Identification of bacterial species by untargeted NMR spectroscopy of the exo-metabolome†
T. L.
Palama‡
a,
I.
Canard
b,
G. J. P.
Rautureau
a,
C.
Mirande
b,
S.
Chatellier
b and
B.
Elena-Herrmann
*a
aUniversité de Lyon, Institut des Sciences Analytiques (CNRS/ENS Lyon/UCB Lyon1), Centre de RMN à Très Hauts Champs, 69100 Villeurbanne, France. E-mail: benedicte.elena@ens-lyon.fr
bbioMérieux, Innovation Unit – Microbiology Research, 38390 La Balme-les-Grottes, France
First published on 16th June 2016
Abstract
Identification of bacterial species is a crucial bottleneck for clinical diagnosis of infectious diseases. Quick and reliable identification is a key factor to provide suitable antibiotherapies and avoid the development of multiple-drug resistance. We propose a novel nuclear magnetic resonance (NMR)-based metabolomics strategy for rapid discrimination and identification of several bacterial species that relies on untargeted metabolic profiling of supernatants from bacterial culture media. We show that six bacterial species (Gram negative: Escherichia coli, Pseudomonas aeruginosa, Proteus mirabilis; Gram positive: Enterococcus faecalis, Staphylococcus aureus, and Staphylococcus saprophyticus) can be well discriminated from multivariate statistical analysis, opening new prospects for NMR applications to microbial clinical diagnosis.
Despite intense developments in microbiology since the first experiments of Louis Pasteur in the 19th century, identification of bacterial species still remains a major challenge in many areas such as clinical diagnosis or industrial microbiological control, motivating a constant search for rapid, precise, accurate and low-cost techniques.1 Improving the characterization of bacterial isolates is of key interest to public health to be able to adapt suitable antibiotherapies to infections and thus prevent the development of multiple-drug resistance.
Bacterial identification by phenotypic approaches is usually limited by time-consuming bacterial growth and biochemical reactions. Nevertheless, technological developments have given birth to automated systems (Vitek2, bioMérieux; BD Phoenix, Becton Dickinson).2,3 Matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (MS) has evolved into a rapid and highly reliable analytical tool for the characterization of micro-organisms in clinical laboratories,4 and other emerging novel diagnostic approaches have been applied to bacterial studies like genomic analysis,5,6 biochemical sensors,7–10 optical scattering technology,3 mass spectrometry,11 or infrared12 and Raman13–15 spectroscopy. Likewise, NMR spectroscopy has been proposed to study bacteria.16 NMR is a powerful technique that has the advantages of being nondestructive, intrinsically quantitative and information-rich with respect to the determination of molecular structures, especially in the context of complex mixture analyses such as bio-fluids. NMR has thus been successfully used to investigate intra- and extracellular bacterial composition17–21 or metabolic pathways.22–24 Metabolic footprinting approaches, i.e. extracellular metabolomics analyses that depict the detailed uptake and excretion of nutrients,25,26 can provide powerful avenues for microbial clinical diagnosis. The feasibility of microorganism strain differentiation from untargeted metabolic footprinting has been demonstrated using MS for the characterization of yeast mutants.27
As regards bacterial identification, several NMR-based strategies have been proposed. Tailored analysis for the detection of given species in culture media or in bio-fluids from urinary tract infection (UTI) patients has been described, which relies on the targeted detection of one or several metabolite markers.28–30 Gupta and coworkers have also proposed NMR targeted profiling of four metabolites in culture media as a method to discriminate Gram positive vs. negative bacteria,31 but that does not deliver precise identification of the present bacterial species. Unsupervised approaches have also been described for strain classification that so far relied on the analysis of the intracellular bacterial content, either exploiting low resolution NMR profiles of bacterial isolate suspensions,32 or high resolution fingerprints recorded on cell extracts.33
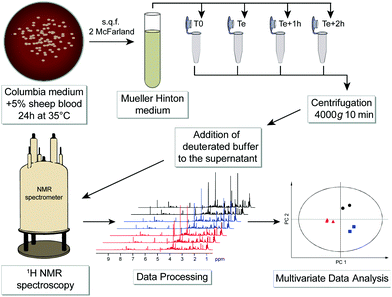
Advances in NMR technology and automation have contributed to promote NMR as a method of choice for such untargeted metabolomic investigations, aiming at a comprehensive and quantitative analysis of all metabolites present in biological samples.34 We propose here a novel and generic strategy to identify bacterial species by untargeted investigation of their exo-metabolome, i.e. the metabolite footprints detected in culture media. A schematic workflow of this NMR-based metabolomics approach is depicted in Fig. 1.
Six bacterial species (Gram negative: Escherichia coli, Pseudomonas aeruginosa, Proteus mirabilis; Gram positive: Enterococcus faecalis, Staphylococcus aureus, and Staphylococcus saprophyticus) that are most frequently responsible for urinary tract infections were cultured without agitation in a liquid Mueller Hinton medium until their specific exponential phase times. The list of strains and detailed protocol are given in Table S1, ESI.†
Samples preparation for NMR analysis was performed as follows: 60 μL of a mono-potassium phosphate solution (1.25 M KH2PO4 in D2O with 2 mM NaN3 and 0.1% trimethylsilyl propionic acid sodium salt (TSP), pH 7.4) were added to 540 μL of centrifuged medium (4000g for 10 min). 1H NMR profiles were acquired for each sample at 600 MHz. The detailed experimental procedures for NMR data acquisition and processing are provided in the ESI.† A total number of 576 samples were profiled in this study. A representative 1H NMR spectrum of an E. coli supernatant culture media sample is detailed in Fig. 2, and representative NMR profiles for all species and the reference culture medium are shown in Fig. S2, ESI.† Metabolite identification was achieved by comparing the spectra with international reference databases,35 and confirmed using two-dimensional NMR spectroscopy. The 43 identified metabolites are listed in Table S3, ESI.†
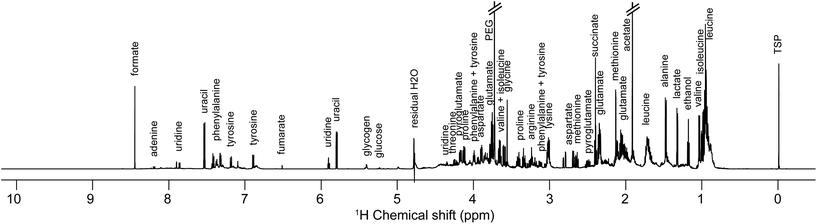 | ||
| Fig. 2 Representative one-dimensional 1H NMR spectrum of an Escherichia coli sample (culture supernatant) at exponential growth, i.e. after 1.5 hours of culture in a Mueller Hinton medium. | ||
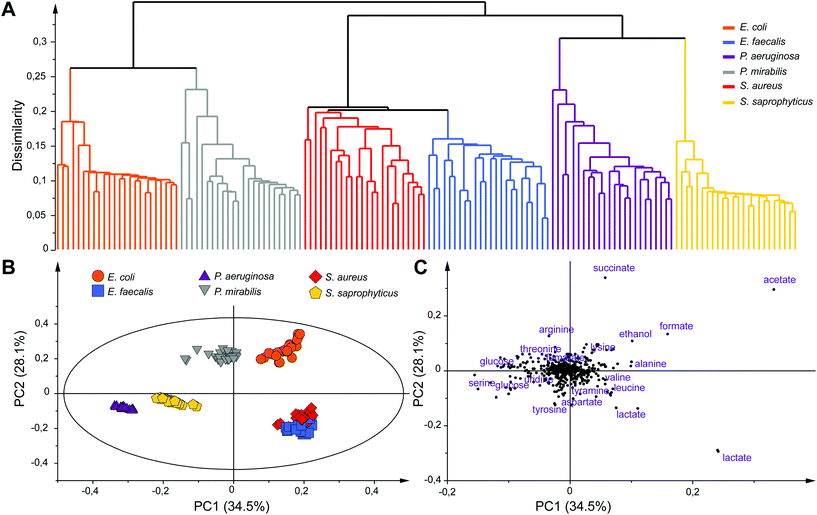
Unsupervised multivariate data analyses of the NMR profiles, including principal component analysis (PCA) and hierarchical clustering analysis (HCA), were applied to evaluate the datasets and discriminate sample classes. A strong species-based discrimination was observed at different time points after the bacterial mid exponential phase was reached (Te, Te + 1 h and Te + 2 h). The results for the earliest time-point, more relevant for efficient bacterial identification are presented in Fig. 3. HCA was carried out using Euclidean distances and a single linkage clustering method, and the corresponding tree-based representation displays a clear clustering of the observations (Fig. 3A). Strain samples of the same species cluster together into six well-defined branches of the dendrogram. Interestingly, additional levels of similarities are detected between species such as the P. aeruginosa and S. saprophyticus groups that merge together to form a new branch. Similar proximities are observed between S. aureus and E. faecalis, or E. coli and P. mirabilis, the later two consistently belonging to the same Enterobacteriaceae family.
To further analyze the discrimination between bacterial species, a PCA model (N = 144) is presented in Fig. 3B and C. The first two principal components of this model explain 62.6% of the variance within the dataset. A clear discrimination of the species groups can be observed on the score plot (Fig. 3B) except for E. faecalis and S. aureus samples that are both overlapping but are still distinct from the rest of the species. Separation between E. faecalis and S. aureus samples occurs along the third principal component (Fig. S4, ESI†).
Analysis of the PCA loading plot indicates that these two species have the highest content of lactate and a higher amount of acetate but less glucose compared to the other four species. Analysis of the score and loading plots also shows that E. coli and P. mirabilis samples have a higher amount of acetate, formate and succinate while a greater concentration of glucose and serine can be found in P. aeruginosa samples. The major discriminating metabolites in the PCA model belong to the glycolysis and Krebs cycle, the key pathways for glucose or other sugars to enter energetic and anabolic pathways. A consistent differentiation of bacterial species was obtained at Te + 1 h and Te + 2 h, while samples at all time points were also well separated from those collected at T0 (Fig. S5, ESI†). We note that an equivalent discrimination of bacterial species is obtained when considering a fixed collection time point after the start of bacterial cultures (e.g. T0 + 3h) for all bacterial species, independently of their respective growth status (data not shown). Our results show that the concentration of end-products of fermentation in the culture media (acetate, lactate, formate, succinate and ethanol) under anaerobic conditions significantly contribute to the discrimination of bacterial species, notably discriminating groups of strains sharing common sugar fermentation mechanisms, e.g. S. aureus/E. faecalis vs. E. coli/P. mirabilis, producing respectively either lactate or succinate among other end-products. We note that these five metabolites have already been identified as broad markers of UTI in the urine of patients against controls,31,36,37 while lactate, succinate and formate were shown to discriminate Gram positive and negative infections.31 Additional microbial-mammalian co-metabolites were also shown to characterize E. coli infection in urines.30 Here, robust individual discrimination of the six studied species is based on the full metabolic profiles considered in our multivariate models, notably involving a range of other metabolites such as tyramine, aspartate, leucine, alanine, arginine, threonine, lysine and serine. Further analyses of the data will be carried out in the future to provide a comprehensive picture of the metabolic pathways involved in this bacterial discrimination.
The approach introduced here demonstrates the potential of NMR footprinting, i.e. relying on observation of the exo-metabolome, for rapid bacterial species discrimination. We have exploited a generic culture medium that does not restrict the proposed strategy to a limited set of bacterial species. The rapid and functional approach proposed here could therefore be straightforwardly extended to broader selections of bacterial species and/or strains, and provide direct methods for bacterial identification from a single culture footprint. While the method was demonstrated for cultures of individual strains, we expect in the presence of mixtures or contamination the metabolic footprint to reflect the combined presence of several microorganisms through singular coordinates on the multivariate models, distinct from individual species footprints. We also note that the complexity of this type of Mueller-Hinton medium is likely to induce systematic differences between commercial broths, and therefore a larger scale model involving multiple batches of cultures should be constructed in the future to provide a robust reference database for unambiguous identification of individual bacterial cultures. The proposed approach opens up new prospects for accurate and cost-effective microbial clinical diagnosis.
Notes and references
- P.-E. Fournier, M. Drancourt, P. Colson, J.-M. Rolain, B. La Scola and D. Raoult, Nat. Rev. Microbiol., 2013, 11, 574–585 CrossRef CAS PubMed.
- P. P. Bourbeau and N. A. Ledeboer, J. Clin. Microbiol., 2013, 51, 1658–1665 CrossRef PubMed.
- P. R. Marcoux, M. Dupoy, A. Cuer, J. L. Kodja, A. Lefebvre, F. Licari, R. Louvet, A. Narassiguin and F. Mallard, Appl. Microbiol. Biotechnol., 2014, 98, 2243–2254 CrossRef CAS PubMed.
- P. Seng, J.-M. Rolain, P. E. Fournier, B. La Scola, M. Drancourt and D. Raoult, Future Microbiol., 2010, 5, 1733–1754 CrossRef CAS PubMed.
- X. Didelot, R. Bowden, D. J. Wilson, T. E. A. Peto and D. W. Crook, Nat. Rev. Genet., 2012, 13, 601–612 CrossRef CAS PubMed.
- J. J. Farrell, A. M. Hujer, R. Sampath and R. A. Bonomo, Expert Rev. Mol. Diagn., 2015, 15, 349–360 CrossRef CAS PubMed.
- R. L. Phillips, O. R. Miranda, C. C. You, V. M. Rotello and U. H. F. Bunz, Angew. Chem., Int. Ed., 2008, 47, 2590–2594 CrossRef CAS PubMed.
- A. Duarte, A. Chworos, S. F. Flagan, G. Hanrahan and G. C. Bazan, J. Am. Chem. Soc., 2010, 132, 12562–12564 CrossRef CAS PubMed.
- J. R. Carey, K. S. Susick, K. I. Hulkower, J. A. Imlay, K. R. C. Imlay, C. K. Ingison, J. B. Ponder, A. Sen and A. E. Wittrig, J. Am. Chem. Soc., 2011, 133, 7571–7576 CrossRef CAS PubMed.
- W. W. Chen, Q. Z. Li, W. S. Zheng, F. Hu, G. X. Zhang, Z. Wang, D. Q. Zhang and X. Y. Jiang, Angew. Chem., Int. Ed., 2014, 53, 13734–13739 CrossRef CAS PubMed.
- A. Fox, J. Clin. Microbiol., 2006, 44, 2677–2680 CrossRef CAS PubMed.
- H. AlRabiah, E. Correa, M. Upton and R. Goodacre, Analyst, 2013, 138, 1363–1369 RSC.
- L. Ashton, K. Lau, C. L. Winder and R. Goodacre, Future Microbiol., 2011, 6, 991–997 CrossRef CAS PubMed.
- D. F. Willemse-Erix, M. J. Scholtes-Timmerman, J. W. Jachtenberg, W. B. van Leeuwen, D. Horst-Kreft, T. C. Bakker Schut, R. H. Deurenberg, G. J. Puppels, A. van Belkum, M. C. Vos and K. Maquelin, J. Clin. Microbiol., 2009, 47, 652–659 CrossRef PubMed.
- I. Espagnon, D. Ostrovskii, R. Mathey, M. Dupoy, P. L. Joly, A. Novelli-Rousseau, F. Pinston, O. Gal, F. Mallard and D. F. Leroux, J. Biomed. Opt., 2014, 19, 027004 CrossRef PubMed.
- J. P. Grivet and A. M. Delort, Prog. Nucl. Magn. Reson. Spectrosc., 2009, 54, 1–53 CrossRef CAS.
- Y. F. Ye, L. M. Zhang, Y. P. An, F. H. Hao and H. R. Tang, Chin. J. Anal. Chem., 2011, 39, 1186–1194 CAS.
- H. Meyer, M. Liebeke and M. Lalk, Anal. Biochem., 2010, 401, 250–259 CrossRef CAS PubMed.
- K. L. Resmer and R. L. White, Mol. BioSyst., 2011, 7, 2220–2227 RSC.
- G. Zandomeneghi, K. Ilg, M. Aebi and B. H. Meier, J. Am. Chem. Soc., 2012, 134, 17513–17519 CrossRef CAS PubMed.
- H. Kusch and S. Engelmann, Int. J. Med. Microbiol., 2014, 304, 133–141 CrossRef CAS PubMed.
- M. R. Sadykov, B. Zhang, S. Halouska, J. L. Nelson, L. W. Kreimer, Y. F. Zhu, R. Powers and G. A. Somerville, J. Biol. Chem., 2010, 285, 36616–36624 CrossRef CAS PubMed.
- T. A. Bartholomeusz, R. Molinie, F. Mesnard, R. J. Robins and A. Roscher, Cron. Chim., 2008, 11, 457–464 CrossRef CAS.
- S. Meier, P. R. Jensen and J. O. Duus, FEBS Lett., 2011, 585, 3133–3138 CrossRef CAS PubMed.
- V. Behrends, H. D. Williams and J. G. Bundy, Methods Mol. Biol., 2014, 1149, 281–292 CrossRef CAS PubMed.
- V. Mapelli, L. Olsson and J. Nielsen, Trends Biotechnol., 2008, 26, 490–497 CrossRef CAS PubMed.
- J. Allen, H. M. Davey, D. Broadhurst, J. K. Heald, J. J. Rowland, S. G. Oliver and D. B. Kell, Nat. Biotechnol., 2003, 21, 692–696 CrossRef CAS PubMed.
- A. Gupta, M. Dwivedi, G. A. N. Gowda, A. Ayyagari, A. A. Mahdi, M. Bhandari and C. L. Khetrapal, NMR Biomed., 2005, 18, 293–299 CrossRef CAS PubMed.
- A. Gupta, M. Dwivedi, G. A. N. Gowda, A. A. Mahdi, A. Jain, A. Ayyagari, R. Roy, M. Bhandari and C. L. Khetrapal, NMR Biomed., 2006, 19, 1055–1061 CrossRef CAS PubMed.
- C.-W. Lam, C.-Y. Law, K.-H. Sze and K. K.-W. To, Clin. Chim. Acta, 2015, 438, 24–28 CrossRef CAS PubMed.
- A. Gupta, M. Dwivedi, A. A. Mahdi, C. L. Khetrapal and M. Bhandari, J. Proteome Res., 2012, 11, 1844–1854 CrossRef CAS PubMed.
- R. Bourne, U. Himmelreich, A. Sharma, C. Mountford and T. Sorrell, J. Clin. Microbiol., 2001, 39, 2916–2923 CrossRef CAS PubMed.
- J. G. Bundy, T. L. Willey, R. S. Castell, D. J. Ellar and K. M. Brindle, FEMS Microbiol. Lett., 2005, 242, 127–136 CrossRef PubMed.
- J. C. Lindon, E. Holmes and J. K. Nicholson, Anal. Chem., 2003, 75, 384A–391A CrossRef CAS PubMed.
- D. S. Wishart, C. Knox, A. C. Guo, R. Eisner, N. Young, B. Gautam, D. D. Hau, N. Psychogios, E. Dong, S. Bouatra, R. Mandal, I. Sinelnikov, J. G. Xia, L. Jia, J. A. Cruz, E. Lim, C. A. Sobsey, S. Shrivastava, P. Huang, P. Liu, L. Fang, J. Peng, R. Fradette, D. Cheng, D. Tzur, M. Clements, A. Lewis, A. De Souza, A. Zuniga, M. Dawe, Y. P. Xiong, D. Clive, R. Greiner, A. Nazyrova, R. Shaykhutdinov, L. Li, H. J. Vogel and I. Forsythe, Nucleic Acids Res., 2009, 37, D603–D610 CrossRef CAS PubMed.
- C.-W. Lam, C.-Y. Law, K. K.-W. To, S. K.-K. Cheung, K.-C. Lee, K.-H. Sze, K.-F. Leung and K.-Y. Yuen, Clin. Chim. Acta, 2014, 436, 217–223 CrossRef CAS PubMed.
- E. Nevedomskaya, T. Pacchiarotta, A. Artemov, A. Meissner, C. Nieuwkoop, J. T. Dissel, O. A. Mayboroda and A. M. Deelder, Metabolomics, 2012, 8, 1227–1235 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: List of the bacterial species, detailed protocols for NMR data acquisition and processing, table of 43 metabolites identified in the culture media, and additional multivariate models. See DOI: 10.1039/c6an00393a |
| ‡ Present address: Laboratoire d'Ingénierie des Systèmes Biologiques et des Procédés – INSA. 135 Avenue de Rangueil. 31077 Toulouse Cedex 4, France. |
| This journal is © The Royal Society of Chemistry 2016 |