Toxicity of engineered metal oxide nanomaterials mediated by nano–bio–eco–interactions: a review and perspective
Xiaojia
He
a,
Winfred G.
Aker
a,
Peter P.
Fu
b and
Huey-Min
Hwang
*a
aJackson State University, Jackson, Mississippi, USA. E-mail: huey-min.hwang@jsums.edu
bNational Center for Toxicological Research, Arkansas, USA
First published on 25th September 2015
Abstract
Along with the expanding use of engineered metal oxide nanomaterials (MONMs), there is a growing concern over their unintentional adverse toxicological effects on human health and the environment upon release and exposure. It is inevitable that biota will be exposed to nanomaterials, through intentional administration or inadvertent contact under such circumstances. Therefore, a thorough investigation of the potential nanotoxicity of MONMs at the nano–bio–eco interface is urgently needed. In general, nanomaterials interact with their surrounding environments, biotic and abiotic, immediately upon introduction into the environment. The behavior and fate of MONMs are influenced by the dynamics of the environment. Thus, understanding the interactions at the nano–bio–eco interface is necessary for selecting and designing MONMs with minimum adverse impacts. Despite the limitations of currently available techniques, careful characterization of nanomaterials and the choosing of methodologies that promote further risk assessment promise more reliable and accurate data output. Conventional toxicological analysis techniques lack the power to handle the large datasets generated from in vitro/in vivo observations. This paper provides a comprehensive review of the recent experimental and theoretical studies on the toxicity of MONMs mediated by two-way or three-way interactions. In the Perspectives, we also call for more open collaborations between industry, academia, and research labs to facilitate nanotoxicological studies focused specifically on interactions at the nano–bio–eco interface, leading to safe and effective nanotechnology for commercial, environmental, and medicinal use.
Nano impactThe behavior and fate of metal oxide nanomaterials (MONMs) are influenced by the dynamic interactions among the different compartments in natural environments. Thus, understanding the interactions at the nano–bio–eco interface is necessary for selecting and designing MONMs with minimum adverse impacts. This paper provides a comprehensive review of the recent experimental and theoretical studies on the toxicity of MONMs mediated by two-way or three-way interactions. In the Perspectives, we also call for more open collaborations between industry, academia, and research labs to facilitate nanotoxicological studies focused specifically on interactions at the nano–bio–eco interface, leading to safe and effective nanotechnology for commercial, environmental, and medicinal use. |
1. Introduction
Nanotechnology is one of the rapidly developing and important research fields in the 21st century. The commercial use of nanomaterials for various novel applications is increasing drastically. The increasing use of manufactured nanomaterials in commercial products and environmental applications has substantially advanced our tactics against barriers in environmental and biomedical practices.1–3 Manufactured metal oxide nanomaterials (MONMs), the theme of this review, are among the most widely used types of engineered nanomaterials. The metal elements on the periodic chart are capable of forming a large diversity of oxide compounds. They can adopt a vast number of structural geometries with electronic structures that exhibit metallic, semiconductor or insulator characteristics.4 Conservative market estimates for the production of MONMs in 2020 are 1,663,168 tons, rising from 270,041 tons in 2012.5 In the long run, it is anticipated that improvements in thermal, mechanical, and other physiochemical properties of engineered nanomaterials, including MONMs, will fundamentally change the way they are used and associated risk assessments must keep their pace. Ensuring the safe use of engineered nanomaterials requires conscious and continuing efforts to establish and adhere to protocols in production, application, and disposal of these synthetic chemicals. Precautions and early actions have to be taken by researchers and scientists, as well as regulatory authorities, in order to minimize the potential hazards and maximize the benefits to humans.It is inevitable that, during their manufacture, use, and disposal, engineered MONMs are released into natural environments. Their appearance in soil, water and air could harm both environmental biota and humans upon exposure. Considerably unknown risks associated with MONMs have raised concerns from both public and authorities. Although some currently reported data suggest that very low concentrations of MONMs present in natural environments do no significant harm to biota, there still exists a huge knowledge gap with regard to the physicochemical properties of MONMs and their impact on environmental and human health. As reviewed and suggested in the existing literature,6–13 the overall life cycle-associated environmental impacts of those synthetic chemicals have to be cautiously addressed. Much of the published literature suggests that upon interaction with surrounding elements (chemicals, bacteria, biological contaminants, etc.) present in the environment physically and chemically, their behavior and fate can be drastically altered, leading to unpredictable outcomes; therefore, one should consider the dynamics of particular environments when making nanotoxicological assessments. Indeed, increased care has been taken in assessing their physical and chemical alteration in various environments for comparison with their intrinsic properties. For these and other reasons, thorough characterization of MONMs, taking into account the conditions of the particular environment under study is essentially indispensable.
Safe handling and disposal of nanomaterials nowadays receives increasing attention from both public and governmental authorities.14,15 It has been recognized that, among the 30 industrialized countries of the Organization for Economic Co-operation and Development (OECD), the United States, England, Germany, European commission and Australia have developed documents of good practices for the safety of manufactured nanomaterials.16,17 Additionally, under the regulation of Registration, Evaluation, Authorisation and Restriction of Chemicals (REACH)18–20 and International Organization for Standardization (ISO),21,22 generic recommendations for exposure assessment and risk characterization of nanomaterials were addressed.23 However, there are still many organizations working with nanomaterials currently using conventional chemical safety methods through the life cycle of nanomaterials, especially in the process of disposal. Disposal is closely related to environmental health once MONMs are released or discharged, and it eventually affects human health.
Notably, issues gaining an increased attention are the establishment of toxicological profiles of engineered nanomaterials with regard to the nano–bio–eco interface, which entails the cataloging of interactions among nanomaterials, biotic, and abiotic environments. Physicochemical properties at the nanoscale afford those artificial nanomaterials to be highly reactive compared to conventional counterparts. Thus, the bioavailability and toxicity of nanomaterials can be altered at the nano–bio–eco interface. The control over the physicochemical properties of nanomaterials makes the design and application of novel nanomaterials possible in a green and sustainable way. We have long been interested in studying the nanotoxicity of nanomaterials (metal oxide nanomaterials in particular) to human and ecological environments, with the research involving biological and computational studies.6,8,24–37 In this review article, we summarize the findings of the studies that have shown MONMs interacting with their immediate environments at the nano–bio–eco interface. The mechanisms of their toxicity are briefly discussed. In addition, this review highlights currently advanced toxicological analysis techniques in quantitative or qualitative approaches. The correlation between nano–bio–eco interactions and nanotoxicology is then further discussed with an emphasis on the quantitative structure–activity relationship (QSAR).
2. Nano–bio–eco interactions
2.1. Nano–eco interactions
The term “nano–eco interactions” here refers to the interactions at the environment–nanomaterial interface, particularly in aquatic ecosystems8 and terrestrial environments.38 Nano–eco interactions generally involve physicochemical interactions with abiotic environments (e.g., surfactants, dissolved organic matter or DOM39). It becomes clear that physicochemical properties of nanomaterials are very likely to be altered once they are released or discharged. First and foremost, the aggregation/agglomeration status of MONMs can be substantially affected by various factors, including pH, ionic strength, organic matter (DOM in aqueous columns), surfactants, temperature, and even clay content in soil matrices. This leads to the alteration of their behavior and fate in the environment. The interactions between MONMs and colloids can be electrostatic/steric40 or related to collision efficiency.41 At any given pH, an increase in ionic strength can increase aggregation profoundly.42 It has been suggested that the aggregation rate of TiO2 nanoparticles within porous media can be quite comparable to that of deposition, at ratios of porous media surfaces (collectors) to nanoparticle surface areas as high as 40.43 In addition, one report indicates that the transition from the reaction to diffusion-limited aggregation occurs at an electrophoretic mobility from around −2 to −0.8 μm s−1 V−1 cm.41 Numerous studies have demonstrated that MONMs exist as aggregates/agglomerates in water41,44 and soil matrices.45,46 Moreover, aggregation/agglomeration status could not only determine the mobility of nanomaterials, but also largely influence their bioavailability.Second, the surface chemistry of MONMs can be changed upon release or discharge. The high specific surface area of MONMs may result in strong adhesion to colloids, e.g., minerals and organic matter, in water and soil columns,47 leading to the alteration of their surface properties. For instance, the surface adsorption of phosphate to CeO2 nanoparticles leads to a significant reduction in their non-equilibrium retention (Kr) values upon addition of phosphate to soils.48 Similarly, Xu et al. (2012) also observed that ZnO and CuO nanoparticles can bind various constituents such as Na, Ca, P, and Cl from biological environments to form an ion corona, as shown in Fig. 1, with or without the addition of a biological environment.49 Surface charge and charge density may also be altered by the addition of organic matter and by ionic strength. Generally, an increase in organic matter may result in a domination of the charge of the organic matter at the surface of MONMs; and the increase of ionic strength can neutralize the surface charge of MONMs, for instance, TiO2, ZnO and CeO2.41 The alteration of the surface chemistry could in turn change the aggregation/agglomeration status of MONMs. More importantly, modifying a surface with organic matter may also affect the potential nanotoxicity by altering reactive oxygen species (ROS) production. It was suggested that humic acid (HA) accounts for the prevention of adhesion and inhibition of ROS generation, thus leading to reduced nanotoxicity.50
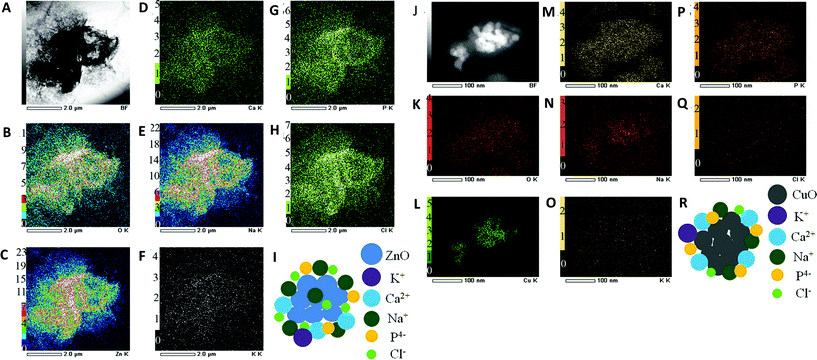 | ||
| Fig. 1 Formation of ZnO nanoparticle complexes without fetal bovine serum (FBS) (A–I) and CuO nanoparticle complexes with FBS (J–R) in high-glucose Dulbecco's modified Eagle's medium (DMEM). TEM image (A) and dark-field TEM image (J) where the elemental maps were obtained; (B, K) TEM/EDS O-K map; (C, L) TEM/EDS Zn-K map; (D, M) TEM/EDS Ca-K map; (E, N) TEM/EDS Na-K map; (F, O) TEM/EDS K-K map; (G, P) TEM/EDS P-K map; (H, Q) TEM/EDS Cl-K map; (I, R) a simple model of ZnO biocomplexes. Permission obtained from Sci. Rep., 2012, 2, 406.49 Copyright Nature Publishing Group. | ||
Additionally, metal ion dissolution can also be greatly affected by pH, ionic strength, and organic matter. It has been shown that the dissolution of ZnO nanoparticles is enhanced at both low and high pH,51,52 as well as high ionic strength.52 However, natural organic matter either enhances or reduces ZnO dissolution, depending on their chemical composition and concentration.51,52 For instance, the presence of citric acid significantly enhanced the extent of Zn2+ release,53 similar to the case of an elevated Cu2+ release from CuO nanoparticles in the presence of Suwannee river fulvic acid.54 Moreover, Zn2+ release can also be affected via ion trapping by an organic matter complex, thereby resulting in decreased toxicity.55
Notably, in addition to chemical factors, physical factors such as sunlight may trigger photocatalytic activity.30,56–58 Light irradiation may substantially affect the physicochemical properties of MONMs in various ways. First, light irradiation may accelerate metal ion dissolution of MONMs. For instance, it is quite well known that the dissolution of ZnO NPs can be enhanced by UV or solar irradiation, which in turn leads to alteration of nanotoxicity.59,60 Second, the crystallinity of MONMs can also be altered. It was reported that photoinduced phase transition (from anatase to rutile) of TiO2 nanoparticles is initiated by intragap irradiation.61 In addition, energy transition occurs within MONMs or their complexes upon the absorption of radiant energy. Photoinduced electron transfer in quantum dot–metal oxide nanoparticle junctions was also reported.62,63 It is also well known that light irradiation can initiate and enhance ROS formation in MONMs.64
2.2. Nano–bio interactions
In addition to nano–eco interactions, understanding the interaction of MONMs with macromolecules, tissues, and organs in biological systems in vitro and in vivo (Fig. 2) will permit us to design a safe nanomaterial for different application scenarios. Intuitively, nano–bio interactions often refer to biotic biomolecule–nanomaterial interactions.8,65 Nano–bio interactions are rather much more complex than those at the nano–eco interface. One should note that protein coronas play an important role in determining the behavior and fate of nanomaterials in biological environments. This particular phenomenon may result in denaturation of proteins, particle wrapping, and biocatalytic processes.66–68 It should also be noted that nano–bio interactions may lead to phase transformations, free energy releases, particle aggregation, restructuring and dissolution at the nanomaterial surface.69 Highly abundant proteins such as immunoglobulin G (IgG), fibrinogen, apolipoproteins, serum albumin, serotransferrin, prothrombin, alpha-fetoprotein, and kininogen-1 are commonly found on MONMs.70,71 The formation of nanomaterial–protein coronas is hydrodynamically associated with the physicochemical properties of nanomaterials, involving different adsorption mechanisms such as entropy-driven binding.72 The thickness of the protein coronas is related to various factors such as protein concentration, particle size, and surface properties of the particle.73 It was reported that all tested MONMs, including Fe3O4, CoO, and CeO2, form a hard protein corona through a dynamic process with different temporal patterns of the protein corona formation, suggesting a possible fingerprint for nanoparticle identification.74 The surface properties of MONMs profoundly affect the formation of protein coronas. It was suggested that surface coatings with negative and neutral surface charges adsorb more serum proteins than the positively charged ones, leading to a higher blood circulation time.75 Similar results were also reported in a metallic nanoparticle–protein complex.76 Thus, the colloidal stability of MONMs can also be possibly altered along with the evolution of the protein corona.70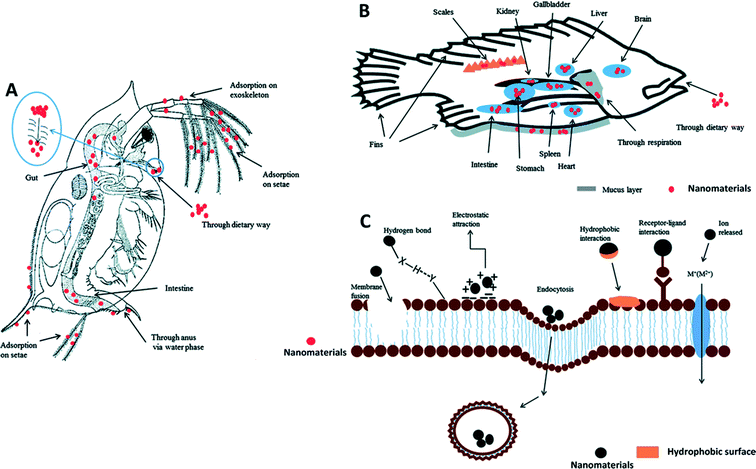 | ||
| Fig. 2 Example of how nanomaterials interact with living organisms at the nano–bio interface. Uptake and distribution of nanomaterials are illustrated in daphnia (A) and fish (B). The interactions between nanomaterials and cell membrane are illustrated in (C). Receptor–ligand interactions, hydrophobic interactions, electrostatic attractions and hydrogen bonds are often involved in the adsorption of nanomaterials onto the cell membrane. Membrane fusion and endocytosis may occur during the internalization of nanomaterials. Metal ions released from dissolvable metal oxide nanomaterials can be transported into the cells via certain membrane channels. Permission obtained from Environ. Sci.: Processes Impacts, 2013, 15, 145–160.77 Copyright Royal Society of Chemistry. | ||
In addition, the nature and complexity of the protein coronas formed on the surface of MONMs can impact the distribution of nanomaterials in a biological system. Generally, there are two types of protein coronas: soft and hard coronas. Nanomaterials that adsorb proteins with low affinity form soft protein coronas, and nanomaterials with tightly bound proteins form hard protein coronas. Thus, it is expected that hard coronas can directly interact with nanomaterials, whereas soft coronas interact with nanomaterials through protein–protein interactions with hard coronas. As we have discussed above, surface coating normally allows the formation of soft coronas with weak coronal covering.78 It was also observed that the tightly bound proteins occur only on MONMs with negatively charged surfaces after the strong protein elution.75
The interaction between nanomaterials and lipids is also critical in determining their behavior and fate in biological systems. It was found that the entrapment of superparamagnetic maghemite nanoparticles (γ-Fe2O3) in lipid bilayers reduces the lipid transition temperature and increases the membrane fluidity of all three types of lipid vesicles, including 1-stearoyl-2-oleoyl-sn-glycero-3-phosphocholine (SOPC), 1-stearoyl-2-oleoyl-sn-glycero-3-phosphocholine and 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (SOPC-POPS).79 They suggested that the negatively charged SOPC-POPS mixture is more predominant in this process due to high density encapsulation of nanoparticles via electrostatic interaction with positively charged γ-Fe2O3 nanoparticles.79 Besides, the interaction of nanomaterials with nucleic acid has also been studied. Recently, Magro et al. (2015) reported that γ-Fe2O3 nanoparticles interact chemically and electrically with DNA by direct covalent binding.80 Reversible electron transfer at the interface between γ-Fe2O3 nanoparticles and DNA, as well as the generation of holes on the DNA bases, were observed. The interaction may be affected by the nucleic acid length, presence of terminal phosphates, and types of DNA (dsDNA and ssDNA).81
2.3. Nano–bio–eco interactions
Studies on nano–bio interactions or nano–eco interactions have been relatively well covered. However, by comparison it was found that nano–bio–eco interactions remain as a critical topic worthy of additional research in the area of nanotoxicology.31,82–85 One should not ignore the fact that nano–bio and nano–eco interactions are often intertwined. For instance, ionic strength can affect protein corona formation on SPIONs.75 Characterizing the nano–bio–eco interactions requires understanding of biotic and abiotic dynamics of the surrounding environments and their interaction with nanomaterials (see Fig. 3). The dynamic microenvironments could result in uncertainty to the fate and behavior of nanomaterials at the cellular level. For example, the simultaneous interactions among a nanomaterial, ingredients of the abiotic environment (e.g. DOM, solar irradiation8), and a component of the target biota in a suspension medium can greatly modify the environmental fate of the nanomaterial and the nanotoxicological response of the cellular system, as a result of alteration of the physicochemical properties of the biological system and the nanomaterial at the boundaries of the suspension. In addition, the quantitative characterization of such nano–bio–eco interactions could foster the development of better MONMs with low hazardous risks,8,29,69,86 and ultimately, enable their wider applications in medicine, commercial products, and environmental protection. It is worthy to note that while statistical analysis of studies at the nano–bio–eco interface can mathematically indicate interactions between or among factorially-arranged biological and abiotic variables in an ecosystem experiment, interpretation of those interactions in an ecological sense is not an easy task.31 A series of more carefully controlled experiments is needed to reveal the separate nano–bio–eco interactions.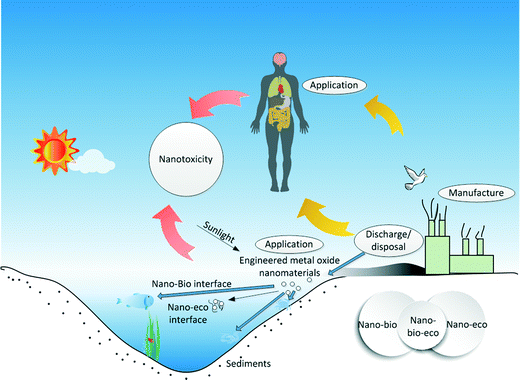 | ||
| Fig. 3 Illustration of the three-way nano–bio–eco interactions during the manufacture, use, and disposal of metal oxide nanomaterials. | ||
Besides the above mentioned issues, nanotoxicologists also face technical challenges while conducting ecotoxicity tests of engineered nanomaterials. The challenges include the transformations of studied engineered materials in environmental test media (e.g., aggregation, dissolution, and other interaction of small molecules) and modes of nanomaterial interference (e.g., adsorption to the test assay components and generation ROS).83,85 Therefore, combined knowledge and skills in the areas of physics, chemistry, and biology of nanomaterials are needed for improving the accuracy of the future toxicological assessments.
3. Toxicity of engineered metal oxide nanomaterials mediated by nano–bio–eco–interactions
3.1. Mechanisms of nanotoxicity
Overproduction of ROS and the consequent production of oxidative stress induced by nanomaterials is a predominant mechanism leading to nanotoxicity.87 It has been reported that oxidative DNA damage is associated with mutagenesis, carcinogenesis, and aging-related diseases in humans.88,89The level of ROS generation induced by nanoparticles is dependent on the physical and chemical nature of the nanoparticles and the surrounding environment and may proceed through different mechanisms.87 The critical chemical and physical properties of engineered nanomaterials, including MONMS, which lead to the generation of ROS and nanotoxicity, include molecular size, shape, oxidation status, surface area, bonded surface species, surface coating, solubility, and degree of aggregation and agglomeration.88,89
It has been demonstrated that nanomaterials induce toxicity mediated by ROS in many biological systems, such as human erythrocytes and skin fibroblasts etc.87 Apparently, nano–eco interactions occur. As aforementioned in the Section “Nano–eco interactions”, engineered nanomaterials, including MONMs, involve physicochemical interactions with abiotic environments. All the above-described changes are the critical factors that lead to the generation of ROS, and thus afford nanotoxicity through nano–eco interactions. It becomes clear that nano–bio–eco interactions can easily mediate nanotoxicity.
Although there are reports claiming that there is no clear evidence of harm with regard to the current low discharge/release levels of nanomaterials (measured or measure-based predicted), it is well recognized that there is a knowledge gap in the behavior and fate of nanomaterials in the dynamic environments that they may encounter.90 Thus, their potential hazard to biological systems urgently needs to be understood and projected. In this context, nanotoxicology has been widely studied in recent years. Various mechanisms of nanotoxicity have been proposed and published. Oxidative stress via overproduction of ROS is regarded as one of the major underlying causes of cellular damage and death.87 Other possible mechanisms include dissolution of metal ions,91 physical damage via direct contact, etc. Internalization of nanomaterials in an organism may also lead to intracellular responses/alterations. Generally, all those factors do not act individually; instead, a combination of multiple factors may be involved in any process. For instance, increasing the solution pH, and the amount of HPO42− and DOM can reduce the availability and/or dissolution of Zn2+ from ZnO nanoparticles, thus reducing the nanotoxicity.92 Below, we briefly illustrate the principles that apply to studying nanotoxicology.
3.2. Approaches to studying nanotoxicology
Recently, an integrative approach involving a microbeam mapping technique of Synchrotron Radiation X-Ray Fluorescence Analysis (SRXRF) was developed for studying the microdistribution of TiO2 nanoparticles.95 Wang and co-workers highlighted this approach with an absolute detection limit of 10−12 to 10−15 g in vivo,95 as shown in Fig. 4. Similarly, with a view to redesign safe nanoparticles, Vranic et al. (2013) reported an alternative method using innovative imaging flow cytometry in conjunction with confocal microscopy to identify the physicochemical characteristics of the SiO2 nanoparticles involved in their uptake,96 as shown in Fig. 5. However, quantitative approaches in evaluating the biodistribution of nanomaterials are still largely limited at the nanoscale.
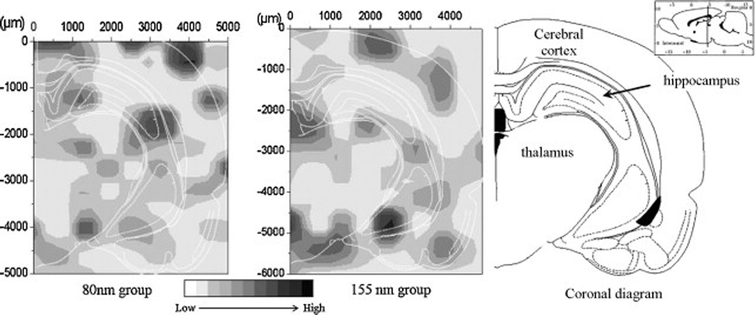 | ||
| Fig. 4 SRXRF mapping of Ti-element distribution in the brain sections at 30 days after intranasal instillation of the differently-sized TiO2 particles.95 Permission obtained from Toxicol. Lett., 2008, 183(1–3), 72–80.95 Copyright Elsevier B.V. | ||
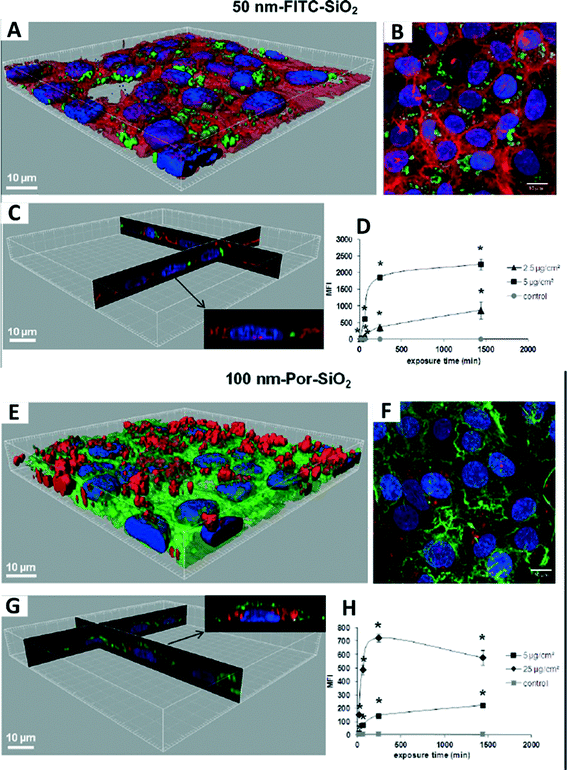 | ||
| Fig. 5 Interaction of 50 nm FITC-SiO2 (A–D) and 100 nm Por-SiO2 (E–H) nanoparticles with human lung adenocarcinoma (NCI-H292) cells. (A, E) 3D reconstruction of a confocal analysis of the cells exposed to SiO2 NPs. Blue: DAPI-stained nuclei, red: TRITC-phalloidin-stained actin filaments, green: FITC/Por-labelled SiO2 nanoparticles. (B, F) A projection of all images acquired in the stack of (A, E). (C, G) 3D reconstruction of x, z and y, z-slices of the corresponding regions on (A, E). The insert shows one selected representative cell. (D, H) Cells were exposed to nanoparticles, followed by flow cytometry (FCM) analysis of median fluorescence intensity (MFI). *p < 0.05, significantly different from the previous time point. Permission obtained from Part. Fibre Toxicol., 2013, 10(1), 2.96 Copyright BioMed Central Ltd. | ||
A single-nanoparticle detection method involving single-nanoparticle plasmonic microscopy and spectroscopy (dark-field optical microscopy and spectroscopy, DFOMS) and ultrasensitive in vivo assay (cleavage-stage zebrafish embryos, critical aquatic species) has been developed to study transport and toxicity of single silver97 and single gold98 nanoparticles on embryonic developments. This technique may also be used for MONMs. Li et al. (2012) successfully developed metal oxide nanoparticle-enhanced Raman scattering (MONERS) that can be applied for direct tracking and understanding of nano–bio–eco interactions at the single nanoparticle level.99 They monitored the photocatalytic decomposition of methylene blue (MB) by TiO2 NPs (P25, Degussa) using MONERS, suggesting its capability of direct molecular observation and understanding of chemical processes at a metal oxide interface. Notably, recent advances in hyperspectral microscopy with enhanced dark-field optical microscopy and hyperspectral imaging (HSI) enable the rapid identification of materials at the micro- and nanoscales with a detection limit of 10–15 nm for MONMs.100–103 For instance, Fig. 6 shows the identification of TiO2 naoparticles in lung tissues. The emerging hyperspectral microscopy exhibits great potential for assessing spatial distribution and spectral characteristics of MONMs in biological and environmental systems, facilitating studies on the fate and transformation of these particles in various environments.101
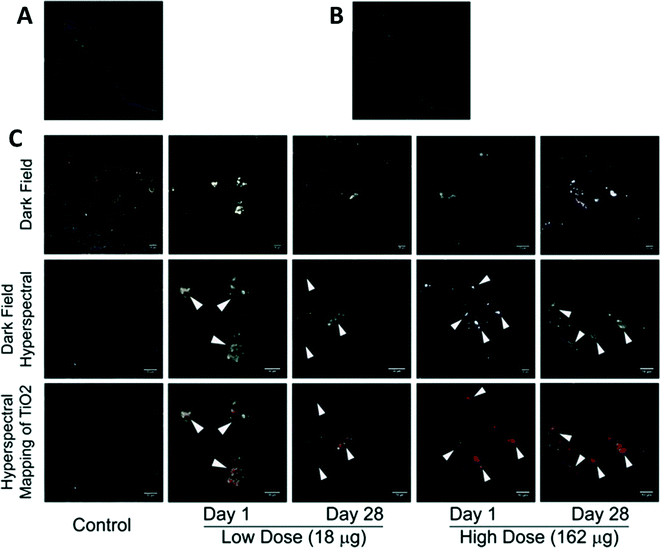 | ||
| Fig. 6 Identification of TiO2 naoparticles in lung tissues. (A) Reference spectral library from TiO2 exposed tissue; (B) reference spectral library from the control tissue; and (C) dark field images from nano-TiO2 exposed tissues (upper panel). Dark field hyperspectral images from TiO2 exposed tissues identifying these nanoparticles, which appeared as aggregates of white inclusions (middle panel). Hyperspectral mapping of TiO2 nanoparticles in these tissues appeared as red dots or aggregates (bottom panel). Permission obtained from Toxicol. Appl. Pharmacol., 2013, 269(3), 250–262.103 Copyright Elsevier B.V. | ||
In addition, studying nanotoxicity at the single cell level is also critical in establishing toxicological profiles quantitatively. Some cell populations, not all of them, may be vulnerable to their exposure to nanomaterials. The cell cycle is also one of the crucial factors underlying nanotoxicity and hence nanotoxicology. In light of this knowledge, nanotoxicity assaying at the single-cell level has been proposed based on flow and scanning image cytometry.104 Furthermore, magnetophoresis, combining fluorescence-based cytotoxicity assaying, is also in practice for assessing the viability and uptake of the single-cellular magnetic nanoparticles (MNPs) simultaneously.105 Notably, the integration of a cell-on-a-chip (CoC) with a microfluidic system has also been proposed for nanotoxicity assessments at the single-cell level.106
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O] mode. However, peaks are shifted to 1571 and 1391 cm−1 upon deprotonation owing to υas[COO−]) and υs[COO−] modes respectively. Moreover, success with ATF-FTIR has also been made to study the photocatalytic peroxidation of E. coli, lipo-polysaccharide (LPS), phosphatidyl-ethanolcholine (PE), and peptidoglycan (PGN) of the E. coli membrane wall on TiO2 porous films,109 and the settlement of Undaria pinnatifida kelp spores on anatase TiO2 films.110 Overall, ATR-FTIR can provide usable information, in a qualitative way, on the surface functionality, which may further imply surface charge, aggregation and sedimentation behaviors, cellular uptake, and ultimately toxicity.
O] mode. However, peaks are shifted to 1571 and 1391 cm−1 upon deprotonation owing to υas[COO−]) and υs[COO−] modes respectively. Moreover, success with ATF-FTIR has also been made to study the photocatalytic peroxidation of E. coli, lipo-polysaccharide (LPS), phosphatidyl-ethanolcholine (PE), and peptidoglycan (PGN) of the E. coli membrane wall on TiO2 porous films,109 and the settlement of Undaria pinnatifida kelp spores on anatase TiO2 films.110 Overall, ATR-FTIR can provide usable information, in a qualitative way, on the surface functionality, which may further imply surface charge, aggregation and sedimentation behaviors, cellular uptake, and ultimately toxicity.
Other instrumentation techniques such as fluorescence microscopy and TEM are also frequently used in fulfilling such needs. Fluorescence microscopy is highly sensitive to specific fluorescent dyes at a certain excitation and emission wavelength. Also, TEM is one of the most efficient ways to identify the internalization of nanomaterials in vivo/in vitro. For example, as shown in Fig. 7, ROS production and biodistribution of TiO2 nanoparticles are revealed in a zebrafish larva on a daily basis.28 Both techniques are highly visualizable, and can be further modified and improved for multiple purposes, particularly enabling semi-quantitative or quantitative analysis. Tai et al. (2012) reported that a microchip nanopipet with a narrow chamber width could be applied to TEM image-based quantitative characterization.111 They successfully developed a nanopipet with a narrow chamber width for sorting nanoparticles from blood and preventing the aggregation of the particles during the preparation process, thus enabling quantitative analysis of their aggregation/agglomeration states and the particle concentration in aqueous solutions. Techniques such as confocal microscopy and flow cytometry can also be used to study particle uptake and subcellular localization in a semi-quantitative approach.112
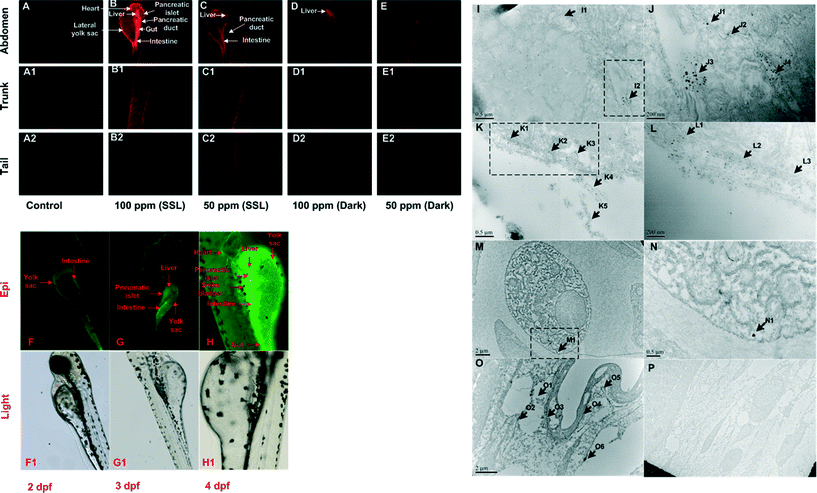 | ||
Fig. 7 Time-dependent biodistribution of TiO2 nanoparticles and their ROS production in zebrafish larva (Danio rerio). (A–E): Dihydroethidium (DHE) detection of superoxide yield at 96 hpf. Epi-fluorescence (F–H) and light microscopy (F1–H1) images of FITC-S-TiO2-treated zebrafish larvae at 2, 3, and 4 dpf. (I–P): TEM of S-TiO2 NPs (100 ppm)-treated embryos (120 hpf). Image (J), (L) and (N) are higher magnification images of NPs in the rectangular region of images (I), (K) and (M), respectively. Magnification for the images: (I) 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×, (J) 100 000×, (J) 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×, (K) 40 000×, (K) 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×, (L) 100 000×, (L) 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×, (M) 15 000×, (M) 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×, (N) 30 000×, (N) 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×, (O) 12 000×, (O) 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000× and (P) 5000×. *hpf: hours post fertilization. dpf: days post fertilization. FTIC: fluorescein isothiocyanate. Permission obtained from Nanotoxicology, 2014, 8(S1), 185–195.28 Copyright Informa Plc. 000× and (P) 5000×. *hpf: hours post fertilization. dpf: days post fertilization. FTIC: fluorescein isothiocyanate. Permission obtained from Nanotoxicology, 2014, 8(S1), 185–195.28 Copyright Informa Plc. | ||
Notably, the integration of the microscope and the Raman spectrometer now allows rapid and easy sample collection, preparation, and analysis in a qualitative way.113 In addition, developing a reliable qualitative method may provide a prototype that can be advanced for quantitative use. For instance, both DFOMS and Hyperspectral Imaging System are developed on the basis of optical microscopy.
3.3. Correlation between nano–bio–eco interactions and nanotoxicology
-to study the translocation/distribution of nanomaterials in biotic/abiotic systems;
-to study the exposure route to biota and humans;
-to identify and monitor the quantity of released nanomaterials in environments;
-and to study the relationship between physicochemical properties of nanomaterials and nanotoxicity with various biological endpoints.
It is prudent to characterize the physicochemical properties of nanomaterials thoroughly prior to any further studies.114,115 Indeed, measurements of particle characteristics in pure water may vary tremendously from those in cell culture media or water samples from lakes/rivers, etc. The alteration of their physicochemical properties tends to change their distribution and behavior in biotic/abiotic environments. Of course, the exposure route also matters most.116,117 The action mechanism and outcome may vary substantially with different routes. The exposure route often includes inhalation, direct contact (i.e. penetration through skin), and ingestion. Exposure may also occur in drug delivery and treatment. Meanwhile, the identification and monitoring of the release/discharge of nanomaterials is paramount in giving validation and credence to nanotoxicology in the long run.118,119 Ultimately, our goal is to generate a group of computational models that can correlate and explain the relationship between the physicochemical properties of nanomaterials and their toxicity, on the solid foundation of experimental researches.
| Tested MONMs | Modeling techniques | Descriptors | Description system | Correlation efficient (represented by (R2, RMSE), if applicable) | Biological model | Reference | |
|---|---|---|---|---|---|---|---|
| Correlation (training set) | Correlation (validation set) | ||||||
| a Density functional theory (DFT), decision treeboost (DTB), decision tree forest (DTF), genetic algorithm-multiple linear regression (GA-MLR), Human Umbilical Vein Endothelial Cells (HUVEC), “liquid drop” model (LDM), multiple linear regression (MLR), Molecular Operating Environment (MOE), spin–lattice relaxivity (R1) and spin–spin relaxivity (R2), Random Forest (RF), root mean square error (RMSE), standard error of estimation (SEE), standard error of external prediction (SEP), simplified molecular input-line entry system (SMILES), self-organizing map (SOM), and support vector machine (SVM). | |||||||
| 17 Metal oxide NPs | Monte Carlo | SMILES-based optimal descriptors | SMILES | R 2 = 0.90–0.94 | R 2 = 0.73–0.98 | E. coli | 135 |
| 17 Metal oxide NPs | DTB | Oxygen percent, molar refractivity and polar surface area | Molecular descriptor | (0.955, 0.11) | E. coli | 133 | |
| DTF | (0.896, 0.19) | ||||||
| 17 Metal oxide NPs | MLR | Enthalpy of formation (ΔHMen+) | DFT | (0.85, 0.20) | (0.83, 0.19) | E. coli | 36 |
| 17 Metal oxide NPs | Monte Carlo | SMILES-based optimal descriptors | SMILES | R 2 = 0.83–0.96 | E. coli | 134 | |
| 17 Metal oxide NPs | Absolute electronegativity | DFT | F = 33.83, R2 = 0.87 (dark exposure) | E. coli | 29 | ||
| Molar heat capacity and average of the alpha and beta LUMO energies | F = 20.51, R2 = 0.804 (photo exposure) | ||||||
| 18 Metal oxides | RF regression | van der Waals interactions, electronegativity and metal–ligand binding characteristics | LDM-based descriptors | (0.96, 0.10) | (0.93,0.13) | HaCaT cells | 136 |
| (0.92,0.12) | (0.78,0.32) | E. coli | |||||
| 18 Metal oxides | GA-MLR | ΔHfc: enthalpy of formation of metal oxide nanocluster representing a fragment of the surface and χc the Mulliken's electronegativity of the cluster | 27 Nanodescriptors including 16 quantum-mechanical descriptors and 11 image descriptors | R 2 = 0.93, RMSE = 0.12 | E. coli and HaCaT cells | 137 | |
| TiO2 | MLR and LDA | Engineered size, size in ultrapure water, size in PBS, and concentration in ultrapure water | General descriptor | With R2 up to 0.77 | Rat L2 lung epithelial cells and rat lung alveolar macrophages | 130 | |
| ZnO | Engineered size, size in ultrapure water, size in PBS, and size in CCM | R 2 = 0.94–0.99 | |||||
| ZnO and TiO2 | Monte Carlo | Optimal descriptor | Squasi-SMILES | R 2 = 0.78–0.92 | R 2 = 0.67–0.83 | Human lung epithelial cells | 135 |
| TiO2 | Monte Carlo | Optimal descriptor | SMILES-based optimal descriptors | (0.9639, 0.049); (0.9893, 0.025); (0.9792, 0.049) | (0.9263, 0.123); (0.8959, 0.118); (0.9647, 0.066) | Human lung epithelial cells | 141 |
| 17 Metal oxide NPs | MLR | Metal electronegativity (χ), the charge of the metal cation corresponding to a given oxide (χox), atomic number and valence electron number of the metal | SMILES | (0.81–0.90, 0.16–0.22) | (0.73–0.96, 0.15–0.26) | E. coli | 150 |
| PLSR | (0.73–0.87, 0.19–0.27) | (0.70–0.96, 0.17–0.29) | |||||
| 15 Metal oxide nanoparticles | PM6 method | Spherical size of nanoparticles and the weighted energy of the highest occupied molecular orbital | Microscopic-image-based and theory-based (calculated) descriptors | R 2 = 0.82–0.94 | NA | 151 | |
| 9 Metal oxide nanoparticles | Logistic regression | Atomisation energy, period of the nanoparticle metal, primary size, and volume fraction | Molecular, chemical and physical information and different concentrations | Accuracy >95% | Bronchial epithelial (BEAS-2B) cells | 128 | |
| 24 Metal oxide nanoparticles | SVM | Conduction band energy and ionic index | An initial pool of 30 NP descriptors | Accuracy ~94% and confidence level of 80% | Human bronchial epithelial (BEAS-2B) and murine myeloid (RAW 264.7) cells | 121 | |
| 24 Metal oxide nanoparticles | Monte Carlo | Optimal descriptors | SMILES | Best model with R2 = 0.8824, RMSE 0.214 for calibration set; and R2 = 0.7809, RMSE = 0.348 for validation set | Human bronchial epithelial cells (BEAS-2B) | 140 | |
| 24 Metal oxide nanoparticles | Markov Chain Monte Carlo (MCMC) | Conduction band energies, dissolution | General descriptor | NA | Human bronchial epithelial cells (BEAS-2B) | 144 | |
| SiO2 | Monte Carlo | Mathematical functions of size and concentration | Optimal descriptors | R 2 = 0.9837, s = 2.53 %, F = 483 | R 2 = 0.9269, s = 7.94 % | Human lung fibroblasts | 139 |
| 70 metal oxide nanoparticles | NA | Band energy | Reactivity descriptors | Accuracies of ca. 99% in both training and prediction sets | NA | 129 | |
| 41 Nanoparticles with 6 metal oxide nanoparticles | Perturbation approach | Molar volume, polarizability, size | Molar volume (V), electronegativity (E), polarizability (P), and nanoparticle size (L) | Accuracy > 93% for both training and prediction sets | 15 Mammalian cell lines, including A549 human cells | 143 | |
| 11 Metal oxide and 7 metallic nanoparticles | MLR and LDA | Four types of 9-variable descriptors | Molar volume (V), electronegativity (E), polarizability (P), and nanoparticle size (L) | Accuracies of ca. 99% in both training and prediction sets | Multiple bioindicators, including D. magna, P. subcapitata, D. rerio, etc | 142 | |
| 48 Fe2O3 and Fe3O4 metal oxide nanoparticles and 3 CdSe quantum dots core with various coating combinations | MLR, and sparse linear modeling and feature selection, MLR-EM | Relaxivities (R1 and R2) and the zeta potential | A set of 691 molecular descriptors | Training set R2 = 0.81; test set regression coefficient R2 = 0.86; SEE = 3.6; and SEP = 3.3 | Endothelial and smooth muscle cells, monocytes, and hepatocytes | 131 | |
| Nonlinear Bayesian regularized artificial neural network methods | Training set R2 = 0.80; test set R2 = 0.90; SEE = 2.8; and SEP = 2.9 | ||||||
| 109 Nanoparticles sharing a superparamagnetic core and dextran coating | Linear | 11 Descriptors | Derived from a set of 124 chemically interpretable descriptors | Training set R2 = 0.74; test set R2 = 0.63; SEE = 0.34; and SEP = 0.36 | Significant variation in HUVEC | ||
| Non-linear | Training set R2 = 0.70; test set R2 = 0.66; SEE = 0.30; and SEP = 0.33 | ||||||
| Linear | 19 Descriptors | Training set R2 = 0.76; test set R2 = 0.79; SEE = 0.19; and SEP = 0.24 | Significant variation in PaCa2 cells | ||||
| Non-linear | Training set R2 = 0.77; test set R2 = 0.54; SEE = 0.15; and SEP = 0.28 | ||||||
| 109 Nanoparticles sharing a superparamagnetic core and dextran coating | kNN QSAR models | Lipophilicity, van der Waals surface area, molecular refractivity, electrostatic descriptors | 2-D MOE descriptors | Coefficients of correlation Rabs2 ranged from 0.65 to 0.80 for external sets | Significant variation in PaCa2 cell line | 127 | |
| 48 Fe2O3 and Fe3O4 metal oxide nanoparticles and 3 CdSe quantum dots core with various coating combinations | SVM | Size, relaxivities, and zeta potential | Molecular descriptors | External prediction accuracies of 56 − 88% for the five independent external validation sets, with the mean external accuracy of 73% | Endothelial and smooth muscle cells, monocytes, and hepatocytes | ||
| 44 Iron oxide core nanoparticles | H4 class definition and naive Bayesian classifier (NBC) model | Spin–lattice relaxivity and zeta potential | Molecular descriptors | Classification accuracy > 78% | Aorta endothelial, vascular smooth muscle, hepatocyte, and monocyte/macrophage | 132 | |
| ‘hit’ (i.e., significant bioactivity) identification analysis and SOM based consensus clustering | Primary size, spin–lattice and spin-spin relaxivities, and zeta potentials | ||||||
Oftentimes, nanotoxicological profiles vary not only on biological models, but also on biological endpoints. Till now, across a wide range of biological endpoints, a considerable number of studies have been conducted to evaluate the impact of MONM exposure with regard to QSAR modeling. The majority of laboratory studies on biological endpoints are mainly in vitro studies, including linear/log-linear regression models of EC50/LC50 cytotoxicity,29,36,133,134,136,137 the concentration of nanoparticles leading to 50% reduction in cell viability (TC50),143 damage to cellular membranes (units L−1) via lactate dehydrogenase (LDH) release,130,135,140,141 oxidative stress,121 intracellular calcium flux,121 mitochondrial membrane potential,121,132,138 surface membrane permeability,121 cytotoxic inhibition ratio with MTT assay,139 cell apoptosis,131,132 ATP content,132,138 apoptosis,138 reducing equivalents,132,138 plasma membrane leakage,128 and cell membrane damage via propidium iodide uptake.140,144 A single indicator may not be sufficient sometimes; therefore, multiple cell types at multiple doses and with multiple endpoints may provide a more comprehensive view of the biological effects resulting from certain nanomaterials.138 Although QSAR with in vitro studies can imply some correlations with in vivo observations, QSAR with a direct observation in vivo can further promote predicting nanotoxicity with high accuracy and reliability.
One should not ignore that sufficiently large nanotoxicity datasets can be rapidly acquired with the advance in High Throughput Screening (HTS) assay.145 For instance, ten independent toxicity-related signaling pathways associated with murine macrophage cell line exposed to a library of MONMs can be readily obtained via HTS assay.146 Later, those data can either be classified through the use of certain computational techniques such as SOM,147 or further analyzed via QSAR modeling.128 Such hierarchical ranking and clustering of MONMs based on HTS basically provide an enormous in vitro profile network for further testing in vivo.121,138 In addition, HTS has shown promising potential for us to perform in vivo hazard risk assessment with high volume datasets.148,149In vivo studies are generally considered to be more definitive regarding nanotoxicity assessment. This is typically valuable in facilitating the establishment and utilization of QSAR models in designing safe nanomaterials. Conventionally, obtaining valid scientific data is quite slow and somewhat objective in some cases. With the help of HTS assay, scientists and researchers can be relieved from intensive lab work and focus more on method development and data analysis. There is a trend in academia for universities and institutions to apply this relatively novel technique in their researches.
Note that the rising popularity of QSAR modeling is essentially associated with a question over their reliable predictions. Thus, various modeling techniques have been acquired and applied in such context. Modeling techniques such as decision tree forest (DTF) and decision treeboost (DTB),133 multiple linear regression (MLR),36,131,142,150 naive Bayesian classifier (NBC) modeling,132 self-organizing map (SOM),132 Random Forest (RF) regression,136 logistic regression (LR),128 k-nearest neighbor (k-NN),127 partial least square regression (PLSR),150 support vector machines (SVM),121,127 ensemble learning (EL),133 linear discriminant analyses (LDA),130,131,142 sparse linear modeling and feature selection linear modeling,131 and Bayesian regularized artificial neural network methods131 have exhibited great potential in underlying the quantitative relationships between the molecular structures and biological activities of MONMs. It is worthy to note that most of the predictive outcomes generated by those modeling techniques are within the acceptable range.
Not only a range of pristine MONMs have been involved in QSAR studies,29,36,128,133–137,142,150 but also surface functionalization of certain MONMs is also studied and reported.127,131 There are a large number of molecular, chemical and physical descriptors of those MONMs available in databases. Selection of a proper descriptor is the most critical step in generating valid QSAR models with acceptable accuracy. Many descriptors can be obtained readily based on the molecular structure and atomic or group contributions, e.g., molecular weight, van der Waals, surface area, and size. Descriptors that relate to the electronic structure, for instance, molar heat capacity, alpha and beta LUMO energies, and electronegativity, are available from quantum chemical calculations. Currently, the simplified molecular input-line entry system (SMILES),134,135,140,150 “liquid drop” model (LDM)-derived descriptors,136 molecular operating environment (MOE),127 and optimal descriptors139 are the most frequently used databases for descriptor selection.
Ultimately, the aim of the QSAR approach is to predict the toxicological behavior of nanomaterials at the nano–bio–eco interface. With the advance in computational technology, QSAR studies are likely to play a vital role in the design of novel nanomaterials on the basis of acceptable reliability and accuracy. Prediction performances of QSAR models have shown great value in fulfilling such needs. Successful implementation of QSAR can certainly facilitate current progress on nanotoxicology in vitro and in vivo with a reasonable cost. Computational scientists in related disciplines can directly retrieve data from published literature, without being troubled by intensive lab work. Such “collaboration” could eventually not only benefit the research groups mutually, but also move the scientific community forward. One can envision that QSAR as a computational strategy can be a powerful tool in the future.
4. Perspective
It is clear that MONM behavior and fate at the nano–bio–eco interface is much more complicated than the pristine ones regarding the way in which nanomaterials interact with biotic (cells, tissues and organisms) and abiotic (pH, ionic strength, organic matter, etc.) environments. Acquiring key data at the nano–bio–eco interface is crucial in understanding the relationship between the physicochemical properties of nanomaterials and their related toxicity. Although a holistic approach is suggested and demanded in order to fully understand how nanomaterials behave in a connected ecosystem,8 conclusive answers to the question over the threats posed by engineered nanomaterials are perhaps unlikely to be revealed, and will probably need to be assessed on a case-by-case basis within any given context. In addition, numerous data published in the literature sometimes may seem controversial. Not only should we continue building database profiles for existing MONMs, but also start establishing a standard system as a reference. Building up routine methods for nanotoxicity evaluation warrants higher reproducibility and reliability of data.One should recognize that quantitative analysis is always necessary in the quest to understand nanotoxicity at the nano–bio–eco interface. With the help of qualitative approaches, one can observe in a wide view, not limited to some definitive figures. We should also be cautious when it comes to the comparison between in vitro and in vivo observations because failure may often exist. Despite the different levels of complexity between the in vitro and in vivo studies at the nano–bio–eco interface, observations in vitro are less labor-intensive and more cost-efficient in most cases. Such merits also allow assessing nanotoxicity rapidly with sensitive and reliable HTS assay. Toxicological profiles can be readily generated with multiple biological models, multiple biological endpoints, and multiple types of nanomaterials. Collectively, the sum of the responses across in vitro and in vivo experiments can be retrieved and analyzed with QSAR repeatedly over time. Unlike laboratory work, the computational approach has its merits of being reliable and reproducible. One fundamental thing that needs to be handled properly is the provision of a toxicological profile based on reliable experimental studies with as much accuracy as possible. Due to their capability of forming a vast number of structural geometries with an electronic structure, MONMs play an important role in nanotechnology and possess advantages for propelling QSAR model development.90 Nowadays, most QSAR studies still mainly focus on the risk assessment of pristine MONMs, that is, with no consideration of doping or surface modification. The increasing demand on novel MONMs with intentional tailoring forces us to pay more attention to newly developed nanomaterials.
Till now, many challenges that remain before the above mentioned approaches in the study of nanotoxicology can be put into practice, though the rate of progress has been laudable. Further studies are still required, further advances are still occurring, and more remain to be revealed. Greater knowledge of how MONMs interact at the nano–bio–eco interface is also needed. Ultimately, more open collaborations between industry, academia, and research labs need to be formed. Moreover, there is a need to take a broader look at facilitating nanotoxicological studies, and to focus specifically on the interactions at the nano–bio–eco interface, leading to safe and effective nanotechnology-driven MONMs for commercial, environmental and medicinal use.
Acknowledgements
This study was supported by the NSF-CREST program (National Science Foundation-Centers of Research Excellence in Science and Technology) with grant # HRD-0833178. This article is not an official U.S. Food and Drug Administration (FDA) guidance or policy statement. No official support or endorsement by the U.S. FDA is intended or should be inferred. The authors declare no competing financial interest.References
-
J. A. Rodriguez and M. Fernández-García, Synthesis, Properties, and Applications of Oxide Nanomaterials, John Wiley & Sons, Hoboken, NJ, 2007 Search PubMed
.
- O. C. Farokhzad and R. Langer, Adv. Drug Delivery Rev., 2006, 58, 1456–1459 CrossRef CAS PubMed
.
- P. G. Tratnyek and R. L. Johnson, Nano Today, 2006, 1, 44–48 CrossRef
.
-
M. Fernandez-Garcıa and J. A. Rodriguez, Metal Oxide Nanoparticles, in Nanomaterials: Inorganic and Bioinorganic Perspectives, ed. C. M. Lukehart and R. A. Scott, John Wiley & Sons Ltd, 2013 Search PubMed
.
- The Global Market for Metal Oxide Nanoparticles to 2020, Future Markets Inc., 2013, p. 322.
- X. He, W. G. Aker, M.-J. Huang, J. D. Watts and H.-M. Hwang, Curr. Top. Med. Chem., 2015, 15, 1887–1900 CrossRef CAS
.
- L. Treuel, K. A. Eslahian, D. Docter, T. Lang, R. Zellner, K. Nienhaus, G. U. Nienhaus, R. H. Stauber and M. Maskos, Phys. Chem. Chem. Phys., 2014, 16, 15053–15067 RSC
.
- X. He, W. G. Aker, J. Leszczynski and H.-M. Hwang, J. Food Drug Anal., 2014, 22, 128–146 CrossRef CAS PubMed
.
- L. Shang, K. Nienhaus and G. U. Nienhaus, J. Nanobiotechnol., 2014, 12, 5 CrossRef PubMed
.
- R. Grillo, A. H. Rosa and L. F. Fraceto, Chemosphere, 2015, 1119, 608–619 CrossRef PubMed
.
- S. J. Soenen, W. J. Parak, J. Rejman and B. Manshian, Chem. Rev., 2015, 115, 2109–2135 CrossRef CAS PubMed
.
- A. D. Dwivedi and L. Q. Ma, Crit. Rev. Environ. Sci. Technol., 2014, 44, 1679–1739 CrossRef CAS PubMed
.
- N. V. Moos, P. Bowen and V. I. Slaveykova, Environ. Sci.: Nano, 2014, 1, 214–232 RSC
.
- G. Amoabediny, A. Naderi, J. Malakootikhah, M. Koohi, S. Mortazavi, M. Naderi and H. Rashedi, J. Phys.: Conf. Ser., 2009, 170, 012037 CrossRef
.
- British Standard Institute (BSI), Nanotechnologies – Part 2: Guide to safe handling and disposal of manufactured nanomaterials, British Standards Institution, London, 2007. PD 6699-2:2007.
- OECD, Current Developments/Activities on the Safety of Manufactured Nanomaterials, H. a. S. P. S. o. t. S. o. M. N. OECD Environment, Ed. Organisation for Economic Co-operation and Development (OECD), Berlin, Germany, 2007.
- OECD, Important issues on risk assessment of Manufactured nanomaterials, Series on the Safety of Manufactured Nanomaterials No. 33 E. OECD Environment Directorate, Health and Safety Division, Ed. Organisation for Economic Co-operation and Development, Paris, 2012, ENV/JM/MONO(2012),. ENV/JM/MONO(2012)8.
- S. M. Hankin, S. A. K. Peters, C. A. Poland, S. Foss Hansen, J. Holmqvist, B. L. Ross, J. Varet and R. J. Aitken, Specific Advice on Fulfilling Information Requirements for Nanomaterials under REACH (RIP-oN 2) - Final Project Report REACH, Ed. 2011. RNC/RIP-oN2/FPR/1/FINAL.
- R. A. Aitken, A. Bassan, S. Friedrichs, S. M. Hankin, S. F. Hansen, J. Holmqvist, S. A. K. Peters, C. A. Poland and C. L. Tran, Specific Advice on Exposure Assessment and Hazard/Risk Characterisation for Nanomaterials under REACH (RIP-oN 3) - Final Project Report, REACH, Ed. 2011. RNC/RIP-oN3/FPR/1/FINAL.
- Matrix Insight Ltd, Request for services in the context of the FC ENTR/2008/006, lot 3: A Study to support the Impact Assessment of relevant regulatory options for nanomaterials in the framework of REACH, TMKG Ltd, 2014. http://ec.europa.eu/DocsRoom/documents/5826/attachments/1/translations/en/renditions/native
.
- ISO, Nanotechnologies - Occupational risk management applied to engineered nanomaterials -Part 1:Principles and approaches. , International Organization for Standardization, 2012. ISO/TS 12901–1:2012.
- ISO, Nanotechnologies - Health and safety practices in occupational settings relevant to nanotechnologies, 2008. ISO/TR 12885:2008.
- ECHA, Human health and environmental exposure assessment and risk characterisation of nanomaterials - Best practice for REACH registrants, European Chemicals Agency, Helsinki, Finland, 2014. http://echa.europa.eu/documents/10162/5399565/best_practices_human_health_environment_nano_3rd_en.pdf.
-
J. J. Yin, B. Zhao, Q. Xia and P. P. Fu, Electron spin resonance spectroscopy for studying the generation and scavenging
of reactive oxygen species by nanomaterials, in Nanopharmaceuticals: The Potential Application of Nanomaterials, ed. X.-J. Liang, World Scientific Publishing Company, Singapore, 2012, ch. 14 Search PubMed
.
- J.-J. Yin, J. Liu, M. Ehrenshaft, J. E. Roberts, P. P. Fu, R. P. Mason and B. Zhao, Toxicol. Appl. Pharmacol., 2012, 263, 81–88 CrossRef CAS PubMed
.
- P. P. Fu, J. Food Drug Anal., 2014, 22, 1–2 CrossRef PubMed
.
- W. He, W. Wamer, P. P. Fu and J. J. Yin, J. Environ. Sci. Health, Part C: Environ. Carcinog. Ecotoxicol. Rev., 2014, 32, 186–211 CrossRef CAS PubMed
.
- X. He, W. G. Aker and H.-M. Hwang, Nanotoxicology, 2014, 8, 185–195 CrossRef CAS PubMed
.
- K. Pathakoti, M.-J. Huang, J. D. Watts, X. He and H.-M. Hwang, J. Photochem. Photobiol., B, 2014, 130, 234–240 CrossRef CAS PubMed
.
-
H. M. Hwang, P. C. Ray, H. Yu and X. He, Toxicology of designer/engineered metallic nanoparticles, in Sustainable preparation of metal nanoparticles: methods and applications, ed. R. Luque, V. R. S., R. S. Varma, J. H. Clark and G. A. Kraus, Royal Society of Chemistry, Cambridge, UK, 2012 Search PubMed
.
- T. P. Dasari and H.-M. Hwang, J. Environ. Sci., 2013, 25, 1925–1935 CrossRef CAS
.
- T. P. Dasari, K. Pathakoti and H.-M. Hwang, J. Environ. Sci., 2013, 25, 882–888 CrossRef CAS
.
- Y. Wang, W. G. Aker, H.-m. Hwang, C. G. Yedjou, H. Yu and P. B. Tchounwou, Sci. Total Environ., 2011, 409, 4753–4762 CrossRef CAS PubMed
.
- E. Ying and H.-M. Hwang, Sci. Total Environ., 2010, 408, 4475–4481 CrossRef CAS PubMed
.
- X. Hu, S. Cook, P. Wang and H.-m. Hwang, Sci. Total Environ., 2009, 407, 3070–3072 CrossRef CAS PubMed
.
- T. Puzyn, B. Rasulev, A. Gajewicz, X. Hu, T. P. Dasari, A. Michalkova, H.-M. Hwang, A. Toropov, D. Leszczynska and J. Leszczynski, Nat. Nanotechnol., 2011, 6, 175–178 CrossRef CAS PubMed
.
- K. Pathakoti, S. Morrow, C. Han, M. Pelaez, X. He, D. D. Dionysiou and H.-M. Hwang, Environ. Sci. Technol., 2013, 47, 9988–9996 CrossRef CAS PubMed
.
- P. S. Tourinho, C. A. Van Gestel, S. Lofts, C. Svendsen, A. M. Soares and S. Loureiro, Environ. Toxicol. Chem., 2012, 31, 1679–1692 CrossRef CAS PubMed
.
- S. J. Klaine, P. J. Alvarez, G. E. Batley, T. F. Fernandes, R. D. Handy, D. Y. Lyon, S. Mahendra, M. J. McLaughlin and J. R. Lead, Environ. Toxicol. Chem., 2008, 27, 1825–1851 CrossRef CAS PubMed
.
- R. A. French, A. R. Jacobson, B. Kim, S. L. Isley, R. L. Penn and P. C. Baveye, Environ. Sci. Technol., 2009, 43, 1354–1359 CrossRef CAS
.
- A. A. Keller, H. Wang, D. Zhou, H. S. Lenihan, G. Cherr, B. J. Cardinale, R. Miller and Z. Ji, Environ. Sci. Technol., 2010, 44, 1962–1967 CrossRef CAS PubMed
.
- R. F. Domingos, N. Tufenkji and K. J. Wilkinson, Environ. Sci. Technol., 2009, 43, 1282–1286 CrossRef CAS
.
- N. Solovitch, J. R. M. Labille, J. R. M. Rose, P. Chaurand, D. Borschneck, M. R. Wiesner and J.-Y. Bottero, Environ. Sci. Technol., 2010, 44, 4897–4902 CrossRef CAS PubMed
.
- D. Lin, X. Tian, F. Wu and B. Xing, J. Environ. Qual., 2010, 39, 1–13 CrossRef PubMed
.
- M. Simonin, J. Guyonnet, J. M. Martins, M. Ginot and A. Richaume, J. Hazard. Mater., 2015, 283, 529–535 CrossRef CAS PubMed
.
- J. Fang, X.-q. Shan, B. Wen, J.-M. Lin and G. Owens, Environ. Pollut., 2009, 157, 1101–1109 CrossRef CAS PubMed
.
- M. N. Moore, Environ. Int., 2006, 32, 967–976 CrossRef CAS PubMed
.
- G. Cornelis, B. Ryan, M. J. McLaughlin, J. K. Kirby, D. Beak and D. Chittleborough, Environ. Sci. Technol., 2011, 45, 2777–2782 CrossRef CAS PubMed
.
- M. Xu, J. Li, H. Iwai, Q. Mei, D. Fujita, H. Su, H. Chen and N. Hanagata, Sci. Rep., 2012, 2, 406 Search PubMed
.
- D. Lin, J. Ji, Z. Long, K. Yang and F. Wu, Water Res., 2012, 46, 4477–4487 CrossRef CAS PubMed
.
- S.-W. Bian, I. A. Mudunkotuwa, T. Rupasinghe and V. H. Grassian, Langmuir, 2011, 27, 6059–6068 CrossRef CAS PubMed
.
- A. J. Miao, X. Y. Zhang, Z. Luo, C. S. Chen, W. C. Chin, P. H. Santschi and A. Quigg, Environ. Toxicol. Chem., 2010, 29, 2814–2822 CrossRef CAS PubMed
.
- I. A. Mudunkotuwa, T. Rupasinghe, C.-M. Wu and V. H. Grassian, Langmuir, 2011, 28, 396–403 CrossRef PubMed
.
- Z. Wang, J. Li, J. Zhao and B. Xing, Environ. Sci. Technol., 2011, 45, 6032–6040 CrossRef CAS PubMed
.
- M. Li, S. Pokhrel, X. Jin, L. Mädler, R. Damoiseaux and E. M. Hoek, Environ. Sci. Technol., 2010, 45, 755–761 CrossRef PubMed
.
- E. S. Bernhardt, B. P. Colman, M. F. Hochella, B. J. Cardinale, R. M. Nisbet, C. J. Richardson and L. Yin, J. Environ. Qual., 2009, 39, 1954–1965 CrossRef
.
- D. Lin, X. Tian, F. Wu and B. Xing, J. Environ. Qual., 2009, 39, 1896–1908 CrossRef
.
- M. R. Wiesner, G. V. Lowry, K. L. Jones, M. F. Hochella Jr., R. T. D. Giulio, E. Casman and E. S. Bernhardt, Environ. Sci. Technol., 2009, 43, 6458–6462 CrossRef CAS
.
- W.-M. Lee and Y.-J. An, Chemosphere, 2013, 91, 536–544 CrossRef CAS PubMed
.
- H. Ma, L. K. Wallis, S. Diamond, S. Li, J. Canas-Carrell and A. Parra, Environ. Pollut., 2014, 193, 165–172 CrossRef CAS PubMed
.
- P. C. Ricci, C. M. Carbonaro, L. Stagi, M. Salis, A. Casu, S. Enzo and F. Delogu, J. Phys. Chem. C, 2013, 117, 7850–7857 CAS
.
- K. Tvrdy, P. A. Frantsuzov and P. V. Kamat, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 29–34 CrossRef CAS PubMed
.
- B.-R. Hyun, Y.-W. Zhong, A. C. Bartnik, L. Sun, H. D. Abruna, F. W. Wise, J. D. Goodreau, J. R. Matthews, T. M. Leslie and N. F. Borrelli, ACS Nano, 2008, 2, 2206–2212 CrossRef CAS PubMed
.
- Y. Li, W. Zhang, J. Niu and Y. Chen, ACS Nano, 2012, 6, 5164–5173 CrossRef CAS PubMed
.
- L. C. Cheng, X. Jiang, J. Wang, C. Chen and R. S. Liu, Nanoscale, 2013, 7, 3547–3569 RSC
.
- A. E. Nel, L. Mädler, D. Velegol, T. Xia, E. M. V. Hoek, P. Somasundaran, F. Klaessig, V. Castranova and M. Thompson, Nat. Mater., 2009, 8, 543–557 CrossRef CAS PubMed
.
- A. Albanese, P. S. Tang and W. C. W. Chan, Annu. Rev. Biomed. Eng., 2012, 14, 1–16 CrossRef CAS PubMed
.
- M. Xu, J. Li, H. Iwai, Q. Mei, D. Fujita, H. Su, H. Chen and N. Hanagata, Sci. Rep., 2012, 2, 406 Search PubMed
.
- X. R. Xia, N. A. Monteiro-Riviere, S. Mathur, X. Song, L. Xiao, S. J. Oldenberg, B. Fadeel and J. E. Riviere, ACS Nano, 2011, 5, 9074–9081 CrossRef CAS PubMed
.
- M. Safi, J. Courtois, M. Seigneuret, H. Conjeaud and J.-F. Berret, Biomaterials, 2011, 32, 9353–9363 CrossRef CAS PubMed
.
- P. Aggarwal, J. B. Hall, C. B. McLeland, M. A. Dobrovolskaia and S. E. McNeil, Adv. Drug Delivery Rev., 2009, 61, 428–437 CrossRef CAS PubMed
.
- I. Lynch and K. A. Dawson, Nano Today, 2008, 3, 40–47 CrossRef CAS
.
- M. Mahmoudi, M. A. Shokrgozar and S. Behzadi, Nanoscale, 2013, 5, 3240–3244 RSC
.
- E. Casals, T. Pfaller, A. Duschl, G. J. Oostingh and V. F. Puntes, Small, 2011, 7, 3479–3486 CrossRef CAS PubMed
.
- U. Sakulkhu, M. Mahmoudi, L. Maurizi, J. Salaklang and H. Hofmann, Sci. Rep., 2014, 4, 5020 CAS
.
- E. Casals, T. Pfaller, A. Duschl, G. J. Oostingh and V. Puntes, ACS Nano, 2010, 4, 3623–3632 CrossRef CAS PubMed
.
- S. Ma and D. Lin, Environ. Sci.: Processes Impacts, 2013, 15, 145–160 CAS
.
- M. Lundqvist, J. Stigler, T. Cedervall, T. Berggård, M. B. Flanagan, I. Lynch, G. Elia and K. Dawson, ACS Nano, 2011, 5, 7503–7509 CrossRef CAS PubMed
.
-
P. B. Santhosh, S. Penič, J. Genova, A. Iglič, V. Kralj-Iglič and N. P. Ulrih, in A study on the interaction of nanoparticles with lipid membranes and their influence on membrane fluidity, Journal of Physics: Conference Series, IOP Publishing, 2012, p. 012034 Search PubMed
.
- M. Magro, D. Baratella, P. Jakubec, G. Zoppellaro, J. Tucek, C. Aparicio, R. Venerando, G. Sartori, F. Francescato, F. Mion, N. Gabellini, R. Zboril and F. Vianello, Adv. Funct. Mater., 2015, 25, 1822–1831 CrossRef CAS PubMed
.
-
Z. Rice, N. C. Cady and M. Bergkvist, In Terminal phosphate group influence on DNA-TiO2 nanoparticle interactions, MRS Proceedings, Cambridge University Press, 2009, p. 1236-SS05-15 Search PubMed
.
- S. J. Klaine, P. J. J. Alvarez, G. E. Batley, T. E. Fernandes, R. D. Handy, D. Y. Lyon, S. Mahendra, M. J. McLaughlin and J. R. Lead, Environ. Toxicol. Chem., 2008, 27, 1825–1851 CrossRef CAS PubMed
.
- R. D. Handy, G. Cornelis, T. Fernandes, O. Tsyusko, A. Decho, T. Sabo-Attwood, C. Metchalfe, J. A. Steevens, S. J. Klaine, A. A. Koelmans and N. Horne, Environ. Toxicol. Chem., 2012, 31, 15–31 CrossRef CAS PubMed
.
- S. J. Klaine, A. A. Koelmans, N. Horne, S. Carley, R. D. Handy, L. Kapuska, B. Nowack and F. von der Kammer, Environ. Toxicol. Chem., 2012, 31, 3–14 CrossRef CAS PubMed
.
- A. G. Schultz, D. Boyle, D. Chamot, K. J. Ong, K. J. Wilkinson, J. C. McGeer, G. Sunahara and G. G. Goss, Environ. Chem., 2014, 11, 207–226 CrossRef CAS
.
- J. Leszczynski, Nat. Nanotechnol., 2010, 5, 633–634 CrossRef CAS PubMed
.
- P. P. Fu, Q. Xia, H.-M. Hwang, P. C. Ray and H. Yu, J. Food Drug Anal., 2014, 22, 64–75 CrossRef CAS PubMed
.
- A. Nel, T. Xia, L. Madler and N. Li, Science, 2006, 311, 622–627 CrossRef CAS PubMed
.
- T. Xia, M. Kovochich, M. Liong, L. Mädler, B. Gilbert, H. Shi, J. I. Yeh, J. I. Zink and A. E. Nel, ACS Nano, 2008, 2, 2121–2134 CrossRef CAS PubMed
.
- A. B. Djurišić, Y. H. Leung, A. Ng, X. Y. Xu, P. K. Lee and N. Degger, Small, 2015, 11, 26–44 CrossRef PubMed
.
- S. K. Misra, A. Dybowska, D. Berhanu, S. N. Luoma and E. Valsami-Jones, Sci. Total Environ., 2012, 438, 225–232 CrossRef CAS PubMed
.
- M. Li, D. Lin and L. Zhu, Environ. Pollut., 2013, 173, 97–102 CrossRef CAS PubMed
.
- Y.-S. Li and J. S. Church, J. Food Drug Anal., 2014, 22, 29–48 CrossRef CAS PubMed
.
- W. He, Y. Liu, W. G. Wamer and J.-J. Yin, J. Food Drug Anal., 2014, 22, 49–63 CrossRef CAS PubMed
.
- J. Wang, C. Chen, Y. Liu, F. Jiao, W. Li, F. Lao, Y. Li, B. Li, C. Ge, G. Zhou, Y. Gao, Y. Zhao and Z. Chai, Toxicol. Lett., 2008, 183, 72–80 CrossRef CAS PubMed
.
- S. Vranic, N. Boggetto, V. Contremoulins, S. Mornet, N. Reinhardt, F. Marano, A. Baeza-Squiban and S. Boland, Part. Fibre Toxicol., 2013, 10, 2 CrossRef CAS PubMed
.
- K. J. Lee, P. D. Nallathamby, L. M. Browning, T. Desai, P. K. Cherukuri and X.-H. N. Xu, Analyst, 2012, 137, 2973–2986 RSC
.
- L. M. Browning, T. Huang and X.-H. N. Xu, Interface Focus, 2013, 3, 20120098 CrossRef PubMed
.
- L. Li, T. Hutter, A. S. Finnemore, F. M. Huang, J. J. Baumberg, S. R. Elliott, U. Steiner and S. Mahajan, Nano Lett., 2012, 12, 4242–4246 CrossRef CAS PubMed
.
- G. A. Roth, S. Tahiliani, N. Neu-Baker and M. S. A. Brenner, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2015, 7, 565–579 CrossRef CAS PubMed
.
- A. R. Badireddy, M. R. Wiesner and J. Liu, Environ. Sci. Technol., 2012, 46, 10081–10088 CrossRef CAS PubMed
.
- M. Mortimer, A. Gogos, N. Bartolome, A. Kahru, T. D. Bucheli and V. I. Slaveykova, Environ. Sci. Technol., 2014, 48, 8760–8767 CrossRef CAS PubMed
.
- M. Husain, A. T. Saber, C. Guo, N. R. Jacobsen, K. A. Jensen, C. L. Yauk, A. Williams, U. Vogel, H. Wallin and S. Halappanavar, Toxicol. Appl. Pharmacol., 2013, 269, 250–262 CrossRef CAS PubMed
.
- T. Eustaquio and J. F. Leary, Methods Mol. Biol., 2012, 926, 69–85 CAS
.
- C.-Y. Huang, T.-R. Ger, Z.-H. Wei and M.-F. Lai, PLoS One, 2014, 9, e96550 Search PubMed
.
- P. Shah, A. Kaushik, X. Zhu, C. Zhang and C. Z. Li, Analyst (Cambridge, U. K.), 2014, 139, 2088–2098 RSC
.
- I. A. Mudunkotuw, A. A. Minshid and V. H. Grassian, Analyst (Cambridge, U. K.), 2014, 139, 870–881 RSC
.
- I. A. Mudunkotuwa and V. H. Grassian, J. Am. Chem. Soc., 2010, 132, 14986–14994 CrossRef CAS PubMed
.
- J. Kiwi and V. Nadtochenko, Langmuir, 2005, 21, 4631–4641 CrossRef CAS
.
- L. Petrone, R. Easingwood, M. F. Barker and A. J. McQuillan, J. R. Soc., Interface, 2011, 8, 410–422 CrossRef CAS PubMed
.
- L.-A. Tai, Y.-T. Kang, Y.-C. Chen, Y.-C. Wang, Y.-J. Wang, Y.-T. Wu, K.-L. Liu, C.-Y. Wang, Y.-F. Ko, C.-Y. Chen, N.-C. Huang, J.-K. Chen, Y.-F. Hsieh, T.-R. Yew and C.-S. Yang, Anal. Chem., 2012, 84, 6312–6316 CrossRef CAS PubMed
.
- T. D. Santos, J. Varela, I. Lynch, A. Salvati and K. A. Dawson, Small, 2011, 7, 3341–3349 CrossRef PubMed
.
- H. Nakajima, K. Ozaki, T. Hongyo, I. Narama and T. Todo, Nanomedicine, 2011, 7, 881–888 CrossRef CAS PubMed
.
- M. Zhu, G. Nie, H. Meng, T. Xia, A. Nel and Y. Zhao, Acc. Chem. Res., 2012, 46, 622–631 CrossRef PubMed
.
- C. M. Sayes and D. B. Warheit, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2009, 1, 660–670 CrossRef CAS PubMed
.
- G. Oberdörster, A. Maynard, K. Donaldson, V. Castranova, J. Fitzpatrick, K. Ausman, J. Carter, B. Karn, W. Kreyling, D. Lai, S. Olin, N. Monteiro-Riviere, D. Warheit and H. Yang, ILSI Research Foundation/Risk Science Institute Nanomaterial Toxicity Screening Working Group, Part. Fibre Toxicol., 2005, 2, 8 CrossRef PubMed
.
- L. C. Abbott and A. D. Maynard, Risk Analysis, 2010, 30, 1634–1644 CrossRef PubMed
.
- F. Gottschalk and B. Nowack, J. Environ. Monit., 2011, 13, 1145–1155 RSC
.
- B. Collin, M. Auffan, A. C. Johnson, I. Kaur, A.
A. Keller, A. Lazareva, J. R. Lead, X. Ma, R. C. Merrifield, C. Svendsen, J. C. White and J. M. Unrine, Environ. Sci.: Nano, 2014, 1, 533–548 RSC
.
- E. Burello and A. Worth, Nat. Nanotechnol., 2011, 6, 138–139 CrossRef CAS PubMed
.
- R. Liu, H. Y. Zhang, Z. X. Ji, R. Rallo, T. Xia, C. H. Chang, A. Nel and Y. Cohen, Nanoscale, 2013, 5, 5644–5653 RSC
.
- K. A. Clark, R. H. White and E. K. Silbergeld, Regul. Toxicol. Pharmacol., 2011, 59, 361–363 CrossRef CAS PubMed
.
- A. Gajewicz, M. T. Cronin, B. Rasulev, J. Leszczynski and T. Puzyn, Nanotechnology, 2015, 26, 015701 CrossRef PubMed
.
- I. Lynch, C. Weiss and E. Valsami-Jones, Nano Today, 2014, 9, 266–270 CrossRef CAS PubMed
.
- H. Godwin, C. Nameth, D. Avery, L. L. Bergeson, D. Bernard, E. Beryt, W. Boyes, S. Brown, A. J. Clippinger, Y. Cohen, M. Doa, C. O. Hendren, P. Holden, K. Houck, A. B. Kane, F. Klaessig, T. Kodas, R. Landsiedel, I. Lynch, T. Malloy, M. B. Miller, J. Muller, G. Oberdorster, E. J. Petersen, R. C. Pleus, P. Sayre, V. Stone, K. M. Sullivan, J. Tentschert, P. Wallis and A. E. Nel, ACS Nano, 2015, 9, 3409–3417 CrossRef CAS PubMed
.
- J. H. E. Arts, M. Hadi, A. M. Keene, R. Kreiling, D. Lyon, M. Maier, K. Michel, T. Petry, U. G. Sauer, D. Warheit, K. Wiench and R. Landsiedel, Regul. Toxicol. Pharmacol., 2014, 70, 492–506 CrossRef CAS PubMed
.
- D. Fourches, D. Pu, C. Tassa, R. Weissleder, S. Y. Shaw, R. J. Mumper and A. Tropsha, ACS Nano, 2010, 4, 5703–5712 CrossRef CAS PubMed
.
- R. Liu, R. Rallo, S. George, Z. Ji, S. Nair, A. E. Nel and Y. Cohen, Small, 2011, 7, 1118–1126 CrossRef CAS PubMed
.
- E. Burello and A. P. Worth, Nanotoxicology, 2011, 5, 228–235 CrossRef CAS PubMed
.
- C. Sayes and I. Ivanov, Risk Analysis, 2010, 30, 1723–1734 CrossRef PubMed
.
- V. C. Epa, F. R. Burden, C. Tassa, R. Weissleder, S. Shaw and D. A. Winkler, Nano Lett., 2012, 12, 5808–5812 CrossRef CAS PubMed
.
- R. Liu, R. Rallo, R. Weissleder, C. Tassa, S. Shaw and Y. Cohen, Small, 2013, 9, 1842–1852 CrossRef CAS PubMed
.
- K. P. Singh and S. Guptaab, RSC Adv., 2014, 4, 13215–13230 RSC
.
- A. A. Toropov, A. P. Toropova, E. Benfenati, G. Gini, T. Puzyn, D. Leszczynska and J. Leszczynski, Chemosphere, 2012, 89, 1098–1102 CrossRef CAS PubMed
.
- A. P. Toropova, A. A. Toropov, R. Rallo, D. Leszczynska and J. Leszczynski, Ecotoxicol. Environ. Saf., 2015, 112, 39–45 CrossRef CAS PubMed
.
- N. Sizochenko, B. Rasulev, A. Gajewicz, V. Kuz'min, T. Puzync and J. Leszczynski, Nanoscale, 2015, 6, 13986–13993 RSC
.
- A. Gajewicz, N. Schaeublin, B. Rasulev, S. Hussain, D. Leszczynska, T. Puzyn and J. Leszczynski, Nanotoxicology, 2015, 9, 313–325 CrossRef CAS PubMed
.
- S. Y. Shaw, E. C. Westly, M. J. Pittet, A. Subramanian, S. L. Schreiber and R. Weissleder, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 7387–7392 CrossRef CAS PubMed
.
- A. P. Toropova, A. A. Toropov, E. Benfenati and R. Korenstein, J. Nanopart. Res., 2014, 16, 1–7 CrossRef
.
- A. P. Toropova, A. A. Toropov, E. Benfenati, R. Korenstein, D. Leszczynska and J. Leszczynski, Environ. Sci. Pollut. Res., 2015, 22, 745–757 CrossRef CAS PubMed
.
- A. P. Toropova and A. A. Toropov, Chemosphere, 2013, 93, 2650–2655 CrossRef CAS PubMed
.
- V. V. Kleandrova, F. Luan, H. González-Díaz, J. M. Ruso, A. Melo, A. Speck-Planche and M. N. D. S. Cordeiro, Environ. Int., 2014, 73, 288–294 CrossRef CAS PubMed
.
- F. Luan, V. V. Kleandrov, H. González-Díaz, J. M. Ruso, A. Melo, A. Speck-Planche and M. N. D. S. Cordeiro, Nanoscale, 2014, 6, 10623–10630 RSC
.
- T. Patel, D. Telesca, C. Low-Kam, Z. X. Ji, H. Y. Zhang, T. Xia, J. I. Zinc and A. E. Nel, Environ. Ecol. Stat., 2014, 25, 57–68 Search PubMed
.
- C. R. Thomas, S. George, A. M. Horst, Z. Ji, R. J. Miller, J. R. Peralta-Videa, T. Xia, S. Pokhrel, L. Mädler, J. L. Gardea-Torresdey, P. A. Holden, A. A. Keller, H. S. Lenihan, A. E. Nel and J. I. Zink, ACS Nano, 2011, 5, 13–20 CrossRef CAS PubMed
.
- R. Rallo, B. France, R. Liu, S. Nair, S. George, R. Damoiseaux, F. Giralt, A. Nel, K. Bradley and Y. Cohen, Environ. Sci. Technol., 2011, 45, 1695–1702 CrossRef CAS PubMed
.
- S. George, S. Pokhrel, T. Xia, B. Gilbert, Z. Ji, M. Schowalter, A. Rosenauer, R. Damoiseaux, K. A. Bradley, L. Mädler and A. E. Nel, ACS Nano, 2010, 4, 15–29 CrossRef CAS PubMed
.
- H. Meng, T. Xia, S. George and A. E. Nel, ACS Nano, 2009, 3, 1620–1627 CrossRef CAS PubMed
.
- R. Liu, S. Lin, R. Rallo, Y. Zhao, R. Damoiseaux, T. Xia, S. Lin, A. Nel and Y. Cohen, PLoS One, 2012, 7, e35014 CAS
.
- S. Kar, A. Gajewicz, T. Puzyn, K. Roy and J. Leszczynski, Ecotoxicol. Environ. Saf., 2014, 107, 162–169 CrossRef CAS PubMed
.
- A. Mikolajczyk, A. Gajewicz, B. Rasulev, N. Schaeublin, E. Maurer-Gardner, S. Hussain, J. Leszczynski and T. Puzyn, Chem. Mater., 2015, 27, 2400–2407 CrossRef CAS
.
| This journal is © The Royal Society of Chemistry 2015 |




