A novel dynamic pseudo[1]rotaxane based on a mono-biotin-functionalized pillar[5]arene†
Xuan
Wu
,
Mengfei
Ni
,
Wei
Xia
,
Xiao-Yu
Hu
* and
Leyong
Wang
Key Laboratory of Mesoscopic Chemistry of MOE and Collaborative Innovation Center of Chemistry for Life Sciences, School of Chemistry and Chemical Engineering, Nanjing University, Nanjing, 210093, China. E-mail: huxy@nju.edu.cn; Fax: +86 25-83317761; Tel: +86 25-83597090
First published on 19th June 2015
Abstract
A mono-biotin-functionalized pillar[5]arene P1 was synthesized by the click reaction, which could form a stable pseudo[1]rotaxane P1′ in a non-polar or weak-polar solution. Interestingly, the obtained pseudo[1]rotaxane P1′ exhibited a dynamic slow disassembly process within the NMR timescale upon adding a strong-polar solvent or competitive guest. Moreover, this dynamic behavior also had potential application in aqueous solution, which might be used as a switch to turn on or off the bioactivity of the biotin moiety.
Mechanically interlocked molecules (MIMs), for their characteristic topological structures and potential application in molecular machines, have attracted tremendous attention over the past few decades.1–5 Among the various types of MIMs, pseudo[1]rotaxane, as the simplest interlocked structure, where the wheel is threaded by its own axle, exhibits a fast or slow exchange process between the self-included and free structure to external stimuli due to its reversible conversion behavior. So far, many research studies have been conducted on this special structure for its potential applications in constructing molecular muscles,6 molecular switches,7–9 insulated molecular wires,10,11 and fluorescence sensors.12,13
However, most of these reported pseudo[1]rotaxanes were usually based on traditional macrocycles, such as cyclodextrins and crown ethers. Pillararenes as a new generation of macrocyclic molecules14 have received considerable attention and development due to their unique structures, easy modification,15,16 and excellent properties in host–guest chemistry.17–21 Up to now, various pillararene-based interlocked structures have been reported, such as rotaxanes22–27 and catenanes,28–30 whereas there are only a few reports on the formation of pseudo[1]rotaxanes based on this novel host molecule.31–34 Therefore, the construction of pillararene-based pseudo[1]rotaxanes through host–guest interactions, especially with stimuli-responsive dynamic behaviors, is of great interest and importance in the application of constructing controllable molecular machines. Recently, a stable pseudo[1]rotaxane based on an ester group containing pillar[5]arene was successfully constructed by Cao, and showed responsiveness to dihalogen alkanes.31 Moreover, Hou also reported a stable pseudo[1]rotaxane by introducing an amino group to the side chain of pillar[5]arene, which was stabilized by the intramolecular hydrogen bond.32 On the basis of our previous work on the construction of dynamic pseudo[1]rotaxane based on a urea-modified pillar[5]arene33 and another mono-functionalized pillar[5]arene bearing the Boc end group,34 we could conclude that the terminal group played a vital role in the dynamic behavior of the formed pseudo[1]rotaxanes. Therefore, we envision that we can design a novel dynamic pillararene-based pseudo[1]rotaxane which could be synthesized by introducing a bioactive biotin moiety to a side chain of pillar[5]arene via a flexible ethylene glycol chain to achieve its controllable dynamic self-inclusion behavior. Furthermore, in aqueous solution, we speculate that this dynamic behavior might be used as a switch to turn on or off the bioactivity of the biotin moiety.
Herein, a mono-biotin-functionalized pillar[5]arene P1 was successfully prepared by the efficient click reaction, which could form a stable pseudo[1]rotaxane P1′ in a non-polar or weak-polar solution such as CHCl3 or acetone. Moreover, the obtained pseudo[1]rotaxane P1′ showed a dynamic slow disassembly process upon the addition of a competitive guest or increasing the polarity of the solution (Scheme 1). More importantly, this dynamic pseudo[1]rotaxane could be applied as a biotin-based switch in aqueous solution, which might have potential applications in drug delivery systems.
The target molecule P1 was synthesized by means of copper(I) catalyzed azide–alkyne cycloaddition (CuAAC).35,36 The alkynyl-functionalized pillar[5]arene 3 was prepared according to the typical [4 + 1]cyclization of monomers 1 and 2. Meanwhile, the azide-modified biotin derivative 6 was obtained by using commercially available triethylene glycol as the starting material. Firstly, through two steps of a typical reaction, a mono-azide-modified glycol 5 was obtained, and then a condensation reaction between compound 5 and biotin was carried out to give the biotin derivative 6. Finally, the CuAAC reaction between 3 and 6 generated the target molecule P1 in a moderate yield (Scheme 2).
The inclusion behavior of P1 was firstly studied by 1H NMR spectroscopy in different solvents (Fig. 1). From the 1H NMR spectra, the obviously upfield chemical shifts of the protons (Ha–e) on the biotin group could be observed in CDCl3 compared with that in DMSO-d6, indicating the inclusion of the biotin group into the cavity of the pillar[5]arene, which resulted in the formation of the inclusion structure P1′. Similar upfield shift phenomena were also observed from the spectrum using acetone-d6 as the solvent, indicating that the polarity of acetone could not destroy the inclusion structure of P1′. However, the signals of the protons of P1′ in 1H NMR spectra tend to be normal in strong polar solvents, such as DMSO-d6, which meant that the inclusion behavior of P1 was completely destroyed in DMSO-d6. The above results showed that this inclusion structure formed by P1 was stable in non-polar and weak-polar solvents, but it was instable in strong polar solvents. This was consistent with the results in previous studies that a strong-polar solvent would destroy the driving force of the formed inclusion structures.31,33
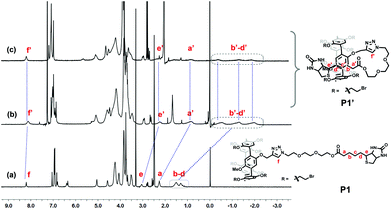 | ||
| Fig. 1 1H NMR spectra (300 MHz, 298 K) of P1 (15 mM) in different solvents: (a) in DMSO-d6, (b) in CDCl3, and (c) in acetone-d6. | ||
To investigate whether the inclusion behavior was intramolecular or intermolecular, the host–guest interaction between 3 and 6 was firstly studied. As shown in Fig. S12 (ESI†), the 1H NMR spectra of 6 with the addition of different equivalents of 3 showed that the chemical shifts of Ha–e on 6 gradually shifted upfield, exhibiting a fast exchange process within the NMR timescale. However, only a slightly upfield shift (0.15 ppm) of protons Ha–e on 6 could be observed when 1.0 equiv. of 3 was added, which indicated that the interaction between 3 and 6 was very weak. Subsequently, variable concentration 1H NMR spectroscopy of P1 in CDCl3 was carried out (Fig. 2). It was found that the aggregates formed by P1 were very stable even in very dilute CDCl3 solution (5 mM). As the concentration increased, the proton resonances did not exhibit obvious changes even at a high concentration of 80 mM, suggesting that P1 did not form intermolecular complexes in CDCl3.37 Therefore, it could be concluded that a stable pseudo[1]rotaxane P1′ was formed in chloroform.
 | ||
| Fig. 2 1H NMR spectra (300 MHz, 298 K) of P1 at various concentrations in CDCl3: (a) 5 mM, (b) 10 mM, (c) 15 mM, (d) 20 mM, (e) 40 mM, (f) 60 mM, and (g) 80 mM. | ||
In addition, diffusion-ordered 1H NMR spectroscopy (DOSY) experiments were performed, and the diffusion coefficients of pseudo[1]rotaxane P1′ in CDCl3 at both low and high concentrations were recorded. From the DOSY spectra (Fig. S13 and S14, ESI†), only one set of signals could be observed at both low concentration (10 mM) and high concentration (45 mM) of P1′, which further indicated that the inclusion behavior of P1 was not intermolecular. Then, the Stokes–Einstein relationship was used to calculate the radius of P1′ in CDCl3, which was calculated to be 9.98 Å (Table S1, ESI†). Furthermore, theoretical calculation was carried out to optimize the structure of P1′ by employing the Gaussian 09 program package.38 The result showed that the calculated diameter of the pseudo[1]rotaxane P1′ was nearly 18.12 Å (r = 9.06 Å) (Fig. S15, ESI†), which was in good agreement with the result obtained from DOSY experiment. Therefore, based on the above observations, we could clearly confirm the self-inclusion behavior of P1 in chloroform, resulting in the formation of pseudo[1]rotaxanes P1′.
Subsequently, we attempted to know the dynamic behavior of this pseudo[1]rotaxane P1′. Initially, the effect of solvent polarity on this disassembly process was investigated. As shown in Fig. 3, it was found that, with the increase of the solvent polarity by gradually adding DMSO-d6 into the CDCl3 solution of P1, the peaks for Ha′–e′ of the self-inclusion structure P1′ disappeared gradually, and meanwhile the peaks for Ha–e of the un-entangled structure P1 appeared and strengthened gradually. This observation suggested that the self-inclusion structure was gradually destroyed, presenting a slow exchange process within the NMR timescale. When the ratio of the mixture DMSO-d6/CDCl3 was increased to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v), most of the self-inclusion structure P1′ was transformed to the un-entangled form P1.
1 (v/v), most of the self-inclusion structure P1′ was transformed to the un-entangled form P1.
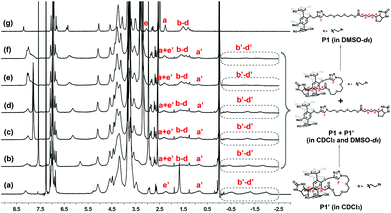 | ||
Fig. 3
1H NMR spectra (300 MHz, 298 K) of P1 (15 mM) in mixed solvents CDCl3/DMSO-d6 with different ratios (v/v): (a) 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0, (b) 5 0, (b) 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, (c) 5 1, (c) 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, (d) 5 2, (d) 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, (e) 5 3, (e) 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, (f) 5 4, (f) 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, and (g) 0 5, and (g) 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5. 5. | ||
A similar dynamic disassembly process of pseudo[1]rotaxane P1′ could also be observed if a competitive guest was added to the P1′ solution. Since hexanedinitrile shows strong host–guest binding affinity with pillar[5]arene,39 herein, hexanedinitrile was chosen as a typical competitive guest to investigate the dynamic behavior of P1′ (Fig. 4). It was found that with the addition of hexanedinitrile to the solution of P1′ in CDCl3, the protons Hb′–d′ of the biotin group attributed to the self-inclusion structure P1′ disappeared gradually, and a new peak assigned to the P1 ⊃ hexanedinitrile complex appeared, which clearly revealed that the self-inclusion structure P1′ was destroyed gradually and a pseudo[2]rotaxane was formed between P1 and hexanedinitrile. Moreover, upon adding hexanedinitrile, the proton Hf′ of the triazole group split into two different sets of peaks, the one in the original position which assigned to the self-inclusion structure P1′ disappeared gradually, while the other that derived from the P1 ⊃ hexanedinitrile complex shifted upfield and strengthened gradually. According to the previous work,13,40 we speculated that hydrogen bonding might be formed between the triazole group and the glycol unit in the pseudo[1]rotaxane P1′, which might play an important role in stabilizing the self-inclusion structure.
As we know biotin is a bioactive molecule, which could be used as a tumor-targeted molecule for drug delivery.41,42 Based on the above results, we noted that the biotin group would slip out from the cavity of pillar[5]arene in P1′ if a competitive guest was added to the solution. So we wondered if this phenomenon could still be observed in aqueous solution. Once such dynamic self-inclusion and disassembly processes could be realized, the formed pseudo[1]rotaxane would be used as a switch to turn on or off the bioactivity of biotin. Along this line, a water-soluble biotin-functionalized pillar[5]arene P2 was synthesized through a simple reaction of P1 with trimethylamine. From the NMR spectra (Fig. 5), it was found that the protons Hb′–d′ on the biotin group shifted up-field obviously, indicated that the self-inclusion structure P2′ could also be formed. Then, sodium caprylate (SC), the competitive guest was added to the P2′ solution. To our delight, a similar disassembly phenomenon like P1′ was also observed, where the biotin group left the cavity of pillar[5]arene in P2′, and the protons H1–7 on SC showed slightly up-field shifts, indicating a host–guest complex between P2 and SC was formed. The above study gave us very useful guideline for our further studies on the design of the biotin-based drug delivery system.
 | ||
| Fig. 5 1H NMR spectra (300 MHz, 298 K, D2O) of (a) P2′ (2 mM), (b) P2′ (2 mM) + SC (20 mM), and (c) SC (20 mM). | ||
Conclusions
In summary, a mono-functionalized pillar[5]arene P1 using biotin as the end group was successfully synthesized through a CuAAC reaction, which could form a stable pseudo[1]rotaxane P1′ in non-polar and weak-polar solutions. However, this self-inclusion structure showed a slow dynamic disassembly process upon adding a strong-polar solvent or competitive guest, resulting in the formation of un-entangled structures. Moreover, this dynamic self-inclusion behavior could be used as a biotin switch to turn on or off the bioactivity of biotin, which gave us constructive guidelines for the following study on the design of biotin-based drug delivery systems.Acknowledgements
We are grateful to the National Natural Science Foundation of China (no. 21202083 and 21472089), and the National Natural Science Foundation of Jiangsu (no. BK20140595).Notes and references
- D. B. Amabilino and J. F. Stoddart, Chem. Rev., 1995, 95, 2725 CrossRef CAS.
- A. Coskun, M. Banaszak, R. D. Astumian, J. F. Stoddart and B. A. Grzybowski, Chem. Soc. Rev., 2012, 41, 19 RSC.
- C. O. Dietrich-Buchecker and J. P. Sauvage, Chem. Rev., 1987, 87, 795 CrossRef CAS.
- D. H. Qu, Q. C. Wang, Q. W. Zhang, X. Ma and H. Tian, Chem. Rev., 2015 DOI:10.1021/cr5006342.
- M. Xue, Y. Yang, X. Chi, X. Yan and F. Huang, Chem. Rev., 2015 DOI:10.1021/cr5005869.
- C. Gao, X. Ma, Q. Zhang, Q. Wang, D. Qu and H. Tian, Org. Biomol. Chem., 2011, 9, 1126 CAS.
- H. Li, X. Li, H. Ågren and D.-H. Qu, Org. Lett., 2014, 16, 4940 CrossRef CAS PubMed.
- Y. Inoue, P. Kuad, Y. Okumura, Y. Takashima, H. Yamaguchi and A. Harada, J. Am. Chem. Soc., 2007, 129, 6396 CrossRef CAS PubMed.
- X. Ma, D. Qu, F. Ji, Q. Wang, L. Zhu, Y. Xu and H. Tian, Chem. Commun., 2007, 1409 RSC.
- J. Terao, Polym. Chem., 2011, 2, 2444 RSC.
- J. Terao, S. Tsuda, Y. Tanaka, K. Okoshi, T. Fujihara, Y. Tsuji and N. Kambe, J. Am. Chem. Soc., 2009, 131, 16004 CrossRef CAS PubMed.
- H. Li, X. Li, Z.-Q. Cao, D.-H. Qu, H. Ågren and H. Tian, ACS Appl. Mater. Interfaces, 2014, 6, 18921 CAS.
- H. Li, H. Zhang, Q. Zhang, Q.-W. Zhang and D.-H. Qu, Org. Lett., 2012, 14, 5900 CrossRef CAS PubMed.
- T. Ogoshi, S. Kanai, S. Fujinami, T.-A. Yamagishi and Y. Nakamoto, J. Am. Chem. Soc., 2008, 130, 5022 CrossRef CAS PubMed.
- Y. Yao, M. Xue, J. Chen, M. Zhang and F. Huang, J. Am. Chem. Soc., 2012, 134, 15712 CrossRef CAS PubMed.
- G. Yu, Y. Ma, C. Han, Y. Yao, G. Tang, Z. Mao, C. Gao and F. Huang, J. Am. Chem. Soc., 2013, 135, 10310 CrossRef CAS PubMed.
- T. Ogoshi and T.-A. Yamagishi, Eur. J. Org. Chem., 2013, 2961 CrossRef CAS PubMed.
- X. Wu, Q. Duan, M. Ni, X. Y. Hu and L. Wang, Chin. J. Org. Chem., 2014, 34, 437 CrossRef.
- M. Xue, Y. Yang, X. Chi, Z. Zhang and F. Huang, Acc. Chem. Res., 2012, 45, 1294 CrossRef CAS PubMed.
- Z. Zhang, Y. Luo, J. Chen, S. Dong, Y. Yu, Z. Ma and F. Huang, Angew. Chem., Int. Ed., 2011, 50, 1397 CrossRef CAS PubMed.
- G. Yu, X. Zhou, Z. Zhang, C. Han, Z. Mao, C. Gao and F. Huang, J. Am. Chem. Soc., 2012, 134, 19489 CrossRef CAS PubMed.
- S. Dong, J. Yuan and F. Huang, Chem. Sci., 2014, 5, 247 RSC.
- X.-Y. Hu, X. Wu, Q. Duan, T. Xiao, C. Lin and L. Wang, Org. Lett., 2012, 14, 4826 CrossRef CAS PubMed.
- C. Ke, N. L. Strutt, H. Li, X. Hou, K. J. Hartlieb, P. R. McGonigal, Z. Ma, J. Iehl, C. L. Stern, C. Cheng, Z. Zhu, N. A. Vermeulen, T. J. Meade, Y. Y. Botros and J. F. Stoddart, J. Am. Chem. Soc., 2013, 135, 17019 CrossRef CAS PubMed.
- T. Ogoshi, R. Iizuka, D. Kotera and T.-A. Yamagishi, Org. Lett., 2014, 17, 350 CrossRef PubMed.
- N. L. Strutt, R. S. Forgan, J. M. Spruell, Y. Y. Botros and J. F. Stoddart, J. Am. Chem. Soc., 2011, 133, 5668 CrossRef CAS PubMed.
- B. Xia and M. Xue, Chem. Commun., 2014, 50, 1021 RSC.
- K. Kitajima, T. Ogoshi and T.-A. Yamagishi, Chem. Commun., 2014, 50, 2925 RSC.
- T. Ogoshi, T. Akutsu, D. Yamafuji, T. Aoki and T.-A. Yamagishi, Angew. Chem., Int. Ed., 2013, 52, 8111 CrossRef CAS PubMed.
- X. Yan, P. Wei, Z. Li, B. Zheng, S. Dong, F. Huang and Q. Zhou, Chem. Commun., 2013, 49, 2512 RSC.
- Y. Chen, D. Cao, L. Wang, M. He, L. Zhou, D. Schollmeyer and H. Meier, Chem. – Eur. J., 2013, 19, 7064 CrossRef CAS PubMed.
- L. Chen, Z. Li, Z. Chen and J. L. Hou, Org. Biomol. Chem., 2013, 11, 248 CAS.
- M. Ni, X. Y. Hu, J. Jiang and L. Wang, Chem. Commun., 2014, 50, 1317 RSC.
- Y. Guan, P. Liu, C. Deng, M. Ni, S. Xiong, C. Lin, X.-Y. Hu, J. Ma and L. Wang, Org. Biomol. Chem., 2014, 12, 1079 CAS.
- V. Aucagne, K. D. Hänni, D. A. Leigh, P. J. Lusby and D. B. Walker, J. Am. Chem. Soc., 2006, 128, 2186 CrossRef CAS PubMed.
- K. D. Hanni and D. A. Leigh, Chem. Soc. Rev., 2010, 39, 1240 RSC.
- M. Miyauchi and A. Harada, J. Am. Chem. Soc., 2004, 126, 11418 CrossRef CAS PubMed.
- L. Liu, Y. Liu, P. Liu, J. Wu, Y. Guan, X. Hu, C. Lin, Y. Yang, X. Sun, J. Ma and L. Wang, Chem. Sci., 2013, 4, 1701 RSC.
- X. Shu, S. Chen, J. Li, Z. Chen, L. Weng, X. Jia and C. Li, Chem. Commun., 2012, 48, 2967 RSC.
- G. Du, E. Moulin, N. Jouault, E. Buhler and N. Giuseppone, Angew. Chem., Int. Ed., 2012, 51, 12504 CrossRef CAS PubMed.
- B. Le Droumaguet, J. Nicolas, D. Brambilla, S. Mura, A. Maksimenko, L. De Kimpe, E. Salvati, C. Zona, C. Airoldi, M. Canovi, M. Gobbi, N. Magali, B. La Ferla, F. Nicotra, W. Scheper, O. Flores, M. Masserini, K. Andrieux and P. Couvreur, ACS Nano, 2012, 6, 5866 CrossRef CAS PubMed.
- Y.-X. Wang, D.-S. Guo, Y.-C. Duan, Y.-J. Wang and Y. Liu, Sci. Rep., 2015, 5, 9901 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Synthetic procedures, characterization, NMR spectra, and calculated results. See DOI: 10.1039/c5qo00159e |
| This journal is © the Partner Organisations 2015 |



