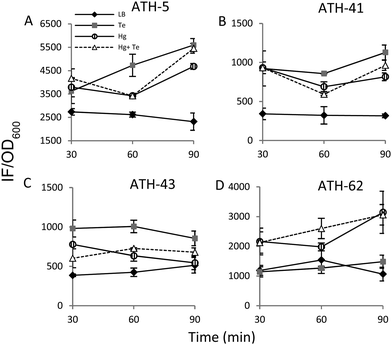
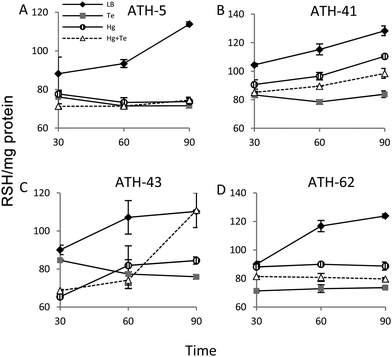
It has been previously demonstrated that both, mercury and tellurite, induce ROS production3,10 and that also show strong affinity for sulfhydryl containing molecules.7,39 In this context, ROS levels were quantified in terms of fluorescence intensity under the same experimental conditions described above for ATH strains (see the Experimental section for details) (Fig. 5). ATH-5 and ATH-41 produced low and rather constant ROS in the absence of toxicant amendment, while toxicant exposure raised ROS levels significantly (Fig. 5A and B). Mercury and mercury/tellurite treatments showed a trend to decrease ROS levels after 30 min of toxicant exposure, increasing afterwards. This was observed in tellurite-exposed ATH-41, but not in ATH-5, where ROS levels exhibited a constant increase (Fig. 5A and B). Interestingly, ATH-43 showed higher ROS levels with tellurite in comparison to mercury and mercury/tellurite exposure (Fig. 5C). Nevertheless, it showed a marked tendency to reduce ROS levels in all metal treatments, almost reaching the ROS content observed under the control conditions with mercury and mercury/tellurite treatments (Fig. 5C). Conversely to Pseudomonas strain results, ATH-62 behaved differently regarding ROS production (Fig. 5D). Tellurite did not affect significantly ROS levels as compared to control treatments. However, mercury and mercury/tellurite exposure resulted in augmented ROS levels (Fig. 5D). These results show that tellurite enhances ROS production in the Pseudomonas isolates, but when combined with mercury a reduction of ROS levels is observed, especially in ATH-43. In the case of ATH-62, a distinct response was evidenced, where mercury did not favor a ROS level decrease in the combined mercury/tellurite treatment.Given the ability of tellurite and mercury to interact with reduced thiols, the RSH content was analyzed (Fig. 6). The RSH content decreased drastically in ATH-5 during all treatments and did not recover the initial levels after 90 min (Fig. 6A). Although there was also a decrease of RSH levels in ATH-41, a general trend to recover them over time was observed (Fig. 6B). ATH-43 also showed an initial reduction of thiol content that in the case of the combined treatment recovered to that exhibited by controls up to 90 min exposure (Fig. 6C). ATH-62 showed a similar response to that observed for ATH-5 where the thiol content decreased after the treatments and remained at low concentrations.
Expression analysis in response to mercury and tellurite in ATH-43
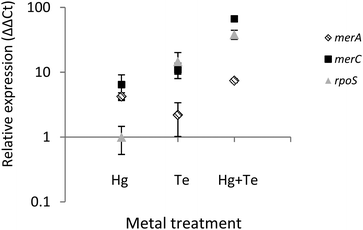
To shed light on the molecular mechanism(s) underlying the mercury-triggered potentiation of tellurite resistance the expression of three selected genes of ATH-43 was analyzed (Fig. 7). The expression patterns of two genes belonging to the mer operon, merA and merC, along with the general stress response gene rpoS was evaluated after 10 min of toxicant exposure. In general, all three genes were upregulated. The sole exception was rpoS where its expression remained unaltered when cells were grown in the presence of mercury. Taken together, qPCR experiments suggest that the combination of mercury and tellurite could generate a synergic effect over the expression of mer genes, and probably other genes involved in the general stress response.Discussion
This work shows that mercury resistance mechanisms from a group of psychrotolerant Antarctic bacteria are involved in tellurite resistance. Four isolates (ATH-5, ATH-41, ATH-43 and ATH-62) were characterized as mercury/tellurite resistant bacteria (Table 1). ATH-5, ATH-41 and ATH-43 were related to Pseudomonas species, while ATH-62 was to the Psychrobacter genus. Phylogenetic trees showed that ATH-5 and ATH-41 were related to psychrotrophic Pseudomonas mandelii CIP 105273T isolated from mineral waters40 and to the arctic isolate Pseudomonas sp. K10 (accession number JF313066) (Fig. 1A). On the other hand, ATH-43 phylogeny was related to P. putida isolated from different ecosystems including water and soil polluted sediments41 (Fig. 1A), while ATH-62 was associated with Psycrobacter species isolated from cold environments like permafrost,42 marine ice43 and Antarctic water,44 among others (Fig. 1B).The mer operon was detected only in the Pseudomonas strains. Phylogenetic relationships among merA genes from ATH-5, ATH-41 and ATH-43 showed that they were related to merA genes found in transposable elements ubiquitous in nature.45 Even though merA from ATH-5 and ATH-41 were related to the merA sequence of Tn5044 found in X. campestris, merA from ATH-41 formed a distinct branch within the phylogenetic tree and could represent a new type of MerA due its low sequence identity with other sequenced merA genes (Fig. 2). Moreover, the mer operon from ATH-5, and not that from ATH-41, contained the accessory gene merC, suggesting that although they are related strains, they do not share the same gene structure of mer operon. On the other hand, merA from ATH-43 was phylogenetically related to merA genes found in other P. putida strains that harbor transposon Tn5041, including the multi-metal resistant strain P. putida SP1.16 Tn5041 and Tn5044 are mercury resistance transposons that are considered as dissemination vehicles of mercury resistance in bacterial populations.46–48 Interestingly, the mer operon from Tn5044 is expressed exclusively at temperatures below 30 °C.46,49 No mer sequences were found in ATH-62, suggesting that this bacterium harbors other molecular mechanism(s) to cope with mercury toxicity. In this line, besides the mer system, other mercury detoxification mechanisms have been described such as reduction of mercury uptake and mercury ion biosorption by cell surface proteins.45,50 Moreover, the mer operon-mediated mercury resistance has been associated with antibiotic determinants since genes encoding them are commonly found in mobile genetic elements such as plasmids and transposons.51 Thus, we hypothesize that the lack of antibiotic resistance observed in ATH-62 may be correlated with the absence of the mer operon. Further studies should be performed to understand the molecular nature of mercury resistance and the intrinsic high tellurite resistance displayed by ATH-62.
Through several experimental approaches such as growth curves, MICs, and growth inhibition zone determinations, it was observed that when ATH-5, ATH-41 and ATH-43 were grown in the presence of mercury, their tellurite resistance was increased (Fig. 3 and 4 and Table 2). While ATH-5 and ATH-41 showed a weak cross-resistance phenotype, ATH-43 exhibited a strong potentiation of tellurite resistance in the presence of mercury. Conversely, ATH-62 showed a totally opposite response, where the presence of mercury did not favor this phenotype, suggesting that the cross-resistance phenotype could be, at least in part, explained by the presence of mer genes.
To date, defining tellurite resistance mechanisms has been limited to some studies that establish involvement of a generalized oxidative stress response,7–9,52 cysteine metabolism,53–55 volatilization of alkylated tellurium compounds,56 and non-enzymatic7,52 and enzymatic reduction. Some of the enzymes exhibiting tellurite-reducing activity belong to the flavoprotein family including glutathione reductase,13 thioredoxin reductase,12 dihydrolipoyl dehydrogenase,14 and alkylhydroperoxide reductase. In this context, preliminary results from our laboratory have shown that MerA encoded in the environmental plasmid pTP657 exhibits tellurite reductase activity, suggesting that it could participate in tellurite reduction (unpublished data). In support of this, recently, Marteyn et al. observed that purified mercuric reductase from Synechocystis PCC6803 can reduce uranyl cations thus breaking the paradigmatic specificity of MerA for mercuric ions.58
To study the molecular response associated with the observed potentiation phenomenon, the reactive oxygen species content was analyzed. It was found that both tellurite and mercury elicited ROS production in all Pseudomonas strains, but not in ATH-62 (Fig. 5). It is well documented that tellurite induces superoxide formation during its reduction to elemental tellurium in several bacterial species.8,9,29 Since ATH-62 displayed the highest tellurite MIC, it may harbor specific tellurite resistance determinants that could be involved in controlling ROS production or detoxification in toxicant-exposed cells. ATH-43 was the only strain that reduced ROS levels up to those observed under control conditions, suggesting that it may possess stronger molecular mechanism(s) that can cope with oxidative stress which would be induced solely in the presence of mercury. Proteomic analysis carried out in E. coli and Corynebacterium glutamicum has shown that mercury triggers molecular responses against oxidative stress such as over-production of superoxide dismutase, cysteine synthase, thioredoxin reductase and glutathione reductase, among others.10,59 Hence, ATH-43 may contain specialized molecular determinants involved in the oxidative stress response which would be strongly elicited by mercury. These mechanisms could be complementary to the mer detoxification system, and as a consequence, also useful for tellurite detoxification.
Both mercury and tellurite decreased the thiol content in all ATH strains (Fig. 6). Hg+2 displays the highest affinity for sulfhydryl groups in nature among heavy metals, and thus its toxicity occurs principally due to oligodynamic effects that causes enzymatic inactivation, thiol depletion and oxidative stress.11,60 On the other hand, tellurite is recognized as a strong oxidizing molecule that also produces rapid intracellular thiol depletion in bacteria, especially of glutathione.7,39,52 Interestingly, only ATH-43 showed thiol levels similar to control conditions when treated with mercury/tellurite (Fig. 6A). This synergic effect may be a consequence of a tellurite-induced activation of still undefined mechanisms promoting thiol restoration. This phenomenon was not observed when ATH-43 was exposed to other thiol-depleting conditions such as cadmium, copper or chromium. Mercury did not cause a rapid thiol depletion in ATH-62 (Fig. 6D), which could suggest that the mercury resistance phenotype in this strain might be related to a reduction of mercury uptake, as occurring in lactic acid bacteria, Weissella viridescens.50
Finally, tellurite induced the expression of merA and merC (Fig. 7). It has been previously shown that heavy metals such as cadmium and zinc can activate the expression of the mer operon in Nitrosomonas europaea by a mechanism that does not involve enzymatic reduction or detoxification of those heavy metals.61,62 In this line, it has been suggested that mer genes could be considered as putative “sentinel genes”, since their expression is highly upregulated in the presence of defined heavy metals and the produced Mer proteins could act as heavy metal chelators.61,62 Although tellurite induces mer genes expression, this appears to be not enough to confer tellurite resistance. Nevertheless, a synergic effect was again observed in terms of merA and merC expression after mercury/tellurite exposure. Up-regulation of merA could be associated with an increased demand of mercury (and probably tellurite) reduction, whereas the heavy metal transporter, merC, could be facilitating the influx of both toxicants thus favoring rapid detoxification, as occurring with cadmium in the transgenic, merC-expressing S. cerevisiae.63 On the other hand, up-regulation of rpoS could be a consequence of a multiple stress condition including oxidative stress as evidenced in E. coli and P. aeruginosa,64,65 during the so-named cross-protection phenotype.66
To date, cross-resistance responses to several toxics have been documented for a number of organisms. Examples include the cadmium-mediated adaptive response conferring cross-protection against high concentrations of hydrogen peroxide and zinc in X. campestris,67,68 as well as hydrogen peroxide confers cross-resistance to cadmium, mercury and arsenite in Chinese Hamster Ovary cells.69 On the other hand, evolutionary engineering microorganisms have shown several cross-resistance phenotypes. Some examples include the cobalt hyper-resistant S. cerevisiae, also displaying high resistance to nickel, zinc and manganese70 and the boron-resistant Bacillus boroniphilus, exhibiting in addition resistance to copper, iron and salt stress.71 Finally, it was communicated that low tellurite concentrations can induce an adaptive response that results in increased tellurite resistance in Proteus mirabilis.72
Further experiments are being carried out in our laboratory to better understand the molecular response that underlies the cross-resistance to tellurite induced by mercury in the ATH-43 strain.
Conclusions
This work shows, for the first time, that the molecular mechanisms underlying bacterial mercury resistance can participate in cross-resistance to tellurite in a group of Pseudomonas isolated from Antarctica. As enzymatic reduction is the most familiar tellurite detoxification mechanism and preliminary results from our laboratory suggest that the flavoprotein mercuric reductase (MerA) displays tellurite reduction activity, this enzyme could be responsible for part of the observed tellurite resistance in mercury-resistant bacteria.Acknowledgements
This work was funded by the doctoral fellowship “Gastos operacionales” #21120114 and Regular Fondecyt #1130362 granted by CONICYT, Chile, to FRR and CCV, respectively.References
- J. J. Harrison, H. Ceri, C. A. Stremick and R. J. Turner, Environ. Microbiol., 2004, 6, 1220–1227 CrossRef CAS PubMed.
- S. Silver and T. Phung le, J. Ind. Microbiol. Biotechnol., 2005, 32, 587–605 CrossRef CAS PubMed.
- J. A. Lemire, J. J. Harrison and R. J. Turner, Nat. Rev. Microbiol., 2013, 11, 371–384 CrossRef CAS PubMed.
- M. Mergeay, D. Nies, H. G. Schlegel, J. Gerits, P. Charles and F. Van Gijsegem, J. Bacteriol., 1985, 162, 328–334 CAS.
- D. E. Taylor, Trends Microbiol., 1999, 7, 111–115 CrossRef CAS PubMed.
- V. B. Mathema, B. C. Thakuri and M. Sillanpaa, Arch. Microbiol., 2011, 193, 837–844 CrossRef CAS PubMed.
- R. J. Turner, J. H. Weiner and D. E. Taylor, Microbiology, 1999, 145, 2549–2557 CrossRef CAS PubMed.
- J. M. Pérez, I. L. Calderón, F. A. Arenas, D. E. Fuentes, G. A. Pradenas, E. L. Fuentes, J. M. Sandoval, M. E. Castro, A. O. Elías and C. C. Vásquez, PLoS One, 2007, 2, e211 Search PubMed.
- V. Tremaroli, S. Fedi and D. Zannoni, Arch. Microbiol., 2007, 187, 127–135 CrossRef CAS PubMed.
- Y. Gao, X. Peng, J. Zhang, J. Zhao, Y. Li, Y. Li, B. Li, Y. Hu and Z. Chai, Metallomics, 2013, 5, 913–919 RSC.
- T. Barkay, S. M. Miller and A. O. Summers, FEMS Microbiol. Rev., 2003, 27, 355–384 CrossRef CAS PubMed.
- M. P. Rigobello, A. Folda, A. Citta, G. Scutari, V. Gandin, A. P. Fernandes, A. K. Rundlof, C. Marzano, M. Bjornstedt and A. Bindoli, Free Radical Biol. Med., 2011, 50, 1620–1629 CrossRef CAS PubMed.
- B. Pugin, F. A. Cornejo, P. Muñoz-Díaz, C. M. Muñoz-Villagrán, J. I. Vargas-Pérez, F. A. Arenas and C. C. Vásquez, Appl. Environ. Microbiol., 2014, 80, 7061–7070 CrossRef PubMed.
- M. E. Castro, R. Molina, W. Díaz, S. E. Pichuantes and C. C. Vásquez, Biochem. Biophys. Res. Commun., 2008, 375, 91–94 CrossRef CAS PubMed.
- V. Tremaroli, M. L. Workentine, A. M. Weljie, H. J. Vogel, H. Ceri, C. Viti, E. Tatti, P. Zhang, A. P. Hynes, R. J. Turner and D. Zannoni, Appl. Environ. Microbiol., 2009, 75, 719–728 CrossRef CAS PubMed.
- W. Zhang, L. Chen and D. Liu, Appl. Microbiol. Biotechnol., 2012, 93, 1305–1314 CrossRef CAS PubMed.
- S. Koc, B. Kabatas and B. Icgen, Bull. Environ. Contam. Toxicol., 2013, 91, 177–183 CrossRef CAS PubMed.
- D. H. Nies, Extremophiles, 2000, 4, 77–82 CrossRef CAS PubMed.
- T. von Rozycki and D. H. Nies, Antonie van Leeuwenhoek, 2009, 96, 115–139 CrossRef CAS PubMed.
- D. Nies, M. Mergeay, B. Friedrich and H. G. Schlegel, J. Bacteriol., 1987, 169, 4865–4868 CAS.
- R. J. Turner, Y. Hou, J. H. Weiner and D. E. Taylor, J. Bacteriol., 1992, 174, 3092–3094 CAS.
- C. Rademacher, M. C. Hoffmann, J. W. Lackmann, R. Moser, Y. Pfander, S. Leimkuhler, F. Narberhaus and B. Masepohl, Biometals, 2012, 25, 995–1008 CrossRef CAS PubMed.
- A. Camacho, C. Rochera, R. Hennebelle, C. Ferrari and A. Quesada, Sci. Total Environ., 2015, 509–510, 145–153 CrossRef CAS PubMed.
- C. Vodopivez, A. Curtosi, E. Villaamil, P. Smichowski, E. Pelletier and W. P. Mac Cormack, Sci. Total Environ., 2015, 502, 375–384 CrossRef CAS PubMed.
- R. Ebinghaus, H. H. Kock, C. Temme, J. W. Einax, A. G. Lowe, A. Richter, J. P. Burrows and W. H. Schroeder, Environ. Sci. Technol., 2002, 36, 1238–1244 CrossRef CAS PubMed.
- H. von Waldow, M. MacLeod, K. Jones, M. Scheringer and K. Hungerbuhler, Environ. Sci. Technol., 2010, 44, 6183–6188 CrossRef CAS PubMed.
- S. Mangano, L. Michaud, C. Caruso and A. Lo Giudice, Polar Biol., 2014, 37, 227–235 CrossRef.
- M. J. De Souza, P. A. Loka Bharathi, S. Nair and D. Chandramohan, BioMetals, 2007, 20, 821–828 CrossRef CAS PubMed.
- F. A. Arenas, B. Pugin, N. A. Henríquez, M. A. Arenas-Salinas, W. A. Díaz-Vásquez, M. F. Pozo, C. M. Muñoz, T. G. Chasteen, J. M. Pérez-Donoso and C. C. Vásquez, Polar Sci., 2014, 8, 40–52 CrossRef.
- W. G. Weisburg, S. M. Barns, D. A. Pelletier and D. J. Lane, J. Bacteriol., 1991, 173, 697–703 CAS.
- A. D. Felske, W. Fehr, B. V. Pauling, H. von Canstein and I. Wagner-Dobler, BMC Microbiol., 2003, 3, 22 CrossRef PubMed.
- N. Saitou and M. Nei, Mol. Biol. Evol., 1987, 4, 406–425 CAS.
- J. Felsenstein, Am. Nat., 1985, 125, 1–15 Search PubMed.
- K. Tamura, G. Stecher, D. Peterson, A. Filipski and S. Kumar, Mol. Biol. Evol., 2013, 30, 2725–2729 CrossRef CAS PubMed.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS PubMed.
- K. J. Livak and T. D. Schmittgen, Methods, 2001, 25, 402–408 CrossRef CAS PubMed.
- J. H. Yoon, C. H. Lee, S. H. Yeo and T. K. Oh, Int. J. Syst. Evol. Microbiol., 2005, 55, 1007–1013 CrossRef CAS PubMed.
- S. M. Andersen, K. Johnsen, J. Sorensen, P. Nielsen and C. S. Jacobsen, Int. J. Syst. Evol. Microbiol., 2000, 50, 1957–1964 CrossRef CAS PubMed.
- R. J. Turner, J. H. Weiner and D. E. Taylor, Microbiology, 1995, 141, 3133–3140 CrossRef CAS PubMed.
- S. Verhille, N. Baida, F. Dabboussi, D. Izard and H. Leclerc, Syst. Appl. Microbiol., 1999, 22, 45–58 CrossRef CAS PubMed.
- V. Subramanian, T. N. Liu, W. K. Yeh, C. M. Serdar, L. P. Wackett and D. T. Gibson, J. Biol. Chem., 1985, 260, 2355–2363 CAS.
- T. A. Vishnivetskaya, M. A. Petrova, J. Urbance, M. Ponder, C. L. Moyer, D. A. Gilichinsky and J. M. Tiedje, Astrobiology, 2006, 6, 400–414 CrossRef CAS PubMed.
- L. A. Romanenko, A. M. Lysenko, M. Rohde, V. V. Mikhailov and E. Stackebrandt, Int. J. Syst. Evol. Microbiol., 2004, 54, 1741–1745 CrossRef CAS PubMed.
- A. Heuchert, F. O. Glockner, R. Amann and U. Fischer, Syst. Appl. Microbiol., 2004, 27, 399–406 CrossRef CAS PubMed.
- A. M. Osborn, K. D. Bruce, P. Strike and D. A. Ritchie, FEMS Microbiol. Rev., 1997, 19, 239–262 CrossRef CAS PubMed.
- G. Kholodii and E. Bogdanova, Genetica, 2002, 115, 233–241 CrossRef CAS PubMed.
- G. Y. Kholodii, O. V. Yurieva, Z. Gorlenko, S. Z. Mindlin, I. A. Bass, O. L. Lomovskaya, A. V. Kopteva and V. G. Nikiforov, Microbiology, 1997, 143, 2549–2556 CrossRef CAS PubMed.
- G. Kholodii, O. Yurieva, S. Mindlin, Z. Gorlenko, V. Rybochkin and V. Nikiforov, Res. Microbiol., 2000, 151, 291–302 CrossRef CAS PubMed.
- G. Kholodii, S. Z. Mindlin, M. Gorlenko Zh, I. A. Bass, E. S. Kaliaeva and V. G. Nikiforov, Genetika, 2000, 36, 459–469 Search PubMed.
- H. Kinoshita, Y. Sohma, F. Ohtake, M. Ishida, Y. Kawai, H. Kitazawa, T. Saito and K. Kimura, Res. Microbiol., 2013, 164, 701–709 CrossRef CAS PubMed.
- C. Cervantes-Vega, J. Chavez, N. A. Córdova, P. de la Mora and J. Amador Velasco, Microbios, 1986, 48, 159–163 CAS.
- R. J. Turner, Y. Aharonowitz, J. H. Weiner and D. E. Taylor, Can. J. Microbiol., 2001, 47, 33–40 CrossRef CAS PubMed.
- D. E. Fuentes, E. L. Fuentes, M. E. Castro, J. M. Pérez, M. A. Araya, T. G. Chasteen, S. E. Pichuantes and C. C. Vásquez, J. Bacteriol., 2007, 189, 8953–8960 CrossRef CAS PubMed.
- C. C. Vásquez, C. P. Saavedra, C. A. Loyola, M. A. Araya and S. Pichuantes, Curr. Microbiol., 2001, 43, 418–423 CrossRef PubMed.
- C. P. Saavedra, M. V. Encinas, M. A. Araya, J. M. Pérez, J. C. Tantaleán, D. E. Fuentes, I. L. Calderón, S. E. Pichuantes and C. C. Vásquez, Biochimie, 2004, 86, 481–485 CrossRef CAS PubMed.
- T. G. Chasteen and R. Bentley, Chem. Rev., 2003, 103, 1–25 CrossRef CAS PubMed.
- K. Smalla, A. S. Haines, K. Jones, E. Krogerrecklenfort, H. Heuer, M. Schloter and C. M. Thomas, Appl. Environ. Microbiol., 2006, 72, 7253–7259 CrossRef CAS PubMed.
- B. Marteyn, S. Sakr, S. Farci, M. Bedhomme, S. Chardonnet, P. Decottignies, S. D. Lemaire, C. Cassier-Chauvat and F. Chauvat, J. Bacteriol., 2013, 195, 4138–4145 CrossRef CAS PubMed.
- A. Onnis-Hayden, H. Weng, M. He, S. Hansen, V. Ilyin, K. Lewis and A. Z. Guc, Environ. Sci. Technol., 2009, 43, 4574–4581 CrossRef CAS PubMed.
- M. Rafati-Rahimzadeh, M. Rafati-Rahimzadeh, S. Kazemi and A. A. Moghadamnia, Daru, J. Fac. Pharm., Tehran Univ. Med. Sci., 2014, 22, 46 CrossRef PubMed.
- S. Park and R. L. Ely, Appl. Environ. Microbiol., 2008, 74, 5475–5482 CrossRef CAS PubMed.
- S. Park and R. L. Ely, Arch. Microbiol., 2008, 189, 541–548 CrossRef CAS PubMed.
- M. Kiyono, K. Miyahara, Y. Sone, H. Pan-Hou, S. Uraguchi, R. Nakamura and K. Sakabe, Appl. Microbiol. Biotechnol., 2010, 86, 753–759 CrossRef CAS PubMed.
- D. Raimunda, T. Padilla-Benavides, S. Vogt, S. Boutigny, K. N. Tomkinson, L. A. Finney and J. M. Arguello, Metallomics, 2013, 5, 144–151 RSC.
- A. S. Gort, D. M. Ferber and J. A. Imlay, Mol. Microbiol., 1999, 32, 179–191 CrossRef CAS PubMed.
- R. Hengge-Aronis, Microbiol. Mol. Biol. Rev., 2002, 66, 373–395 CrossRef CAS PubMed , table of contents.
- P. Banjerdkij, P. Vattanaviboon and S. Mongkolsuk, Curr. Microbiol., 2003, 47, 260–262 CrossRef CAS PubMed.
- P. Banjerdkij, P. Vattanaviboon and S. Mongkolsuk, Appl. Environ. Microbiol., 2005, 71, 1843–1849 CrossRef CAS PubMed.
- O. Cantoni, S. Hussain, A. Guidarelli and F. Cattabeni, Mutat. Res., 1994, 324, 1–6 CAS.
- Z. P. Cakar, C. Alkim, B. Turanli, N. Tokman, S. Akman, M. Sarikaya, C. Tamerler, L. Benbadis and J. M. Francois, J. Biotechnol., 2009, 143, 130–138 CrossRef CAS PubMed.
- M. Sen, U. Yilmaz, A. Baysal, S. Akman and Z. P. Cakar, Antonie van Leeuwenhoek, 2011, 99, 825–835 CrossRef CAS PubMed.
- A. Toptchieva, G. Sisson, L. J. Bryden, D. E. Taylor and P. S. Hoffman, Microbiology, 2003, 149, 1285–1295 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5mt00256g |
| This journal is © The Royal Society of Chemistry 2016 |