 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Correction: Interpretation of type 2 diabetes mellitus relevant GC-MS metabolomics fingerprints by using random forests†
Jian-Hua
Huang
a,
Hua-Lin
Xie
ac,
Jun
Yan
a,
Dong-Sheng
Cao
a,
Hong-Mei
Lu
a,
Qing-Song
Xu
b and
Yi-Zeng
Liang
*a
aResearch Center of Modernization of Traditional Chinese Medicines, Department of Chemistry and Chemical Engineering, Central South University, Changsha 410083, P. R. China. E-mail: yizeng_liang@263.net; Fax: +86-731-88830831; Tel: +86-731-88830831
bSchool of Mathematical Sciences and Computing Technology, Central South University, Changsha 410083, P. R. China
cSchool of Chemistry and Chemical Engineering, Yangtze Normal University, Fuling 408100, P. R. China
First published on 8th February 2016
Abstract
Correction for ‘Interpretation of type 2 diabetes mellitus relevant GC-MS metabolomics fingerprints by using random forests’ by Jian-Hua Huang et al., Anal. Methods, 2013, 5, 4883–4889.
The authors wish to draw the readers' attention to their previous related study, published in Talanta,1 which should have been cited in this Analytical Methods paper.
The authors regret not giving correct attribution to Fig. 4 in the paper and Table 1 in the ESI,† which were reproduced for the readers' information. The figures are reproduced below with the correct copyright permission.
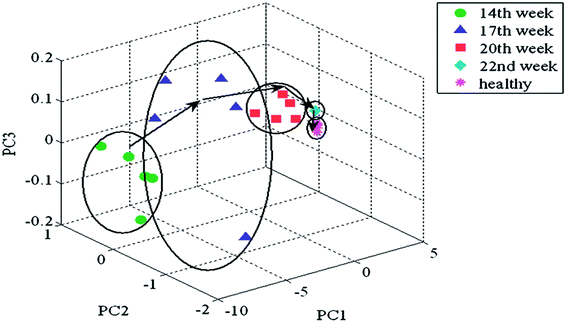 | ||
| Fig. 4 3D-projection plot of metabolic fingerprints from PCA of the first three principal components for the second data set. Reproduced from ref. 2 with permission from Taylor & Francis. | ||
| id | t r a (min) | Endogenous metabolites | C57 | AMPK-male | AMPK-female |
|---|---|---|---|---|---|
| 1 | 5.922 | Aminoethane | 0.2456 ± 0.0705 | 0.1905 ± 0.0567 | 0.1958 ± 0.0551 |
| 2 | 6.593 | Ethylene glycol | 0.0182 ± 0.0020 | 0.0530 ± 0.0428 | 0.0746 ± 0.0626 |
| 3 | 6.84 | N,N-Diethylacetamide | 0.0657 ± 0.0087 | 0.0476 ± 0.0202 | 0.0557 ± 0.0107 |
| 4 | 7.716 | Lactic acid* | 0.0872 ± 0.0374 | 0.0952 ± 0.0592 | 0.1482 ± 0.2155 |
| 5 | 7.934 | Acetic acid | 0.0856 ± 0.0333 | 0.0229 ± 0.0140 | 0.0412 ± 0.0203 |
| 6 | 10.01 | Phosphate | 2.1278 ± 0.9173 | 1.4730 ± 0.7381 | 1.3767 ± 0.9361 |
| 7 | 10.2 | L-Threonine | 0.0173 ± 0.0098 | 0.0108 ± 0.0068 | 0.0096 ± 0.0065 |
| 8 | 10.297 | Phenylacetic acid | 0.0047 ± 0.0023 | 0.0159 ± 0.0103 | 0.0147 ± 0.0097 |
| 9 | 10.382 | Succinic acid* | 0.0311 ± 0.0129 | 0.0098 ± 0.0031 | 0.0119 ± 0.0086 |
| 10 | 10.447 | 1,2-Hydroquinone | 0.0120 ± 0.0072 | 0.0078 ± 0.0047 | 0.0067 ± 0.0039 |
| 11 | 10.503 | Glyceric acid | 0.0961 ± 0.0266 | 0.0400 ± 0.0232 | 0.0183 ± 0.0087 |
| 12 | 10.723 | (R*,R*)-2,3-Dihydroxybutanoic acid | 0.0167 ± 0.0053 | 0.0037 ± 0.0014 | 0.0053 ± 0.0029 |
| 13 | 11.357 | 2,4-Dihyoxybutanoic acid | 0.0147 ± 0.0051 | 0.0155 ± 0.0080 | 0.0166 ± 0.0047 |
| 14 | 11.583 | (R*,S*)-3,4-Dihydroxybutanoic acid | 0.0304 ± 0.0098 | 0.0132 ± 0.0064 | 0.0178 ± 0.0107 |
| 15 | 11.797 | N-(1-Oxobutyl)-glycine | 0.0653 ± 0.0244 | 0.0319 ± 0.0186 | 0.0274 ± 0.0151 |
| 16 | 12.341 | Isovaleroglycine | 0.0356 ± 0.0134 | 0.0160 ± 0.0079 | 0.0107 ± 0.0073 |
| 17 | 12.483 | D-Threitol | 0.0714 ± 0.0273 | 0.0290 ± 0.0130 | 0.0251 ± 0.0151 |
| 18 | 12.645 | N-Crotonylglycine | 0.0240 ± 0.0146 | 0.0207 ± 0.0129 | 0.0148 ± 0.0099 |
| 19 | 12.973, 13.203 | 2,3,4-Trihydroxybutyrate | 0.1276 ± 0.0162 | 0.0631 ± 0.0343 | 0.0412 ± 0.0250 |
| 20 | 14.53 | N-(1-Oxohexyl)-glycine | 0.0960 ± 0.0319 | 0.0421 ± 0.0273 | 0.0232 ± 0.0081 |
| 21 | 14.58 | 3-Hydroxyphenylacetic acid | 0.0326 ± 0.0100 | 0.0140 ± 0.0081 | 0.0134 ± 0.0088 |
| 22 | 14.713 | D-Xylose | 0.0408 ± 0.0150 | 0.0182 ± 0.0044 | 0.0193 ± 0.0053 |
| 23 | 14.823, 15.057 | D-Ribose | 0.0926 ± 0.0370 | 0.0252 ± 0.0142 | 0.0250 ± 0.0179 |
| 24 | 15.509, 15.733 | Arabitol | 0.0287 ± 0.0164 | 0.0283 ± 0.0179 | 0.0278 ± 0.0215 |
| 25 | 16.023 | 6-Deoxy-D-galactose | 0.0336 ± 0.0083 | 0.0177 ± 0.0100 | 0.0149 ± 0.0104 |
| 26 | 16.087 | Mannonic acid | 0.0505 ± 0.0177 | 0.0211 ± 0.0143 | 0.0168 ± 0.0138 |
| 27 | 16.2 | Cis-aconitic acid* | 0.0535 ± 0.0288 | 0.0105 ± 0.0079 | 0.0168 ± 0.0147 |
| 28 | 16.357 | Phosphoric acid | 0.0414 ± 0.0202 | 0.0230 ± 0.0141 | 0.0212 ± 0.0168 |
| 29 | 17.177 | Isocitric acid* | 0.0348 ± 0.0121 | 0.0140 ± 0.0093 | 0.0248 ± 0.0138 |
| 30 | 17.563 | Hippuric acid | 0.0470 ± 0.0126 | 0.0180 ± 0.0074 | 0.0156 ± 0.0096 |
| 31 | 17.85, 17.96 | D-Fructose* | 0.0512 ± 0.0286 | 0.0371 ± 0.0145 | 0.0480 ± 0.0131 |
| 32 | 18.087 | N-Phenyl glycine* | 0.0596 ± 0.0214 | 0.0455 ± 0.0272 | 0.0389 ± 0.0287 |
| 33 | 18.197, 18.147 | D-Glucose* | 0.3785 ± 0.1618 | 0.1741 ± 0.0654 | 0.1859 ± 0.0736 |
| 34 | 18.507 | Altronic acid | 0.0302 ± 0.0069 | 0.0185 ± 0.0100 | 0.0102 ± 0.0074 |
| 35 | 18.577, 18.65 | D-Sorbitol* | 0.0896 ± 0.0269 | 0.0254 ± 0.0187 | 0.0300 ± 0.0275 |
| 36 | 18.983, 19.533 | Galactonic acid | 0.0613 ± 0.0282 | 0.0617 ± 0.0328 | 0.0441 ± 0.0351 |
| 37 | 19.99 | Palmitic acid | 0.0084 ± 0.0009 | 0.0067 ± 0.0017 | 0.0071 ± 0.0025 |
| 38 | 20.403 | Myo-inositol | 0.0347 ± 0.0228 | 0.0097 ± 0.0037 | 0.0134 ± 0.0129 |
| 39 | 25.465 | D-Turanose | 0.0216 ± 0.0138 | 0.0197 ± 0.0090 | 0.0510 ± 0.0099 |
| 40 | 25.653, 25.783 | D-(+)-Lactose monohydrate* | 1.0400 ± 0.3349 | 0.7475 ± 0.2366 | 0.6559 ± 0.3286 |
| 41 | 25.927 | Lactose* | 0.0142 ± 0.0043 | 0.0143 ± 0.0075 | 0.0190 ± 0.0163 |
The Royal Society of Chemistry apologises for these errors and any consequent inconvenience to authors and readers.
References
- J.-H. Huang, R.-H. He, L.-Z. Yi, H.-L. Xie, D.-sheng Cao and Yi-Z. Liang, Talanta, 2013, 110, 1–7 CrossRef CAS PubMed.
- H. Yi, L. Yi, R. He, Q. Lv, X. Ren, Z. Zhang, Y. Liang and J. He, Anal. Lett., 2012, 45(13), 1862–1874 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ay90023b |
| This journal is © The Royal Society of Chemistry 2016 |
