Open Access Article
Open Access ArticleThe ring residue proline 8 is crucial for the thermal stability of the lasso peptide caulosegnin II†
Julian D.
Hegemann
a,
Christopher D.
Fage
a,
Shaozhou
Zhu
a,
Klaus
Harms
a,
Francesco Saverio
Di Leva
b,
Ettore
Novellino
b,
Luciana
Marinelli
b and
Mohamed A.
Marahiel
*a
aDepartment of Chemistry/Biochemistry, LOEWE Center for Synthetic Microbiology, Philipps-Universität Marburg, Hans-Meerwein-Strasse 4, 35032 Marburg, Germany. E-mail: marahiel@staff.uni-marburg.de
bDepartment of Pharmacy, Università di Napoli “Federico II”, Via D. Montesano 49, 80131 Napoli, Italy
First published on 5th February 2016
Abstract
Lasso peptides are fascinating natural products with a unique structural fold that can exhibit tremendous thermal stability. Here, we investigate factors responsible for the thermal stability of caulosegnin II. By employing X-ray crystallography, mutational analysis and molecular dynamics simulations, the ring residue proline 8 was proven to be crucial for thermal stability.
Lasso peptides are intriguing members of the superfamily of ribosomally synthesized and post-translationally modified peptides (RiPPs).1–3 While genome mining studies showed that they are distributed throughout the bacterial domain,4–8 only lasso peptides of proteo- and actinobacterial origin have been isolated and characterized thus far.2,3 Their unique defining feature is a 7–9 residue macrolactam ring that is threaded by the C-terminal tail.1–3,9 The ring is formed between the α-amino group of the first residue (typically Gly, but some systems were shown to feature Ala, Ser or Cys instead8,10) and the carboxylic acid side chain of an Asp or Glu (Fig. S1, ESI†). This unusual fold is stabilized by bulky side chains of residues above and below the ring, so-called plug amino acids (AAs), which trap the threaded structure by sterical means. The fact that this topology is reminiscent of a lariat knot explains the name of this natural product family. Generally, lasso peptides were shown to exhibit a wide range of diverse biological activities, ranging from antimicrobial1,3,10–12 to inhibitory1,3,12 and antagonistic.1,3,13 However, with the recent increase in lasso peptide isolations by genome mining approaches, a large number with unknown functions have been reported as well.4–8
Intriguingly, their compact structures confer a high stability against chemical and proteolytic degradation, which makes them useful scaffolds for the grafting of bioactive peptide epitopes.14 Additionally, lasso peptides can exhibit tremendous stability against thermal denaturation. This resistance against thermal denaturation was assumed to be true for all lasso peptides due to their shared topology as early studies on the lasso peptide microcin J25 revealed that it could even withstand autoclaving at 120 °C.11 Still, recent studies revealed that thermal stability is not intrinsic to all lasso peptides, and since this finding was reported5 more and more heat sensitive lasso peptides have been identified.6–8,15,16
The first lasso peptides that were thoroughly investigated in this regard were the caulosegnins I–III (CsegI–III; Fig. 1).5 These lasso peptides are produced by the same biosynthetic gene cluster and feature either an eight-(CsegI) or nine-residue (CsegII/III) macrolactam ring. Interestingly, while CsegI and CsegIII readily unthread at elevated temperatures, CsegII resists even prolonged incubation at 95 °C. This becomes especially intriguing considering that CsegII and CsegIII share a high level of similarity and utilize both Tyr16 as the lower plug as was previously reported.5 In this study, we explored factors that define the heat resistance of CsegII and thereby were able to identify Pro8 as a key residue responsible for its thermal stability.
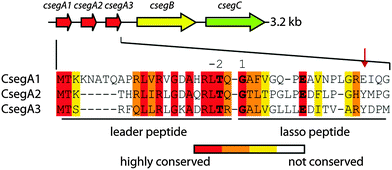 | ||
| Fig. 1 Schematic representation of the caulosegnin biosynthetic gene cluster from Caulobacter segnis ATCC 21756.5 Below, an alignment of all three precursor peptides is shown. Gly1 of the core peptides, the ring forming Glu8/9 and Thr-2 of the leader regions are highlighted. A red arrow marks the lower plug residues. | ||
So far, the main factors that are considered when discussing the thermal stability of lasso peptides are the nature and location of the lower plug AAs in combination with the size of the macrolactam rings. For example, the heat sensitive lasso peptide astexin-1 has a Gly1–Asp9 ring and was reported to be converted into a heat resistant lasso peptide by exchange of its Phe15 plug residue with a bulkier Trp.15 In another study, it was shown that the heat resistant lasso peptide capistruin, also featuring a Gly1–Asp9 ring, could be converted into a heat sensitive one.16 This was achieved by exchanging both the original plug (Arg15) and the neighboring Phe16 with Ala. In this R15A/F16A variant, Phe18 of capistruin is still able to act as lower plug and thereby maintains the lasso fold. Nevertheless, the double Ala substitution apparently causes such an increase in the flexibility of the C-terminal tail that it is now able to unthread at high temperatures. Xanthomonin II is another highly stable lasso peptide comprising a ring of only seven AAs (Gly1–Glu7).9 Due to the small diameter of its ring, any AA with a side chain larger than Ser was found to act as a thermostable plug.9
These studies emphasize the importance of lower plug residues for the thermal stability of a lasso peptide, but the question remains, whether other residues can affect the behavior of a lasso peptide upon exposure to elevated temperatures. This is especially intriguing for CsegII and CsegIII; two lasso peptides with identical ring sizes, identical lower plugs (Tyr16) and similar primary structures.5 Unlike all of the aforementioned compounds, they comprise the largest macrolactam ring (Gly1–Glu9) known for lasso peptides and employ Tyr (besides Trp one of the bulkiest side chains possible) as plugs. Therefore, we proposed that the observed differences in thermal stability are caused by other residues.
To investigate this hypothesis, we generated a Y16W variant of the heat sensitive CsegIII by site-directed ligase-independent mutagenesis (SLIM),17,18 which was isolated and then incubated at 95 °C. Even in this variant, Trp was found to be insufficient to confer thermal stability to the lasso fold, as it still unthreads similarly to the wild type (WT) CsegIII (see Fig. S2, ESI†). In contrast, the according E16W substitution in CsegI, which features only an eight-residue ring, transformed the heat sensitive WT into a heat resistant lasso peptide, again emphasizing the importance of ring size in this context (see Fig. S2, ESI†). Nonetheless, the question remained, which residues other than the plugs may cause the thermal stability of CsegII.
To allow a better estimate which residues could be important for the heat resistance of CsegII (or the lack thereof in CsegIII), we sought to elucidate the 3D structure of at least one of these lasso peptides. For this, both compounds were applied to crystallization screens, which yielded rod-shaped crystals of CsegII that diffracted to a resolution of 0.86 Å and allowed the elucidation of its structure (Fig. 2; see also Fig. S3 (ESI†) for a comparison with the lasso peptide macrolactam rings of the compounds discussed above).
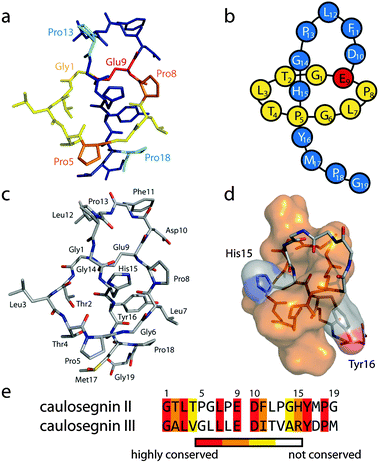 | ||
| Fig. 2 Crystal structure of CsegII (PDB code 5D9E). (a) Stick representation with the tail shown in blue and the ring in yellow. Further emphasized are Pro5 and Pro8 in orange as well as Pro13 and Pro18 in cyan. The ring forming Glu9 is highlighted in red. (b) Schematic representation. (c) Stick representation, colored by elements (carbon in gray, nitrogen in blue, oxygen in red and sulfur in yellow). As can be seen in this depiction, the Met17 thioether was oxidized to a sulfoxide moiety during crystallization. (d) Surface maps of the macrolactam ring (orange) and the His15 and Tyr16 side chains. The crystal structure confirms the mutagenesis based prediction of His15 and Tyr16 as the upper and lower plugs, respectively, as previously reported.5 (e) Alignment of CsegII and CsegIII (36.8% identity, 63.2% similarity). | ||
A defining feature of this structure is the kinks induced by the four Pro residues at positions 5, 8, 13 and 18. In comparison to CsegIII, the abundance of prolines in CsegII (4 out of 19 residues versus 1 out of 19 in CsegIII) and the rather conserved substitution of AAs at other positions led us to hypothesize a crucial role of the prolines for the thermal stability of CsegII.
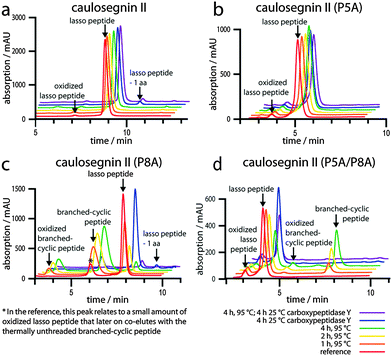
Therefore, a set of CsegII variants was generated by SLIM;17,18 namely P5A, P8A, P13A, P18A, P5A/P8A and P13A/P18A. All were expressed under previously published conditions5 and production was confirmed by analyzing the corresponding pellet extracts via high resolution LC-MS. We then tested the thermal stability of each variant by an assay that combines incubation at 95 °C with subsequent carboxypeptidase Y (cpepY) treatment (Fig. 3).5,6,8,9,15
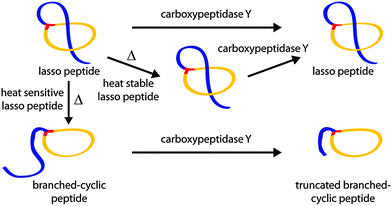 | ||
| Fig. 3 Schematic depiction of the combined thermal and cpepY stability assays.5,6,8,9,15 As the topology of a thermally stable lasso peptide does not change upon heating, it behaves like untreated lasso peptide towards cpepY. Normally, cpepY can remove no or only a few residues from the C-terminal tail if the lasso fold is intact. Conversely, heat sensitive lasso peptides unthread during prolonged incubation at high temperatures and are consequently more susceptible to subsequent cpepY mediated degradation. Therefore, much shorter truncation products (up to the lone macrolactam ring) are detected after treatment with cpepY. | ||
The results of these assays clearly show that WT, P5A, P13A, P18A and P13A/P18A are all heat resistant lasso peptides. In contrast, significant amounts of the P8A and P5A/P8A lasso peptides were converted to new compounds upon incubation at 95 °C. The resulting compounds were furthermore completely degraded to −9 AA truncation products by subsequent cpepY treatment, while the residual lasso peptides were unaffected by this protease.
As the only variants that showed a loss of thermal stability were those carrying a P8A (one of the two prolines in the ring) exchange, we wanted to investigate the lasso peptides featuring substitutions of the ring prolines in more detail. Therefore, we isolated the P5A, P8A and P5A/P8A variants and employed the purified compounds alongside a WT control again for thermal stability and cpepY assays (Fig. 4).
The results of these experiments were in accordance with the pellet extract assays described above and clearly showed the time-dependent unthreading of P8A and P5A/P8A at 95 °C, as well as the subsequent degradation of the resulting branched-cyclic peptides by cpepY. At the same time for WT and P5A, all measurements looked practically identical, independent of the prior treatment. Taken together, these experiments strongly suggest that Pro8 is one of the key residues responsible for the thermal stability of CsegII, while the other prolines seem to be negligible in this regard. To further test this hypothesis, two additional triple substitution variants were generated, namely P5A/P13A/P18A and P8A/P13A/P18A. Both could be produced, although P8A/P13A/P18A was only detectable in trace amounts. Repetition of the combined thermal stability and cpepY assays for these variants revealed that P5A/P13A/P18A is still relatively heat resistant, though after 2 h at 95 °C around 10% of the lasso peptide unthreaded to its branched-cyclic analog. For P8A/P13A/P18A, the interpretation of the data is not so simple due to the low yields of this variant. Still, as the cpepY treatment after incubation at 95 °C decreases the overall amount of the compound to less than 5% compared to the untreated sample, it can be assumed that it also represents a heat sensitive lasso peptide.
To elucidate at atomic level the reasons for the thermal stability of CsegII, extensive computational studies were carried out. 1 μs molecular dynamics (MD) simulations were performed at 368 K on both the WT CsegII crystal structure and its P5A/P8A variant (see ESI† for details), to evaluate how the Pro to Ala ring exchanges might modulate the heat resistance of these lasso peptides. At the end of the calculations, we first measured the flexibility rate of the macrolactam ring in both peptides by calculating the rmsd of residues 1–9 (Fig. S4a, ESI†). Interestingly, in the WT the ring shows a very low rmsd value (0.77 ± 0.13), indicating that it is tightly locked in its native (crystallographic) state. Increased rmsd value and fluctuations (1.18 ± 0.29), corresponding to a higher ring flexibility, are instead observed for the P5A/P8A variant.
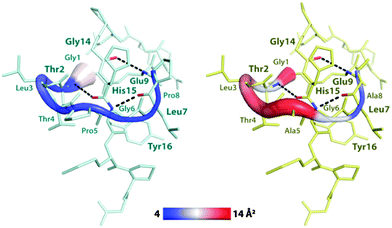
To investigate in more detail how each ring AA influences the peptide's conformational flexibility, we computed their B-factors from simulation trajectories. B-Factor mapping (Fig. 5) shows that in the WT, residues 1 to 5 are the most flexible ones, while residues 6 to 9 appear somehow more rigid. This can be ascribed to the formation of two tight hydrogen bonds (Fig. 5) between the E9 and G14 and the L7 and Y16 residues, which are almost conserved during the whole simulation (Fig. S4b, ESI†). On the other hand, only one hydrogen bond with the C-terminal tail is established by residues 1–5, which is located between T2 and H15 (Fig. 5 and Fig. S4b, ESI†). As expected, all of the AAs in the P5A/P8A variant's ring show higher B-factors compared to the WT peptide, particularly those occupying positions 1 to 5. In fact, the hydrogen bond between T2 and H15 of the P5A/P8A variant is less stable than in the WT, although it is still present during the MD simulation (Fig. S4b, ESI†). Overall, these results indicate that the exchange of Pro to Ala, which is devoid of rigid constraints on the N–Cα bond, increases the ring's flexibility, probably allowing this variant to more easily adopt a conformation suitable for the unthreading of the C-terminal tail. Furthermore, we have shown that while the P5A substitution occurs in a flexible region of the ring, the P8A exchange can induce fluctuations in a more rigid ring fragment, explaining why it is crucial for transforming the heat resistant CsegII to a heat sensitive lasso peptide.
Based on these findings, we were interested if in absence of Pro8, another Pro residue in the more rigid region of the macrolactam ring can fulfill its function and thereby would yield again a heat stable lasso peptide variant. For this, we generated two additional double substitution variants, G6P/P8A and L7P/P8A. While the G6P/P8A variant was not produced, L7P/P8A could be analyzed for thermal stability and cpepY resistance. Interestingly, even though the position of the proline is only shifted by one residue in this variant, the thermal stability was lost nonetheless. This might be explained by the absence of the aforementioned L7–Y16 hydrogen bond due to the L7P replacement and indicates that not only the proline at position 8 alone is essential for thermal stability, but that it might indeed be the concerned rigidity of Pro8 with other factors present in the CsegII scaffold. While this specific way of enabling thermal stability is most likely not present in every other heat stable lasso peptide scaffold, it is interesting to contemplate that these could at least contain similar variations thereof. Hence, it could be feasible that in these lasso peptides incorporation of specific residues at specific positions in the ring could also significantly alter their stability.
Conclusions
In conclusion, we show in this study for the first time that even though the plug residues are important factors with regard to the maintenance of the lasso fold and its thermal stability, the AAs of the macrolactam ring also contribute to the overall stability of lasso peptides. Based on the crystal structure of CsegII and an alignment of CsegII and CsegIII, we proposed that the large number of prolines in CsegII is the reason for its high resistance against thermally induced unthreading. Surprisingly, our experiments revealed that only Pro8 is essential for a thermally stable lasso fold, while the other prolines can be substituted more or less freely with alanine.This is especially highlighted by the observation that the thrice substituted P5A/P13A/P18A variant remains mostly heat stable and even though some slight unthreading at 95 °C can be observed, the unthreading process for this lasso peptide is still much slower than for the variant carrying a single P8A point mutation. Our findings were substantiated by MD simulations showing how the ring of CsegII becomes significantly more flexible upon Pro to Ala exchanges. Thus, the results of this study demonstrate the influence of the macrolactam ring's primary structure on the thermal stability of a lasso peptide and thereby help to better understand the criteria that govern how these natural products behave at elevated temperatures.
Acknowledgements
The Deutsche Forschungsgemeinschaft (DFG) and the LOEWE Center for Synthetic Microbiology (SYNMIKRO) are gratefully acknowledged for financial support. Furthermore, we would also like to thank Holger Steuber, Julian Koch, Bastian Langer and Shawn Walsh for initial assistance with this project and Pavel Afonine for aid with the PHENIX software.Notes and references
- M. O. Maksimov, S. J. Pan and A. James Link, Nat. Prod. Rep., 2012, 29, 996–1006 RSC.
- M. O. Maksimov and A. J. Link, J. Ind. Microbiol. Biotechnol., 2014, 41, 333–344 CrossRef CAS PubMed.
- J. D. Hegemann, M. Zimmermann, X. Xie and M. A. Marahiel, Acc. Chem. Res., 2015, 48, 1909–1919 CrossRef CAS PubMed.
- M. O. Maksimov, I. Pelczer and A. J. Link, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 15223–15228 CrossRef CAS PubMed.
- J. D. Hegemann, M. Zimmermann, X. Xie and M. A. Marahiel, J. Am. Chem. Soc., 2013, 135, 210–222 CrossRef CAS PubMed.
- J. D. Hegemann, M. Zimmermann, S. Zhu, D. Klug and M. A. Marahiel, Biopolymers, 2013, 100, 527–542 CrossRef CAS PubMed.
- M. O. Maksimov and A. J. Link, J. Am. Chem. Soc., 2013, 135, 12038–12047 CrossRef CAS PubMed.
- M. Zimmermann, J. D. Hegemann, X. Xie and M. A. Marahiel, Chem. Sci., 2014, 5, 4032–4043 RSC.
- J. D. Hegemann, M. Zimmermann, S. Zhu, H. Steuber, K. Harms, X. Xie and M. A. Marahiel, Angew. Chem., Int. Ed., 2014, 53, 2230–2234 CrossRef CAS PubMed.
- M. Metelev, J. I. Tietz, J. O. Melby, P. M. Blair, L. Zhu, I. Livnat, K. Severinov and D. A. Mitchell, Chem. Biol., 2015, 22, 241–250 CrossRef CAS PubMed.
- R. A. Salomon and R. N. Farias, J. Bacteriol., 1992, 174, 7428–7435 CAS.
- E. Gavrish, C. S. Sit, S. Cao, O. Kandror, A. Spoering, A. Peoples, L. Ling, A. Fetterman, D. Hughes, A. Bissell, H. Torrey, T. Akopian, A. Mueller, S. Epstein, A. Goldberg, J. Clardy and K. Lewis, Chem. Biol., 2014, 21, 509–518 CrossRef CAS PubMed.
- O. Potterat, K. Wagner, G. Gemmecker, J. Mack, C. Puder, R. Vettermann and R. Streicher, J. Nat. Prod., 2004, 67, 1528–1531 CrossRef CAS PubMed.
- J. D. Hegemann, M. De Simone, M. Zimmermann, T. A. Knappe, X. Xie, F. S. Di Leva, L. Marinelli, E. Novellino, S. Zahler, H. Kessler and M. A. Marahiel, J. Med. Chem., 2014, 57, 5829–5834 CrossRef CAS PubMed.
- M. Zimmermann, J. D. Hegemann, X. Xie and M. A. Marahiel, Chem. Biol., 2013, 20, 558–569 CrossRef CAS PubMed.
- T. A. Knappe, U. Linne, L. Robbel and M. A. Marahiel, Chem. Biol., 2009, 16, 1290–1298 CrossRef CAS PubMed.
- J. Chiu, P. E. March, R. Lee and D. Tillett, Nucleic Acids Res., 2004, 32, e174 CrossRef PubMed.
- J. Chiu, D. Tillett, I. W. Dawes and P. E. March, J. Microbiol. Methods, 2008, 73, 195–198 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Materials and methods, experimental data, supporting figures and tables. See DOI: 10.1039/c6mb00081a |
| This journal is © The Royal Society of Chemistry 2016 |