The self-assembly and secondary structure of peptide amphiphiles determine the membrane permeation activity†
Rie Wakabayashi*a,
Yuko Abea,
Noriho Kamiyaab and
Masahiro Goto*abc
aDepartment of Applied Chemistry, Graduate School of Engineering, Kyushu University, Motooka 744, Nishi-ku, Fukuoka 819-0395, Japan. E-mail: m-goto@mail.cstm.kyushu-u.ac.jp; Fax: +81-802-2810; Tel: +81-92-802-2806
bCenter for Future Chemistry, Kyushu University, Motooka 744, Nishi-ku, Fukuoka 819-0395, Japan
cCenter for Transdermal Drug Delivery, Kyushu University, Motooka 744, Nishi-ku, Fukuoka 819-0395, Japan
First published on 10th July 2014
Abstract
Membrane fusogenic peptides have attracted increasing attention because of their unique biofunctions in membrane translocation and viral infection. Here, we designed GALA-related peptides with palmitoyl tails. Our study indicated that the self-assembling propensity and the secondary structure of these peptide amphiphiles greatly influenced the membrane permeability.
Membrane fusion is a protein-mediated event, which is critical in cellular physiology and viral infection.1–3 Most membrane-active proteins possess fusion peptides. As a common structural feature, these peptides adopt a relatively hydrophobic, amphipathic α-helix when active. Because membrane fusogenic peptides can be useful in bionanotechnology and nanomedicine, not only naturally derived, but de novo designed peptides have been explored extensively.4
GALA is an artificial membrane-active peptide designed by Szoka et al.5,6 The sequence of the GALA peptide (WEAALAEALAEALAEHLAEALAEALEALAA) is composed of repeating sequences of glutamic acid–alanine–leucine–alanine (EALA). The secondary structure of GALA transforms from a random coil to an amphipathic α-helix when the pH is decreased from physiological to slightly acidic conditions (<6). The formation of a pH-responsive α-helix enables the site-specific activation of the peptide at an intracellular acidic membrane called the endosome. This peptide has significant potential as a functional group for drug delivery via carriers because the structure is believed to enhance the endosomal escape and to deliver efficiently encapsulated drugs into cells.7–16
Intensive studies on GALA peptides include the modification of the peptide with different peptide sequences or the conjugation of hydrophobic tails to the terminus. Although GALA-modified peptides with shorter peptide sequences would be synthetically easier, the shorter sequence destabilizes the secondary structure.5,17 The peptides conjugated with hydrophobic tails, also known as peptide amphiphiles (PAs), form supramolecular assemblies, which can stabilize the secondary structure of the peptides.18,19 However, the contribution of the self-assembling propensity combined with the secondary structure to the membrane activity has not been explored. In this study, we have synthesized two different PAs with GALA-derived 8 or 14 amino acid sequences and examined how the structural features affect membrane permeating ability.
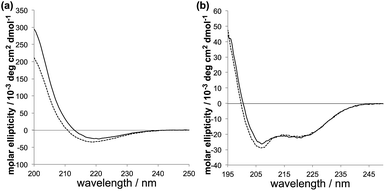
The new PAs have an alanine–leucine–alanine–glutamic acid (ALAE) repeating unit; the palmitoylated peptide sequences are palmitoyl-(ALAE)2 (PA1) and palmitoyl-E(ALAE)3W (PA2) (Scheme 1). In the PA design, the amino acid adjacent to the hydrophobic core plays an important role in the formation of intermolecular hydrogen bonding of PAs.20–22 The relatively hydrophobic alanine for PA1 can be expected to assist the formation of β-sheet. The hydrophilic glutamic acid for PA2 disrupts the intermolecular hydrogen bonding and can lead the peptide to form an α-helix structure, which is found in the original GALA peptide. The secondary structures that PA1 and PA2 formed at physiological and acidic pH were confirmed by the circular dichroism (CD) spectroscopy. The characteristic CD spectra for β-sheet structure were observed for PA1 both at pH 5.5 and 7.5 (Fig. 1a). In contrast to PA1, PA2 formed an α-helix with little difference between spectra recorded at pH 5.5 and 7.5 (Fig. 1b). We thus succeeded in synthesizing GALA-mimic PAs with different secondary structures. It was reported that short peptides with 4–16 amino acids composed of 1–4 repeating units of EALA, (EALA)n, where n = 1–4, showed no stable secondary structure.5,17 We assume that the short peptide sequences of PA1 and PA2 could form stable secondary structures because the palmitoylation of the amino-terminus decreased the conformational freedom of the molecule.18,19
Because the formation of β-sheet has been found to be important for self-assembly of PAs,20–22 PA1 may have higher tendency to assemble than PA2. To investigate the self-aggregation propensity of the peptides, the critical micelle concentration (CMC) was determined by using the pyrene solubilization method.23 The CMC values of PA1 both at pH 5.5 and 7.5 were lower than those of PA2 (Fig. S2†). It is clear that the formation of a β-sheet structure through intramolecular hydrogen bonding is important for self-assembly. Interestingly, PA1 has a higher CMC at pH 5.5 than pH 7.5 (8 μM for pH 5.5 and 2 μM for pH 7.5), whereas PA2 has a similar CMC at pH 5.5 and pH 7.5 (12 μM). This result indicated that the aggregation scheme differs between PA1 and PA2; the balance between hydrophilicity and hydrophobicity is important for the β-sheet forming PA1 by enabling efficient intramolecular hydrogen bonding, whereas the protonation of the glutamate has little influence on aggregation for the α-helix forming PA2. It indicates that the main driving force of assembly for PA2 is the hydrophobic interaction between the palmitoyl tails. Since PA2 forms amphipathic α-helix both at pH 5.5 and 7.5, the hydrophobic interaction between leucine residues may have an additional contribution to the assembly. Although acylated GALA peptides with fatty acids show lower CMC values under acidic conditions because of the protonation of glutamate side chains,19 the protonation of shorter peptide sequences of PA2 may have less influence on aggregation.
The aggregation of PAs was further confirmed by transmission electron microscopy (TEM). Nanofibers with microns lengths were observed in the PA1 solution above the CMC (100 μM) at pH 5.5 and 7.5 (Fig. S4†). Conversely, not a clear nanostructure but only amorphous aggregate was observed for PA2 either at pH 5.5 or 7.5 (data not shown). In PA design, introducing a β-sheet forming peptide represents an effective approach for self-assembly into one-dimensional structures through intramolecular hydrogen bonding along the long axis.20,21 It is clear that the β-sheet structure of PA1 plays an important role in self-assembling into well-defined nanofibers.
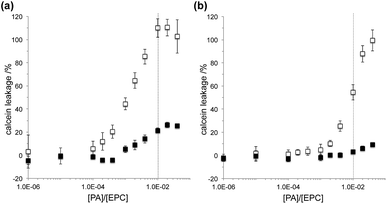
The membrane activity of the PAs was examined by monitoring the leakage of encapsulated calcein from egg PC (EPC) liposomes. PA was added to liposome suspensions containing 100 μM EPC with various PA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EPC ratios. The released calcein increased in proportion to the PA
EPC ratios. The released calcein increased in proportion to the PA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EPC ratio and PA2 showed higher permeation activity than PA1 both at pH 5.5 and 7.5 (Fig. 2). The amphipathic nature of the GALA-based α-helix was shown to be important for the membrane permeation activity.5 It is clear that the formation of an amphipathic α-helix by PA2 is responsible for its membrane permeation activity. The tryptophan residue in the PA2 sequence may have an additional effect on the permeation of membranes.24,25 Although calcein leakage reached >90% in the presence of PA2 with the PA
EPC ratio and PA2 showed higher permeation activity than PA1 both at pH 5.5 and 7.5 (Fig. 2). The amphipathic nature of the GALA-based α-helix was shown to be important for the membrane permeation activity.5 It is clear that the formation of an amphipathic α-helix by PA2 is responsible for its membrane permeation activity. The tryptophan residue in the PA2 sequence may have an additional effect on the permeation of membranes.24,25 Although calcein leakage reached >90% in the presence of PA2 with the PA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EPC ratio of 1
EPC ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 for pH 5.5 and 4
100 for pH 5.5 and 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 for pH 7.5, the leakage reached a plateau at ∼25% for PA1 with a ratio of 1
100 for pH 7.5, the leakage reached a plateau at ∼25% for PA1 with a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 at pH 5.5 and at ∼10% with the ratio of 4
100 at pH 5.5 and at ∼10% with the ratio of 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 at pH 7.5. Suppression of membrane activity of PA1 at higher PA concentrations may be caused by the formation of β-sheet structures and self-assembly of PA1.26 The assembled PA1 nanofibers may bind to liposomes, but the calcein release profiles suggest that not the aggregate but the monomeric PA leads to the perturbation of the bilayer. Unlike PA1, PA2 forms no defined nanostructure and the membrane permeation activity continues to increase in proportion to the PA
100 at pH 7.5. Suppression of membrane activity of PA1 at higher PA concentrations may be caused by the formation of β-sheet structures and self-assembly of PA1.26 The assembled PA1 nanofibers may bind to liposomes, but the calcein release profiles suggest that not the aggregate but the monomeric PA leads to the perturbation of the bilayer. Unlike PA1, PA2 forms no defined nanostructure and the membrane permeation activity continues to increase in proportion to the PA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EPC ratio.
EPC ratio.
For both PAs, higher leakage of calcein was observed at pH 5.5 than pH 7.5. The pH responsive permeation of membranes by the GALA peptide is usually explained by the pH responsive α-helix formation.5,17 The amphipathic nature of the α-helix formed at lower pH facilitates the fusion of the peptide with the membrane to form pores. Given that both PA1 and PA2 show little difference in secondary structures between pH 5.5 and 7.5, this suggests that the higher permeation activity at pH 5.5 may stem from the increase in hydrophobicity at the lower pH rather than the secondary structure. It was shown that PAs composed of the GALA peptide and a lauryl tail led to membrane rupture through a surfactant-like mechanism at physiological pH; but the PA leaked an encapsulated low-molecular-weight dye in a similar manner to that observed for the GALA peptide at pH 5.5.19 To investigate the mechanism how PAs in this study interact with membranes, a dynamic light scattering measurement was performed on liposomal suspensions with or without PA treatment. The scattering light intensity is a function of size of colloidal particles and can be used to monitor the stability of liposomes. When a liposome was treated with Triton X-100, a well-known nonionic surfactant, at a final concentration of 0.1 wt%, the scattering intensity dramatically decreased, which results from a membrane lysis (Fig. S4†). On the other hand, upon addition of PA1 or PA2 with a PA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EPC ratio of 1
EPC ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, the surfactant-like lysis was suppressed and the scattering intensity was nominally preserved both at pH 5.5 and pH 7.5. It is noteworthy that liposomes treated with PA2 scattered >95% intensity of the light even though >99% (pH 5.5) or ∼55% (pH 7.5) of calcein was released from the liposome (Fig. 2a, Table S1†). This behavior suggests that the EPC liposome maintains its structure upon addition of PA2; however, the amphipathic α-helix creates pores within the membrane that release calcein over time. For PA1, a similar mechanism seems feasible at pH 5.5 because >95% intensity of light was detected while ∼20% of calcein was released; however, the mechanism remains unclear at pH 7.5 when most of calcein (∼98%) is kept inside liposomes. Based on those results, we concluded that the calcein leakage induced by PA2 both at pH 5.5 and 7.5 or by PA1 at pH 5.5 was not because of liposome bursts, but because calcein could pass through the membrane presumably through pores formed by the PAs. Although PAs in this study are composed of much shorter peptide sequences (8 and 14 amino acids for PA1 and PA2, respectively) than GALA (30 amino acids), the fundamental nature of the interactions with membranes at pH 5.5 may not differ.
100, the surfactant-like lysis was suppressed and the scattering intensity was nominally preserved both at pH 5.5 and pH 7.5. It is noteworthy that liposomes treated with PA2 scattered >95% intensity of the light even though >99% (pH 5.5) or ∼55% (pH 7.5) of calcein was released from the liposome (Fig. 2a, Table S1†). This behavior suggests that the EPC liposome maintains its structure upon addition of PA2; however, the amphipathic α-helix creates pores within the membrane that release calcein over time. For PA1, a similar mechanism seems feasible at pH 5.5 because >95% intensity of light was detected while ∼20% of calcein was released; however, the mechanism remains unclear at pH 7.5 when most of calcein (∼98%) is kept inside liposomes. Based on those results, we concluded that the calcein leakage induced by PA2 both at pH 5.5 and 7.5 or by PA1 at pH 5.5 was not because of liposome bursts, but because calcein could pass through the membrane presumably through pores formed by the PAs. Although PAs in this study are composed of much shorter peptide sequences (8 and 14 amino acids for PA1 and PA2, respectively) than GALA (30 amino acids), the fundamental nature of the interactions with membranes at pH 5.5 may not differ.
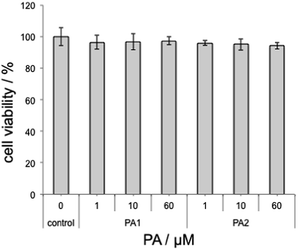
Finally, cytocompatibility of PAs was examined using Chinese Hamster Ovary (CHO) cells. CHO cells were incubated in the presence of PAs for 24 h and the cell proliferation was examined by the WST assay. Both PAs showed negligible cytotoxicity to CHO cells up to 60 μM (Fig. 3). Although PA2 showed a relatively high perturbation activity towards liposomes at physiological pH, the activity was only evidenced for simple synthetic liposomes and not generally for any biological membranes.
Conclusions
In conclusion, we showed that PAs with peptide sequences of 2 or 3 repeating units of ALAE can act as pH-responsive membrane activators. PA2 showed significantly higher membrane permeation activity than PA1, because PA2 formed an amphipathic α-helix and did not self-assemble into a well-defined nanostructure. Our results provide a more productive way of using GALA-related peptides because the molecular design is more versatile and synthetically accessible. The efficacy of PAs when incorporated in a drug delivery carrier is currently ongoing in our laboratory.Acknowledgements
This work was supported by a Grant-in-Aid for Scientific Research (S) 24226019 from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and the Sasakawa Scientific Research Grant from the Japan Science Society. The authors thank Professors Nobuo Kimizuka and Masaaki Morikawa, and Ms Natsuko Ide for the facility support for the TEM measurement.Notes and references
- J. M. White, Annu. Rev. Physiol., 1990, 52, 675 CrossRef CAS PubMed.
- R. Jahn, T. Lang and T. C. Südhof, Cell, 2003, 112, 519 CrossRef CAS.
- W. Wickner and R. Schekman, Nat. Struct. Mol. Biol., 2008, 15, 658 CAS.
- B. Gupta, T. S. Levchenko and V. P. Torchilin, Adv. Drug Delivery Rev., 2005, 57, 637 CrossRef CAS PubMed.
- N. K. Subbarao, R. A. Parente, F. C. Szoka Jr, L. Nadasdi and K. Pongracz, Biochemistry, 1987, 26, 2964 CrossRef CAS.
- W. Li, F. Nicol and F. C. Szoka Jr, Adv. Drug Delivery Rev., 2004, 56, 967 CrossRef CAS PubMed.
- T. Kakudo, S. Chaki, S. Futaki, I. Nakase, K. Akaji, T. Kawakami, K. Maruyama, H. Kamiya and H. Harashima, Biochemistry, 2004, 43, 5618 CrossRef CAS PubMed.
- S. Futaki, Y. Masui, I. Nakase, Y. Sugiura, T. Nakamura, K. Kogure and H. Harashima, J. Gene Med., 2005, 7, 1450 CrossRef CAS PubMed.
- K. Sasaki, K. Kogure, S. Chaki, Y. Nakamura, R. Moriguchi, H. Hamada, R. Danev, K. Nagayama, S. Futaki and H. Harahima, Anal. Bioanal. Chem., 2008, 391, 2717 CrossRef CAS PubMed.
- Y. Sakurai, H. Hatakeyama, H. Akita, M. Oishi, Y. Nagasaki, S. Futaki and H. Harashima, Biol. Pharm. Bull., 2009, 32, 928 CAS.
- S. Kobayashi, I. Nakase, N. Kawabata, H.-H. Yu, S. Pujals, M. Imanishi, E. Giralt and S. Futaki, Bioconjugate Chem., 2009, 20, 953 CrossRef CAS PubMed.
- H. Hatakeyama, E. Ito, H. Akita, M. Oishi, Y. Nagasaki, S. Futaki and H. Harashima, J. Controlled Release, 2009, 139, 127 CrossRef CAS PubMed.
- H. Akita, K. Kogure, R. Moriguchi, Y. Nakamura, T. Higashi, T. Nakamura, S. Serada, M. Fujimoto, T. Naka, S. Futaki and H. Harashima, J. Controlled Release, 2010, 143, 311 CrossRef CAS PubMed.
- I. A. Khalil, Y. Hayashi, R. Mizuno and H. Harashima, J. Controlled Release, 2011, 156, 374 CrossRef CAS PubMed.
- Y. Sakurai, H. Hatakeyama, Y. Sato, H. Akita, K. Takayama, S. Kobayashi, S. Futaki and H. Harashima, Biomaterials, 2011, 32, 5733 CrossRef CAS PubMed.
- F. S. Nouri, X. Wang, M. Dorrani, Z. Karjoo and A. Hatefi, Biomacromolecules, 2013, 14, 2033 CrossRef CAS PubMed.
- R. A. Parente, S. Nir and F. C. Szoka Jr, J. Biol. Chem., 1988, 263, 4724 CAS.
- C. Puyal, L. Maurin, G. Miquel, A. Bienvenüe and J. Philippot, Biochim. Biophys. Acta, 1994, 1195, 259 CrossRef.
- B. F. Lin, D. Missirlis, D. V. Krogstad and M. Tirrell, Biochemistry, 2012, 51, 4658 CrossRef CAS PubMed.
- J. D. Hartgerink, E. Beniash and S. I. Stupp, Science, 2001, 294, 1684 CrossRef CAS PubMed.
- S. E. Paramonov, H.-W. Jun and J. D. Hartgerink, J. Am. Chem. Soc., 2006, 128, 7291 CrossRef CAS PubMed.
- T. Muraoka, H. Cui and S. I. Stupp, J. Am. Chem. Soc., 2008, 130, 2946 CrossRef CAS PubMed.
- K. P. Ananthapadmanabhan, E. D. Goddard, N. J. Turro and P. L. Kuo, Langmuir, 1985, 1, 352 CrossRef PubMed.
- W.-M. Yau, W. C. Wimley, K. Gawrisch and S. H. White, Biochemistry, 1998, 37, 14713 CrossRef CAS PubMed.
- M. R. R. de Planque, B. B. Bonev, J. A. A. Demmers, D. V. Greathouse, R. E. Koeppe II, F. Separovic, A. Watts and J. A. Killian, Biochemistry, 2003, 42, 5341 CrossRef CAS PubMed.
- M. Rafalski, J. D. Lear and W. F. DeGrado, Biochemistry, 1990, 29, 7917 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Materials, methods, and figures. See DOI: 10.1039/c4ra02901a |
| This journal is © The Royal Society of Chemistry 2014 |