Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Graphene oxide-enhanced cytoskeleton imaging and mitosis tracking†
Qian-Ru
Li‡
a,
Jin-Biao
Jiao‡
b,
Li-Li
Li
c,
Xiao-Peng
He
*b,
Yi
Zang
*c,
Tony D.
James
 d,
Guo-Rong
Chen
b,
Lin
Guo
*a and
Jia
Li
*c
d,
Guo-Rong
Chen
b,
Lin
Guo
*a and
Jia
Li
*c
aState Key Laboratory of Virology, College of Life Sciences, Wuhan University, Wuhan, 430072, P. R. China. E-mail: guol@whu.edu.cn
bKey Laboratory for Advanced Materials & Institute of Fine Chemicals, School of Chemistry and Molecular Engineering, East China University of Science and Technology, Shanghai 200237, P. R. China. E-mail: xphe@ecust.edu.cn
cNational Center for Drug Screening, State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, 189 Guo Shoujing Rd., Shanghai 201203, P. R. China. E-mail: yzang@simm.ac.cn; jli@simm.ac.cn
dDepartment of Chemistry, University of Bath, Bath, BA2 7AY, UK
First published on 28th February 2017
Abstract
Here we show that graphene oxide greatly enhances the imaging ability of a peptide probe that selectively targets microtubules of the cytoskeleton, thus enabling the dynamic tracking of mitosis in live cells.
Microtubules, a key component of the cytoskeleton, are composed of polymerized α- and β-tubulin dimers. Their dynamic polymerization is involved in a variety of cellular processes including mitosis, cytokinesis, polarity and vesicular transport.1,2 Microtubule tracks are covered with microtubule-associated proteins (MAPs), which contribute to their stability and thus play important roles in maintaining normal functions of the microtubules. The best-studied members of the MAPs are MAP4, tau and MAP2.3,4
To date, the most widely used microtubule probe is fluorophore-linked taxane.5 However, while the use of drugs as a targeting agent for cell imaging may cause toxicity concerns leading to dysfunction of the cells (resulting in an inability to track the normal physiological processes of live cells), probes based on the more biocompatible and versatile peptides with specific microtubule-binding motifs have been rarely used. In 1988, an 18-amino-acid peptide that targets a microtubule binding site shared by MAP2 and tau was reported.6 By using the major-coat phage display technique, a specific microtubule-binding peptide for MAPs has been identified.7 However, the tubulin-binding affinity of these peptide ligands has only been confirmed in vitro (i.e. using transmission electron microscopy to determine the interaction between affinity-selected tubulin-binding phage and tubulins).7 This is largely due to the poor membrane permeability of some peptides and the lack of reliable imaging materials for spatiotemporal tracking of microtubules in live cells.
Graphene oxide (GO), a two-dimensional material, has recently been extensively used for biomedical applications.8–16 The large surface area of GO and its unique optical properties makes the construction of ligand-attached GO material composites for targeted imaging of cells and intracellular macromolecules possible.16 In particular, we have demonstrated that GO can greatly enhance the imaging ability of fluorophore-tagged ligands for cells that express a selective transmembrane receptor.17,18 Here we show that GO has the ability to enhance the cytoskeleton imaging ability of a membrane–impermeable peptide ligand selective for microtubules, thus enabling the dynamic tracking of mitosis in live cells (Fig. 1).
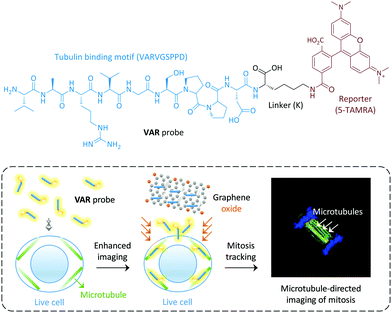 | ||
| Fig. 1 Structure of the peptide-ligand-based VAR probe and schematic illustration of graphene oxide enhanced, microtubule-targeted imaging of live cells and tracking of cell mitosis. | ||
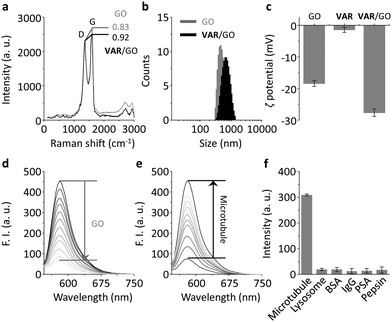
We used a microtubule-selective peptide ligand (VARVGSPPD) obtained by the phage display technique,7 to which a fluorescent reporter (5-carboxytetramethylrhodamine (5-TAMRA)) was introduced through L-arginine (K) (Fig. 1). With the probe in hand, we first tested its interaction with GO (produced by the modified Hummer's method)19,20 in phosphate buffered saline. The morphology of the GO was characterized using transmission electron microscopy (Fig. S1, ESI†). As determined by Raman spectroscopy, the ID/IG (intensity of D band to that of G band) ratio of GO increased with VAR probe (Fig. 2a), suggesting that the attachment of the peptide to GO perturbed the C-sp2-hybridization order of the material surface.11 The size of VAR/GO increased with respect to GO alone as shown by dynamic light scattering (Fig. 2b), and addition of VAR probe to GO significantly changed the zeta potential of the latter (Fig. 2c).
We also used fluorescence spectroscopy to characterize the binding between the probe and material. The fluorescence of the VAR probe gradually decreased with increasing GO (Fig. 2d), which was probably a result of Förster resonance energy transfer between the two closely conjugated species.21 However, the presence of microtubule (taxol-stabilized microtubule that is polymerized by α/β tubulins) gradually recovered the peptide fluorescence (Fig. 2e) with good linearity (Fig. S2a, ESI†). The limit of detection of VAR/GO for microtubule was determined to be as low as 2.4 pM. This ultra-sensitivity suggests a strong binding affinity between the peptide ligand and microtubules, setting a basis for the facile in vitro screening of other microtubule binders using the VAR/GO material system. A kinetic assay indicated that the fluorescence recovery of the VAR probe with microtubule quickly reached equilibrium within 15 min (Fig. S2b, ESI†). The fact that the presence of a range of unselective proteins did not enhance the fluorescence of the material composite suggests a good biospecificity of the peptide ligand (Fig. 2f and Fig. S2c, ESI†). To further corroborate the selective ligand–microtubule binding, a competition assay was performed. In the presence of GO and microtubule, the addition of increasing free peptide ligand (VARVGSPPD) gradually decreased the fluorescence of the VAR probe (Fig. S3, ESI†), suggesting the specific association between the probe and protein.
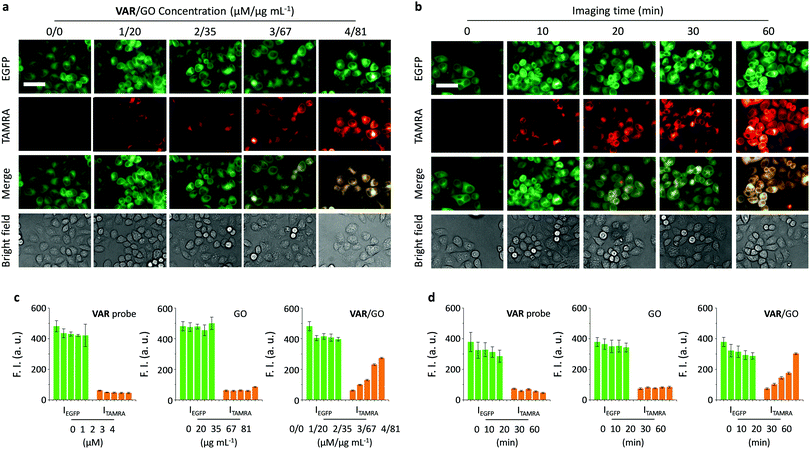
Having determined the specific binding between the VAR probe and microtubule as facilitated by GO, we then set out to test the imaging ability of the probe for the cell cytoskeleton. HeLa cells expressing EGFP-α tubulin (EGFP = enhanced green fluorescence protein), established in a previous report, were utilised.22 Using EGFP fluorescence as a reference, we first observed that treatment of just the VAR probe with the cells did not lead to any obvious concentration-dependent production of probe (TAMRA) fluorescence intracellularly (Fig. S4, ESI†), suggesting poor cell permeability of the peptide probe. In contrast, the presence of GO significantly enhanced the imaging ability of the VAR probe in both a dose (Fig. 3a and c) and time-dependent manner (Fig. 3b and d). Both the GO material and VAR probe were determined to be not toxic to HeLa with increasing concentrations (Fig. S5, ESI†).
Furthermore, we used an alternative cell line (human embryonic kidney 293T) transfected with EGFP-α-tubulin to investigate the generality of the developed method. TAMRA fluorescence was only observed for the VAR/GO ensemble and not the free VAR probe and GO (Fig. S6, ESI†). These observations are comparable to those observed for HeLa cells, indicating the broad applicability of GO for enhancing microtubule-targeted imaging of peptide probes. These results are in agreement with the excellent intracellular delivery ability of GO for small-molecular drugs, peptides and aptamers.16,18,23,24,31–34
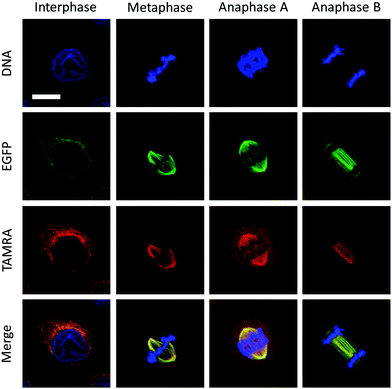
To test whether the VAR probe could bind dynamic mitotic microtubules intracellularly, a confocal laser scanning microscope was used. We first determined that the EGFP-α-tubulin fluorescence localized well with that of TAMRA, suggesting that the probe was probably bound to the polymerized tubulins (Fig. 4). In addition, we also determined that the probe could be used for tracking the mitosis of HeLa (Fig. 4). During interphase, cells were adhered to the coverslip with a flat shape, whereas upon entering mitotic phase, mitotic apparatuses were dynamically organized by microtubules and microtubule associated proteins, including the spindle during metaphase and the central spindle during anaphase. These mitotic apparatuses were clearly visualized by the VAR probe.
In summary, we have shown that GO enhances the cytoskeleton imaging ability of a cell-impermeable peptide ligand for microtubules, thus enabling the dynamic tracking of mitosis in live cells. This research provides insights into the use of 2D materials for enabling imaging and spatiotemporal tracking of cell fate by peptide probes. Our ongoing research will focus on the use of other 2D materials25–30 for peptide-based cytoskeleton imaging.
This research is supported by the 973 project (2013CB733700), the National Natural Science Foundation of China (21572058 and 21576088), the Science and Technology Commission of Shanghai Municipality (15540723800), the Shanghai Rising-Star Program (16QA1401400) (to X.-P. He) and the Shanghai Science and Technology Development Funds (14YF1413300). The Catalysis And Sensing for our Environment (CASE) net-work is thanked for research exchange opportunities. T. D. J. thanks ECUST for a guest professorship.
Notes and references
- H. de Forges, A. Bouissou and F. Perez, Int. J. Biochem. Cell Biol., 2011, 44, 266 CrossRef PubMed.
- M. Stiess and F. Bradke, Dev. Neurobiol., 2011, 71, 430 CrossRef CAS PubMed.
- L. Dehmelt and S. Halpain, Genome Biol., 2005, 6, 204 CrossRef PubMed.
- S. Halpain and L. Dehmelt, Genome Biol., 2006, 7, 224 CrossRef PubMed.
- G. Lukinavičius, L. Reymond, E. D'Este, A. Masharina, F. Göttfert, H. Ta, A. Güther, M. Fournier, S. Rizzo, H. Waldmann, C. Blaukopf, C. Sommer, D. W. Gerlich, H. D. Arndt, S. W. Hell and K. Johnsson, Nat. Methods, 2014, 11, 731 CrossRef PubMed.
- S. A. Lewis, D. H. Wang and N. J. Cowan, Science, 1988, 242, 936 CrossRef CAS PubMed.
- B. Cao and C. Mao, Biomacromolecules, 2009, 10, 555 CrossRef CAS PubMed.
- K. Kostarelos and K. S. Novoselov, Science, 2014, 344, 261 CrossRef CAS PubMed.
- K. Yang, S. Zhang, G. Zhang, X. Sun, S.-T. Lee and Z. Liu, Nano Lett., 2010, 10, 3318 CrossRef CAS PubMed.
- L. Feng, L. Wu and X. Qu, Adv. Mater., 2013, 25, 168 CrossRef CAS PubMed.
- H.-L. Zhang, X.-L. Wei, Y. Zang, J.-Y. Cao, S. Liu, X.-P. He, Q. Chen, Y.-T. Long, J. Li, G.-R. Chen and K. Chen, Adv. Mater., 2013, 25, 4097 CrossRef CAS PubMed.
- (a) D.-K. Ji, G.-R. Chen, X.-P. He and H. Tian, Adv. Funct. Mater., 2015, 25, 3483 CrossRef CAS; (b) D. Xie, X.-Q. Feng, X.-L. Hu, L. Liu, Z. Ye, J. Cao, G.-R. Chen, X.-P. He and Y.-T. Long, ACS Appl. Mater. Interfaces, 2016, 8, 25137 CrossRef CAS PubMed; (c) L. Cui, B.-W. Zhu, S. Qu, X.-P. He and G.-R. Chen, Dyes Pigm., 2015, 121, 312 CrossRef CAS; (d) X.-P. He, Q. Deng, L. Cai, C.-Z. Wang, Y. Zang, J. Li, G.-R. Chen and H. Tian, ACS Appl. Mater. Interfaces, 2014, 6, 5379 CrossRef CAS PubMed; (e) X. Sun, B. Zhu, D.-K. Ji, Q. Chen, X.-P. He, G.-R. Chen and T. D. James, ACS Appl. Mater. Interfaces, 2014, 6, 10078 CrossRef CAS PubMed.
- K. Yang, L. Feng, H. Hong, W. Cai and Z. Liu, Nat. Protoc., 2013, 8, 2392 CrossRef CAS PubMed.
- C. Chung, Y.-K. Kim, D. Shin, S.-R. Ryoo, B. H. Hong and D.-H. Min, Acc. Chem. Res., 2013, 46, 2211 CrossRef CAS PubMed.
- V. Georgakilas, J. N. Tiwari, K. C. Kemp, J. A. Perman, A. B. Bourlinos, K. S. Kim and R. Zboril, Chem. Rev., 2016, 116, 5464 CrossRef CAS PubMed.
- For a tutorial review, see: X.-P. He, Y. Zang, T. D. James, J. Li and G.-R. Chen, Chem. Soc. Rev., 2015, 44, 4239 RSC.
- D.-K. Ji, Y. Zhang, X.-P. He and G.-R. Chen, J. Mater. Chem. B, 2015, 3, 6656 RSC.
- D.-K. Ji, Y. Zhang, Y. Zang, W. Liu, X. Zhang, J. Li, G.-R. Chen, T. D. James and X.-P. He, J. Mater. Chem. B, 2015, 3, 9182 RSC.
- N. I. Kovtyukhova, P. J. Olivier, B. R. Martin, T. E. Mallouk, S. A. Chizhik and E. V. Buzaneva, Chem. Mater., 1999, 11, 771 CrossRef CAS.
- D. R. Dreyer, S. Park, C. W. Bielawski and R. S. Rouff, Chem. Soc. Rev., 2010, 39, 228 RSC.
- E. Morales-Naráez and A. Merkoçi, Adv. Mater., 2012, 24, 3298 CrossRef PubMed.
- T. Chinen, P. Liu, S. Shioda, J. Pagel, B. Cerikan, T.-C. Lin, O. Gruss, Y. Hayashi, H. Takeno, T. Shima, Y. Okada, I. Hayakawa, Y. Hayashi, H. Kigoshi, T. Usui and E. Schiebel, Nat. Commun., 2015, 6, 8722 CrossRef CAS PubMed.
- Y. Wang, Z. Li, D. Hu, C.-T. Lin, J. Li and Y. Lin, J. Am. Chem. Soc., 2010, 132, 9274 CrossRef CAS PubMed.
- K. Yang, L. Feng, X. Shi and Z. Liu, Chem. Soc. Rev., 2013, 42, 530 RSC.
- D.-K. Ji, Y. Zhang, Y. Zang, J. Li, G.-R. Chen, X.-P. He and H. Tian, Adv. Mater., 2016, 28, 9356 CrossRef CAS PubMed.
- X.-P. He and H. Tian, Small, 2016, 12, 144 CrossRef CAS PubMed.
- M. Wahiba, X.-Q. Feng, Y. Zang, T. D. James, J. Li, G.-R. Chen and X.-P. He, Chem. Commun., 2016, 52, 11689 RSC.
- D. Xie, D.-K. Ji, Y. Zhang, J. Cao, H. Zheng, L. Liu, Y. Zang, J. Li, G.-R. Chen, T. D. James and X.-P. He, Chem. Commun., 2016, 52, 9418 RSC.
- Y.-H. Ma, W.-T. Dou, Y.-F. Pan, L.-W. Dong, Y.-X. Tan, X.-P. He, H. Tian and H.-Y. Wang, Adv. Mater., 2017, 29, 1604253 CrossRef PubMed.
- S. Guo, J. Chen, B.-Y. Cai, W.-W. Chen, Y.-F. Li, X. Sun, G.-R. Chen, X.-P. He and T. D. James, Mater. Chem. Front., 2017, 1, 61 RSC.
- L. Li, J. Feng, H. Liu, Q. Li, L. Tong and B. Tang, Chem. Sci., 2016, 7, 1940 RSC.
- H. Ji, Y. Guan, L. Wu, J. Ren, D. Miyoshi, N. Sugimoto and X. Qu, Chem. Commun., 2015, 51, 1479 RSC.
- Z. Xu, S. Zhu, M. Wang, Y. Li, P. Shi and X. Huang, ACS Appl. Mater. Interfaces, 2015, 7, 1355 CAS.
- M. Hong, L. Xu, Q. Xue, L. Li and B. Tang, Anal. Chem., 2016, 88, 12177 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Additional figures and experimental section. See DOI: 10.1039/c7cc01019b |
| ‡ Equal contribution. |
| This journal is © The Royal Society of Chemistry 2017 |