Delivery of dual miRNA through CD44-targeted mesoporous silica nanoparticles for enhanced and effective triple-negative breast cancer therapy†
Manisha
Ahir
a,
Priyanka
Upadhyay
a,
Avijit
Ghosh
a,
Sushmita
Sarker
a,
Saurav
Bhattacharya
a,
Payal
Gupta
b,
Swatilekha
Ghosh
c,
Sreya
Chattopadhyay
b and
Arghya
Adhikary
 *a
*a
aCenter for Research in Nanoscience and Nanotechnology, Technology Campus, University of Calcutta, JD-2, Sector-III, Salt Lake City, Kolkata 700106, India. E-mail: adhikaryarghya@gmail.com; Tel: +91 9830428550
bDepartment of Physiology, University of Calcutta, 92 Acharya Prafulla Chandra Road, Kolkata, 700009, WB, India
cAmity School of Biotechnology, Amity University, Kolkata, Major Arterial Road (South-East), Action Area II, Newtown, Kolkata, West Bengal 700135, India
First published on 6th April 2020
Abstract
The development of new therapeutic strategies to target triple-negative breast cancer (TNBC) is in much demand to overcome the roadblocks associated with the existing treatment procedures. In this regard, therapies targeting the CD44 receptor have drawn attention for more than a decade. MicroRNAs (miRNAs) modulate post-transcriptional gene regulation and thus, the correction of specific miRNA alterations using miRNA mimics or antagomiRs is an emerging strategy to normalize the genetic regulation in the tumor microenvironment. It has been acknowledged that miR-34a is downregulated and miR-10b is upregulated in TNBC, which promotes tumorigenesis and metastatic dissemination. However, there are a few barriers related to miRNA delivery. Herein, we have introduced tailored mesoporous silica nanoparticles (MSNs) for the co-delivery of miR-34a-mimic and antisense-miR-10b. MSN was functionalized with a cationic basic side chain and then loaded with the dual combination to overexpress miR-34a and downregulate miR-10b simultaneously. Finally, the loaded MSNs were coated with an hyaluronic acid-appended PEG-PLGA polymer for specific targeting. The cellular uptake, release profile, and subsequent effect in TNBC cells were evaluated. In vitro and in vivo studies demonstrated high specificity in TNBC tumor targeting, leading to efficient tumor growth inhibition as well as the retardation of metastasis, which affirmed the clinical application potential of the system.
1. Introduction
Triple-negative breast cancer (TNBC) accounts for about 10–20% of all the diverse subtypes of breast cancers and has been negatively tested for the estrogen receptor (ER−), the progesterone receptor (PR−), and the human epidermal development factor receptor 2 (HER2−).1 The survival rate of patients experiencing TNBC is less due to the lack of commercially available targeted therapies.2,3 Hence, designing a new therapeutic approach for TNBC might be useful for improved treatment. Small non-coding endogenous RNA molecules, known as microRNAs, regulate genes and are situated at the introns of protein-coding genes, the introns of non-coding genes, or the exons of non-coding genes.4,5 An investigation by Ferracin et al. revealed that there are some signature microRNAs that control human breast cancer6 and their deregulation is associated with diverse processes, including their roles as oncogenes (oncomiRs) or tumor silencers adding to tumorigenesis;7 furthermore, they play a role in the modulation of cancer properties, such as cell death, metastasis, and stemness.8,9 Thinking about these classes of microRNAs, Weinberg's group showed that in breast cancer, miR-10b assumes an exceptionally critical role by starting the tumor attack by focusing on the homeobox D10 (HOXD-10) gene.10 In order to come up with an alternative option, a prerequisite approach could be by using miRNA mimics or antagomirs.11 It is hypothesized that we could make some alterations to these specific miRNA that could lead to gene regulatory network normalization and thus could reverse the phenotype in cancerous cells. Similarly, studies have also provided insights into another category of microRNA that plays a significant role in the suppression of cancer. One such tumor suppressor microRNA is miR-34a, whose expression analysis verified that the miR-34a/c expression is significantly decreased in metastatic breast cancer cells and human primary breast tumors with lymph node metastases.10,12 Despite significant potential, there remain some serious difficulties associated with the systemic delivery of miRNAs to tumors due to their poor cellular uptake, low stability, and rapid clearance from the systemic circulation.13 The lack of a targeted delivery vehicle for the protected delivery of miRNAs is also a significant drawback in miRNAs-mediated therapy.14 Porous silica nanoparticles contain a highly ordered porosity that provides a high surface area, large pore volume, favorable biocompatibility, thermal stability, and easy surface modification, making them suitable for the simultaneous delivery of bioactive compounds or small nucleotides.15–17 In research, MSN was chosen as a carrier for the easy encapsulation of miRNA by simple physical diffusion methods. The MSN core was further surface modified by attachment of a basic side chain cationic spacer. Furthermore, MSN can be coated with cationic polymers, including PEI, PEI-cyclodextrin, and PDMAEMA, for cellular entrance. However they lack cancer cells specificity.18–21On the other hand, poly(lactic-co-glycolic acid) copolymer (PLGA) is commonly used for hosting several FDA-approved therapeutics, owing to its biodegradability and biocompatibility.22–24 PEG-PLGA can act as a steric layer to protect the miRNAs against blood enzymes, to prevent the aggregation of cationic nanoparticles, to enhance blood safety, and to prevent an adverse immune response to the nanoparticles. Hence we hypothesized that PEG-PLGA coating on the MSN surface can allow it to act as a protective and improved delivery system.25–29 There are a few delivery frameworks that have reported this type of shielding effect of the nucleic acids from degradation.29 Several miRNAs, such as miR-29b, miR-10b, and miR-155, have previously been delivered using different nanoparticles and have shown efficacy against different types of cancer.30–32 On the other hand, CD44 is a cell attachment layer glycoprotein overexpressed on the surface of TNBC cells, in contrast with normal cells. CD44 has been shown to be elevated in multiple diseases, including cancer, making it a potential diagnostic biomarker and an ideal receptor for targeted drug-delivery systems. Therefore, hyaluronic acid (HA), a polysaccharide, was supplied with a PLGA polymer coating at the tumor micro environment due to its high affinity toward CD44.33,34
Keeping all this in mind, efforts have been devoted to the synthesis of HA-adorned PLGA-coated miRNA (anti-miR-10b and miR-34a mimic)-loaded-mesoporous silica nanoparticles (MSN) and toward the determination of its anti-tumorigenic effect against TNBC. Remarkably, we found that, upon surface modification (basic cationic side chain), MSN was able to load dual miRNAs with enhanced efficiency. Strongly cationic particles are known to interact more favorably with cell membranes, thereby enhancing their intracellular uptake. Once decorated, these nanoparticles can get easily internalized inside the cells; thereby the respective microRNAs are able to terminate cancer cells by causing cell death. Additionally, our strategy successfully restricted the migration of TNBC cells. We validated our findings in BALB/c mice model and studied the biodistribution of nanoparticles as well. Our results are promising and we hope that the development of such alternative therapeutic strategies for TNBC treatment may lead to clinical translation, which might improve the survival rates of patients worldwide by restricting cancer relapse and metastasis.
2. Materials and methods
2.1 Chemicals
Tetraethyl orthosilicate (TEOS), cetyltrimethylammonium bromide (CTAB) diethanolamine (DEA), aminopropyl-triethoxysilane (APTES), poly(D,L-lactide-co-glycolide) (PLGA Mw 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–38
000–38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000), 2,2′-(ethylenedioxy)bis(ethylamine) [mol. wt 148.20], hyaluronic acid sodium salt mol. wt 8000–15
000), 2,2′-(ethylenedioxy)bis(ethylamine) [mol. wt 148.20], hyaluronic acid sodium salt mol. wt 8000–15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, 2,2′ (ethylenedioxy)bis(ethylamine)-N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC), N-hydroxysuccinimide (NHS), and absolute ethanol were purchased from Sigma-Aldrich (Germany). The dialysis membrane (MWCO 2000 Da, 12
000, 2,2′ (ethylenedioxy)bis(ethylamine)-N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC), N-hydroxysuccinimide (NHS), and absolute ethanol were purchased from Sigma-Aldrich (Germany). The dialysis membrane (MWCO 2000 Da, 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Da) was purchased from Himedia (Mumbai, India).
000 Da) was purchased from Himedia (Mumbai, India).
2.2 Synthesis of cationic amine-functionalized mesoporous silica nanoparticle (MSN-NMe3)+I−
First, 40 mg of amine-functionalized mesoporous silica nanoparticles (MSN-NH2) were sonicated in a probe sonicator and then stirred in dichloromethane at 1000 rpm to obtain a white suspension of MSN-NH2. Then, 1 mL of methyl iodide (excess CH3I) was added to the reaction mixture. Then, the above solution was stirred for 48 h at 500 rpm in an argon atmosphere. The fractionation of MSN may be accomplished using centrifugal filter fractionation or sequential centrifugal filtration. Sequential centrifugal filtration can remove unreacted mesoporous silica nanoparticles.Silica nanoparticles have a high dispersibility in water or ethanol solvent. In the case of silane containing long carbon chains, the made silica NPs have a higher dispersibility due to the existence of long carbon chains. Therefore, the dispersibility of MSN-NH2 is greater than that of free MSN. On the other hand, the cationic nature of the amine functionalization reduces its dispersibility. Hence, sequential centrifugal filtration may remove unreacted amine-functionalized mesoporous silica nanoparticles.
2.3 Preparation of MSN-loaded miRNAs
About 50 mg (MSN-NMe3)+I− was added into 5 mL DEPC water solution containing miRNAs (5 μg mL−1) and stirred for 2 h at 4 °C. The obtained MSNs-miRNAs were then collected by centrifugation and washed with deionized water.2.4 Preparation of the HA-PEG-PLGA triblock copolymer
First, 400 mg of HA sodium salt was dissolved in Millipore water (20 ml), and dialyzed against a dilute HCl solution (pH ∼3.5) using a dialysis tube of MWCO 2 kDa (Sigma, USA), for 12 h each, followed by lyophilization. To prepare the HA-PEG-PLGA conjugate, the COOH groups of HA were activated by treating with EDC and NHS for 2 h in DMSO. Then, PLGA-PEG-NH2 copolymer was added to the activated HA and stirred overnight to obtain nanoparticles decorated with HA. After completion of the reaction, the reactant was purified by successive dialysis (MWCO 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Da) against an excess of 0.1 M NaCl, 25% (v/v) ethanol solution, and then deionized water. The solution was then lyophilized and stored at −20 °C until further use. The product was characterized by 1H NMR.
000 Da) against an excess of 0.1 M NaCl, 25% (v/v) ethanol solution, and then deionized water. The solution was then lyophilized and stored at −20 °C until further use. The product was characterized by 1H NMR.
2.5 Preparation of MSN-miRNAs @HA-PLGA by the W1/O/W2 double emulsion method
HA-PEG-PLGA copolymer was dissolved in dichloromethane–DMSO and sonicated for 5 min to obtain a clear solution. MSN or micro-RNA-loaded MSN was dispersed in 1 mL DEPC/water. These two solutions were mixed by injection and sonicated for 2 min at 100 W to form a microemulsion. Then, the microemulsion was added into 2% PVA DEPC/water by sparing and sonicated in a probe sonicator for 10 min. After the organic solvents were removed in a vacuum, the composite nanoparticles were obtained by repeated centrifugation and washing with DEPC/water. Custom RNA oligos 1.0 μmol, HPLC purified grade, namely hsa-miR-34a-5p (5′-UGGCAGUGUCUUAGCUGGUUGU-3′) and anti-sequence miR-10b-5p (5′-CACAAAUUCGGUUCUACAGGGUA-3′), were synthesized and purchased from Metabion.2.6 Physicochemical characterization of the nanoparticles
The size and morphology of the nanoparticles were observed by dynamic light scattering (DLS), transmission electron microscopy (TEM) (JEM-2100HR-TEM, JEOL, Japan), and field emission scanning electron microscopy (FESEM) (JSM-7600F, JEOL, Japan). Fourier-transform infrared spectroscopy (FTIR, JASCO, Japan) was used to determine the chemical properties of the NPs. For 1H NMR analysis, the synthesized NPs were dissolved in D2O, and analyzed by a 300 MHz NMR spectrometer (Bruker AC 400 superconducting system, Switzerland). Thermogravimetric analysis (TGA) of the synthesized nanoparticles was done on a TG/DTA analyzer (Perkin Elmer, USA) under a nitrogen atmosphere at a heating rate of 20 °C min−1.2.7 Cell culture and PBMC isolation
Human mammary carcinoma cell lines (MDAMB-231 and MDAMB-468) were acquired from the National Centre for Cell Science (NCCS, Pune, India). These specific cell lines were maintained in Dulbecco's modified eagle medium (DMEM) consisting of 100 mL L−1 of fetal bovine serum (FBS) at 5% CO2. The 4T1 murine mammary carcinoma cells were a kind gift from Prof. P. C. Sen's lab and maintained in RPMI with 10% FBS. A layer of human peripheral blood mononuclear cells (PMBC) was isolated and cultured in RPMI-1640 with 10% FBS.2.8 In vitro controlled release of miRNAs from the NPs
To examine the release of miR-34a from the NPs, the NPs were suspended in 2 ml of deionized water and fixed in two particular dialysis tubes with a sub-atomic weight cut-off of 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Da for miR-34a. Two separate sets were used for the dual miRNA NPs and HA-Dual miRNA NPs, respectively. The dialysis tubes were then drenched in a 10 ml PBS cushion dialysis arrangement at 37 °C under direct shaking utilizing a water-washing consistent temperature vibrator.
000 Da for miR-34a. Two separate sets were used for the dual miRNA NPs and HA-Dual miRNA NPs, respectively. The dialysis tubes were then drenched in a 10 ml PBS cushion dialysis arrangement at 37 °C under direct shaking utilizing a water-washing consistent temperature vibrator.
2.9 Fluorescent labeling of miRNA and microscopic analysis
First, 2 mM Cy5 was tagged to microRNA-34a and FAM labeling reagent to anti-micro-10b and incubated at room temperature (21 °C ± 1 °C) for 2 h. The labeled RNA molecules were purified by standard procedures. The Cy5-labeled as well as FAM-labeled RNA molecules were aliquoted as 10 μM and stored at −80 °C before the experiments. To obtain the cellular uptake images of RNA-nanoparticles, we used an Invitrogen fluorescent microscope. A 20× NA objective was used. The FAM and Cy5 fluorophores were excited by 488 nm and 633 nm lasers simultaneously. The emission band of 493–628 nm for the FAM-labeled miRNA channel and 638–759 nm bands for the Cy5-labeled miRNA channel were recorded simultaneously to detect the specific localization of miRNA within the cells.2.10 MTT assay
In order to determine the toxic effects of MSN loaded with microRNAs on PBMC cells, they were harvested, counted, and transferred to 96-well plates and incubated for 24 h prior to the addition of the nanoparticles. The MSN loaded with microRNAs was applied in various concentrations, and the treated cells were incubated for another 24 h. An MTT study was performed to determine the cellular viability by measuring the reading at 575 nm.2.11 Determination of cellular apoptosis by Annexin V
To determine the presence of apoptosis and necrosis, cell death was analyzed by Annexin V and PI staining as per the kit protocol (BD Biosciences) and further analyzed using a flow cytometer (BD Biosciences, San Diego, CA, USA).2.12 Measurement of intracellular ROS production
First, 2 μL of H2DCFDA working solution was used at a concentration of 2 μM. The cell suspensions were incubated for 20 min at 37 °C. After that, FACS analyses of the samples were carried out using FACSVerse at 488 nm excitation and 520 nm emission using the FACSuite software.2.13 JC-1 staining mitochondrial membrane potential
MDAMB-231 cells were treated with the nanoparticles in a six-well plate. The mitochondrial membrane potential was monitored using a fluorescent cationic dye JC-1 mitochondrial membrane potential detection kit (Biotium, Hayward, CA, USA). The samples were then observed under a fluorescence microscope (EVOS FL, Invitrogen).2.14 Immunostaining assay
The cells were fixed with 3.7% formaldehyde and permeabilized with 0.1% Triton X-100. Cells were then incubated with anti-cytochrome-c (Santa Cruz Biotechnologies, USA), followed by RITC-conjugated secondary antibody (Sigma-Aldrich, Germany). The nucleus was stained with DAPI (Invitrogen, Carlsbad, CA, USA). The samples were mounted on clean glass slides using Prolong Gold Antifade Reagent (Invitrogen, Carlsbad, CA, USA) and visualized under both fluorescence microscopes (EVOS FL, Invitrogen and BD Pathway 855).2.15 RNA isolation and quantitative real-time PCR study
Total RNAs were isolated from cells using miRVANA kit (Invitrogen, Carlsbad, CA, USA). The integrity and purity of total RNAs and small RNAs were verified by an ND1000 spectrophotometer (Thermo Scientific). Also, 1 μg of total RNA was reverse-transcribed using the transcriptor first strand cDNA synthesis kit and oligo (dT)18 primer (Biobharti, India). In the case of miRNA detection, the miR-specific stem-loop primers were used along with the same cDNA synthesis kit. qRT-PCR was performed with a 50 μg sample using Light cycler 96 (Roche) and SYBR green ready mix (Roche) and specific primers (Tables S1 and S2†). snRNA U6 and 18S rRNA were used as internal controls for miRNA and mRNA quantification, respectively. Gene and miRNA expression was defined from the threshold cycle (Cq), and the relative expression levels were calculated by using the 2−ΔCq method after normalization with reference to the expression of housekeeping genes (18S rRNA or U6).352.16 Western blot analysis
After appropriate treatment, the cells were harvested to prepare a whole cell lysate using RIPA buffer (Sigma-Aldrich, Germany) with a protease inhibitor mixture. Protein estimation was done using a bicinchoninic acid (BCA) assay kit (Merck, Germany). An equal amount of proteins was separated by SDS-PAGE and transferred onto a PVDF membrane (Millipore) and then incubated with primary antibodies, like anti-Bcl-2, Bax, HOXD-10, RhoC, Cyt C, and β-actin (Santa Cruz Biotechnologies, USA) and ALP-conjugated secondary antibodies. The blots were developed with NBT/BCIP (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). The results are expressed as the fold change relative to the control after normalization to β-actin.
1). The results are expressed as the fold change relative to the control after normalization to β-actin.
2.17 Mammosphere formation assay and CD44+/CD24− staining
First, 1 × 105 cells per mL were seeded on ultralow attachment plates (Corning, NY, USA) in serum-free DMEM/F12 media supplemented with 10 ng mL−1 b-FGF, 20 ng mL−1 EGF, 1× ITS (insulin + transferrin + selenium), and 1× B27. The nanoparticles were administered during mammosphere seeding on day 1 and MFE was calculated on day 6 by counting under an EVOS FL Invitrogen microscope at 40× magnification. In parallel identical sets, mammospheres were cultured and treated with the nanoparticles on day 5. On day 6, human CD44-FITC (5 μL) and CD24-PE (5 μL), (BD Biosciences) labeled cells were then washed in PBS to eliminate any unbound antibody, and data were acquired using a BD FACS Verse flow cytometer.362.18 Cell migration study
Cell migration was determined by means of a unidirectional wound healing assay. For this assay, 0.5–2.5 μl tips were used to scratch the monolayer of cells to form a unidirectional wound in cells grown to confluency in 12-well plates. The cultures were incubated at 37 °C and photographed immediately and monitored with a phase-contrast microscope. The experiments were performed in triplicate and were counted double-blind by at least two investigators. Also, a migration assay was done using cell culture inserts (BD Biosciences, Sparks, MD, USA). After treatment, the cells in the insert were washed with PBS, fixed with 3.7% formaldehyde, and then permeabilized using methanol and stained with Giemsa stain for 30 min. Cells present in the lower part of the inserts were determined by counting the cells in five microscopic fields per well, and the extent of migration was expressed as an average number of cells per microscopic field.2.19 Tumor regression in an ex ovo model
To detect the migration of human cancer cells through the CAM layer of a chick embryo, we performed a semi-quantitative chorioallantoic membrane (CAM) assay following the defined protocol.37,38 On days 10 and 12, the chick embryos were administered with the nanoparticles intravenously at a dosage of 15 μg mL−1, while the control set was left untreated. On day 16, to examine the migrated human cells through the CAM layer of chick tissue, the embryos were sacrificed and primer specific RT PCR was performed for the human Alu sequences against chick GAPDH as the internal control. Also the relative expressions of Bcl-2, HoxD-10 mRNAs, and miR-34a, miR-10b were estimated by real-time PCR, while intracellular metastasis was checked in tumor tissue isolated from the control and treated set.2.20 In vivo study
Female BALB/c mice were procured from registered animal distributor Chittaranjan National Cancer Institute (CNCI; 37, S. P. Mukherjee Road, Kolkata-700026, India), (Reg no.: 175/99/CPCSEA) and allowed to acclimatize in the laboratory environment of the animal house of the Department of Physiology, University of Calcutta, Kolkata, according to the institutional guideline of the Animal Ethical Committee (IAEC) (Ethical Clearance no. IAEC-V/P/SC-05/2019 dated 07.08.2019). To perform the in vivo experiment, all the animals were divided into three groups, where each group consisted of four mice (n = 4): group I, untreated control possessing 4T1 induced tumor in mammary fat pads; group II, dual miRNA NPs-treated set (25 mg per kg bodyweight dosage); group III, HA-Dual miRNA NPs-treated set (15 mg per kg bodyweight dosage). On every alternative day of treatment, the tumor volume was measured as well as the tumor weight following sacrifice of the mice.2.21 In vivo biodistribution profile of the nanoparticles by ICP-AES
Inductively coupled plasma atomic emission spectroscopy (ICP-AES) is a type of emission spectroscopy that is used to perform elemental analysis and is also used for detection of the presence of any silica. Dual miRNA NPs and HA-Dual miRNA NPs were injected intra-peritoneally at a dose of 25 mg kg−1 and 15 mg per kg of bodyweight each, respectively. This was followed by dissection of the animals at 24 h and 10 days post-injection and the organs and tumors were expunged. Tumor samples were prepared as per the protocol described in Ahir et al.39 Then, analyses of the pre-injection solutions, organs, and tumor samples for the presence of silica were carried out at S.G.S Pvt. Ltd, Kolkata, India.2.22 In vivo toxicity assessment of the nanoparticles
BALB/c animals treated with both dual miRNA NPs (25 mg kg−1 of bodyweight) and HA-Dual miRNA NPs (15 mg kg−1 of bodyweight) were used to study the following parameters.2.23 Estimation of the serum specific toxicity markers (AST, ALT, ALP, LDH)
Blood of the anaesthesia-induced mice was collected followed by incubation at 4 °C for 1 h at a 45° angle. To estimate certain serum specific toxicity markers, AST (aspartate transaminase), ALT (alanine transaminase), ALP (alkaline phosphatase), LDH (lactate dehydrogenase), and clear serum, excluding the blood clot, was collected by centrifugation at 2000 rpm for 10 min at 4 °C and further the serum was diagnosed using a commercially available diagnostic kit procured from Span Diagnostics, following the manufacturer's protocol.2.24 Analysis of the oxidative stress-based biochemical parameters
In hepatic PMS, the activity of superoxide dismutase (SOD) enzyme was measured by a spectrophotometer and is expressed here as the units mg−1 of protein. The activity of catalase (CAT) enzyme was assayed using the spectrophotometer at a wavelength of 240 nm as per claimed in previously described protocols.39,40 In hepatic PMS, the level of lipid peroxidation was estimated by measuring the concentration of malonaldehyde (MDA).412.25 Statistics study
All of the experiments were performed at least in triplicate. The data are presented as the mean ± SD. The statistical significance was analyzed by one-way analysis of variance (ANOVA). The differences were considered to be statistically significant when the p values were less than 0.05 (p < 0.05).3. Results and discussion
3.1 Synthesis and characterization of the hyaluronic acid-coated MSN
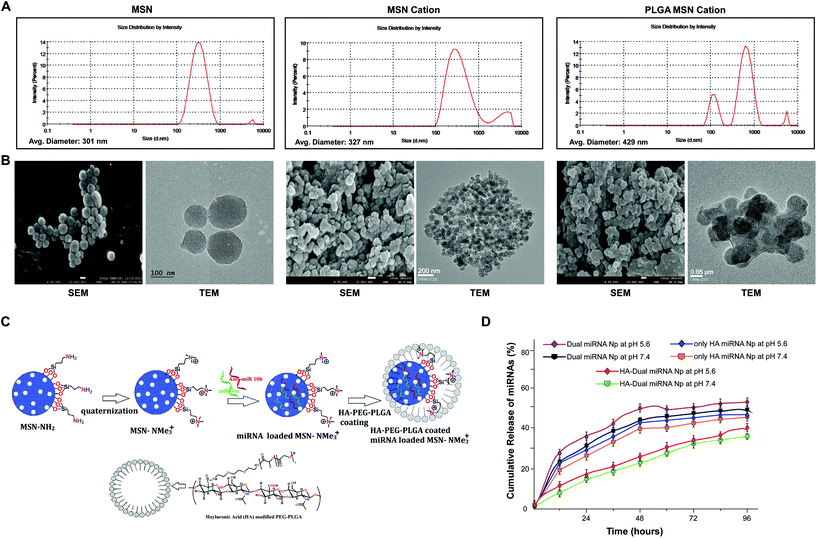
Porous MSN was synthesized and functionalized by adding a side chain –NH2 group. After quaternization of the amine functional group, the resulting MSN could effectively bind miRNA to them. Further, the PLGA-HA coating enhanced the stability, sustained and targeted the release of miRNAs at the desired site of action, i.e., CD44 marker enriched cancer cells. A dynamic light scattering study determined that the average hydrodynamic diameter was in the range of 300–400 nm (Fig. 1A). The zeta potential was determined to be −24.20 ± 3.76 mV (Table S4†).27 SEM and TEM analyses showed the presence of pores within the MSN and the size was found to lie within 80–120 nm. The size of cationic-MSNs was 110–130 nm. TEM analysis also showed a prominent coating of PLGA over the MSN, which was quite visible in the magnified view. However, after coating with HA-PEG-PLGA the size increased to 180–220 nm. The reaction scheme is explained as shown in Fig. 1C and S1.†To further study the modification of nanoparticles, FTIR and 1H NMR spectra studies were also performed (Fig. S2 and S3†). The FTIR spectrum of MSN showed a strong spectrum signal around 1100 cm−1 (corresponding to the Si–O spectrum). It was evident from the results that MSN-NH2 exhibited a positive group at around 1600 cm−1 due to the presence of an NH2 group. Interestingly, cationic MSN also exhibited positive functional groups. The thermal stability and coating amount of polymer were analyzed by TGA and DTGA (first derivative) curves and our data clearly showed multiple weight loss steps for HA-PEG-PLGA, as shown and explained in Fig. S4.† We also calculated the loading of microRNAs in all the sets and measured the absorption spectra at 260 nm and chose the following values for our experimental studies. The present delivery system showed a high loading efficiency toward negatively charged miRNAs (Table 1). We performed a comparative study, which demonstrated that among the different modifications of mesoporous silica nanoparticles, the cationic group showed the highest binding with the microRNAs (Table 1). Initially it was found that the loading of dual miRNAs to MSN was very poor. Among the different functionalized MSNs, the neutral surface-functionalized mesoporous silica nanoparticles (MSN-CH2-CH2-CH3) exhibited less binding of microRNAs. Anionic-functionalized mesoporous silica nanoparticles (MSN-SO3H) exhibited almost no binding to microRNAs because of the presence of a negatively charged surface. Finally, MSNs functionalized with elongated basic cationic side chains had a significant loading of miRNAs. The miRNAs electrostatically adsorb onto the positively charged nanoparticles. The presence of a cationic group enhances the electrostatic interactions between anionic microRNAs and cationic surface-functionalized mesoporous silica nanoparticles (MSN-NCH3)+I− and increases the binding of microRNAs to it, thus giving a maximum concentration reading of 578.01 ng μL−1. The concentration readings corresponding to the incorporation of microRNAs are demonstrated in Table S3.†
| Sample | Absorbance (A260/280) | Concentration (ng μL−1) | Encapsulation of miRNAsa (%) |
|---|---|---|---|
| a Effects of the parameters on the encapsulating efficiencies of miRNAs. Results are the mean ± SD (n = 3). | |||
| MSN cation | 1.94 | 53.80 ± 0.01 | |
| MSN cation + 10b | 1.95 | 578.01 ± 0.01 | 90% |
| MSN cation + 34a | 1.90 | 502.85 ± 0.02 | 89% |
| Dual miRNA NPs | 1.90 | 511.35 ± 0.02 | — |
| HA-Dual miRNA NPs | 1.77 | 339.85 ± 0.01 | 84% |
| HA-PEG-PLGA | 1.08 | 19.994 ± 0.02 | — |
3.2 In vitro release kinetics, cellular uptake, and cytotoxicity of miRNA through nanoparticles
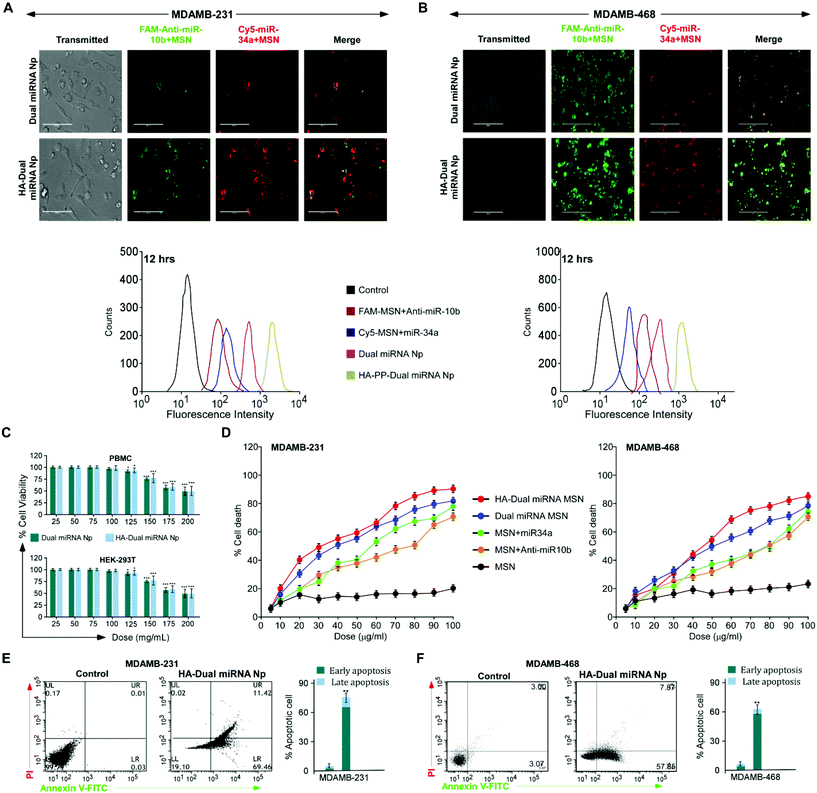
To evaluate the stability of the miRNAs-loaded NPs in a real biological system, a competitive release profiling of miRNAs from each of the NPs were carried out at different pHs. Compared to the fast in vitro release of loaded miRNAs from MSN NPs, the HA-coated NPs exhibited a sustained release at pH 7.4 and 5.6, respectively. The accumulative miRNA release in the dual miRNA MSN was around 35% at 12 h, 50% at 24 h, and stayed for up to 48 h only at pH 5.6. A similar release pattern was also observed at pH 7.4. From the figure of cumulative release (Fig. 1D), it could also be observed that only the HA-coated MSN loaded with miRNAs followed a somewhat better pattern corresponding to the non-coated MSN with dual miRNAs, but not much significant controlled release was observed at both pH 5.6 and 7.4. On the other hand, in the case of HA-targeted PEG-PLGA-coated NPs (HA-Dual miRNA NPs) a more sustained release of miRNA was observed. In the case of HA-Dual miRNA NPs, after 12 h (pH 5.6), only 10% of the miRNAs were released from the NPs, which ultimately went up to 30% after 48 h. A similar release profile was observed at pH 7.4. Within the tumor and lysosomal microenvironment, acidic conditions prevail, so our HA-coating study would enhance tumor therapy within the system. Hence, we can state that with the shielding effect of the HA coating, the release of miRNA from the HA-Dual miRNA NPs was even slower than that from the dual miRNA NPs (Fig. 1D).FAM (carboxyfluorescein)-tagged antimiR-10b and Cy5-tagged miR-34a were used to confirm intracellular internalization in MDAMB-231 and MDAMB-468. As compared to free miRNAs, those loaded with MSN showed better internalization within cells. The merged image gives a detailed view of accumulation of both the miRNAs inside the cells when delivered with MSN in both cell lines. Interestingly, enhanced internalization was observed in the HA-Dual miRNA NPs-treated sets due to the overexpression of CD44 in TNBC cells42,43 (Fig. 2A & B upper panel). To confirm the findings, we performed flow cytometry in both MDAMB-231 and MDAMB-468 cells and found that HA-Dual miRNA NPs-treated cells exhibited the highest fluorescence intensity in contrast to non-targeted nanoparticles, whereas the dual miRNA NPs sets showed the least fluorescence intensity (Fig. 2A & B lower panel).
Next, to investigate the effect of the miRNA-loaded nanoparticles on cell viability in MDAMB-231 and MDAMB-468 cells, an MTT assay was performed. Cells treated with HA-Dual miRNA NPs exhibited a significant percentage of cell death, which was found to be higher than for dual miRNA NPs (Fig. 2D). Contrastingly, a lower percentage of cell death was observed in MSN + miR34/MSN + antimiR-10b-treated cells. Furthermore, as no significant cell death was recorded in both cases of our nanoparticles for PBMC and HEK293T (Fig. 2C), it was concluded that our nanoparticles showed toxicity toward cancer cells but left the normal cells. Treatment with nanoparticles in both the cell lines depicted a condensation of the nuclei and DNA fragmentation, which are morphological changes considered to be features of early apoptosis (Fig. S5†). Next, we validated the results and found that HA-Dual miRNA NPs were able to cause an increase in apoptosis as compared to the non-targeted ones and control sets in both MDAMB-231 and MDAMB-468 cells (Fig. 2E).
3.3 Elucidation of the cytotoxic effect of nanoparticles on MDAMB-231 cells
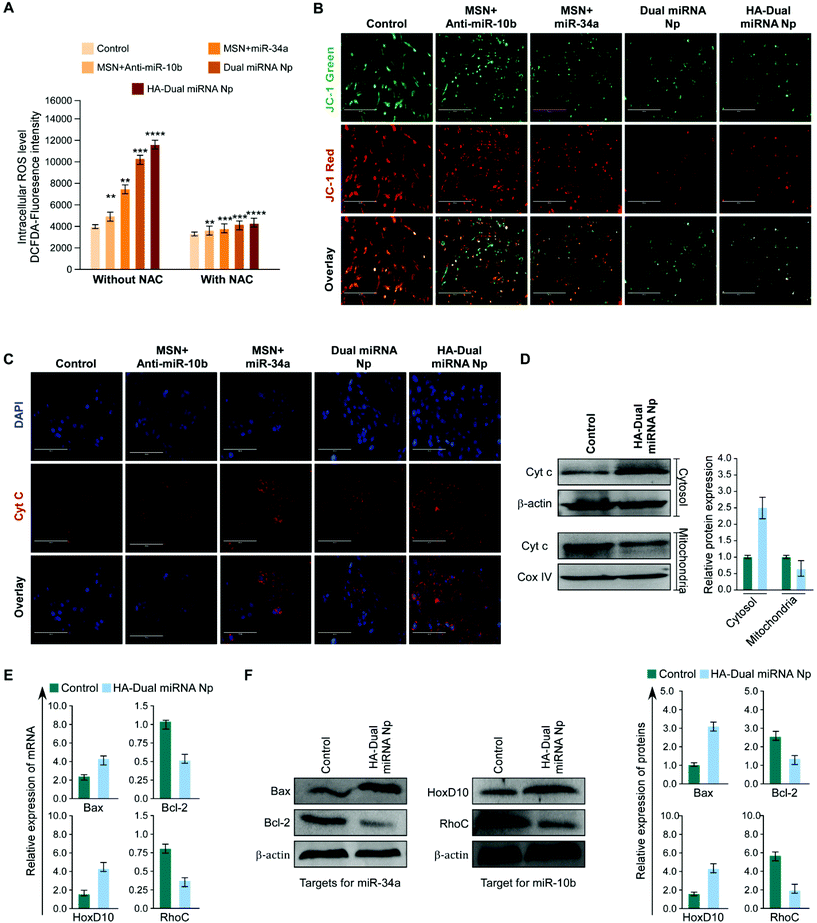
In order to estimate ROS generation in cells treated with nanoparticles, a DCFH-DA fluorescence assay was performed. We observed that the miR-34a + MSN loaded sets showed a high amount of ROS generation than MSN + antimiR-10b treated sets, whereas the maximum intensity was shown by HA-Dual miRNA NPs-treated cells. In parallel and identical sets, the cells were pre-treated with NAC and an observable decrease in fluorescence in all the sets was observed (Fig. 3A). Moreover to check whether mitochondrial depolarization plays any role or not, MDAMB-231 cells were treated with the predicted treatments and stained with JC-1 dye. We observed that the red to green fluorescence ratio decreased subsequently from the control to the single-loaded MSN to dual miRNA NPs, with a minimum intensity in the HA-Dual miRNA NPs-treated cells (Fig. 3B). Additionally, we also checked for the expression of cytochrome C (Cyt C), a prominent marker of apoptosis. Immunofluorescence imaging highlighted an increased expression of Cyt C in the cytosol (Fig. 3C), which was further validated from our western blotting data (Fig. 3D).TNBC cells are found to express significantly higher miR-10b and in contrast less miR-34a. Several reports support that the overexpression of miR-10b in non-metastatic breast cancer cells enables them to acquire invasive and metastatic behaviors. The miR-10b induced by Twist proceeds to inhibit translation of the messenger RNA encoding homeobox D10, resulting in an increased expression of the well-characterized pro-metastatic gene RHOC.44 In addition miR-34a is capable of regulating the proliferation, apoptosis, invasion, and migration of tumor cells by targeting multiple genes, indicating its important benefit in gene therapy. Different reports support the fact that the tumor suppressor miR-34a regulates Bcl-2 and plays a significant role in causing apoptotic cell death in the MDAMB-231 cell line.12,45,46
Once inside the cells, we explicated the effect of HA-Dual miRNA NPs on the intracellular mir-10b and mir-34a status using RT-PCR. It was found that the expression levels of miR-10b decreased and miR-34a increased upon HA-Dual miRNA NPs treatment. These results for the change in expression of miRNAs were in accordance with our study with specific miRNA 34a mimics and miR-10b inhibitors in MDAMB-231 cells (Fig. S6†). We also observed that in the HA-Dual miRNA NPs-treatment sets, the expression of Bcl-2 and RhoC had declined, whereas the expression of HOXD10 was increased with respect to the control sets at both the mRNA and protein levels (Fig. 3E & F). Our findings suggested that HA-Dual miRNA NPs regulated the expression of the above-mentioned miRNA, and suppressed its direct target and in turn deactivated the metastatic gene and activated the apoptotic genes leading to the suppression of TNBC cell invasion, metastasis, and inducing apoptosis.
3.4 Validation of the anti-cancer effect in mouse mammary carcinoma cells, 4T1
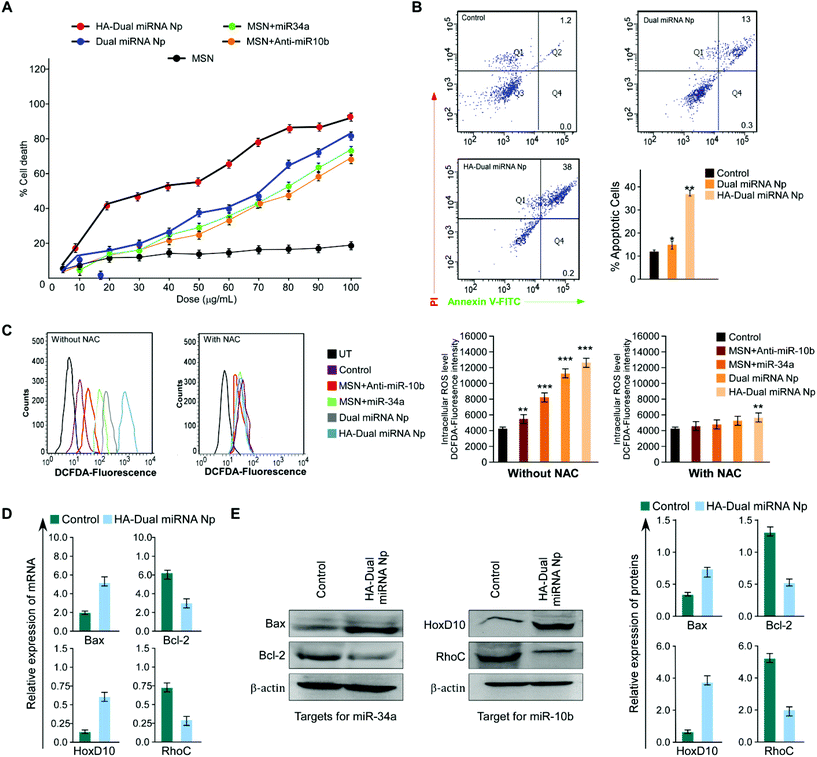
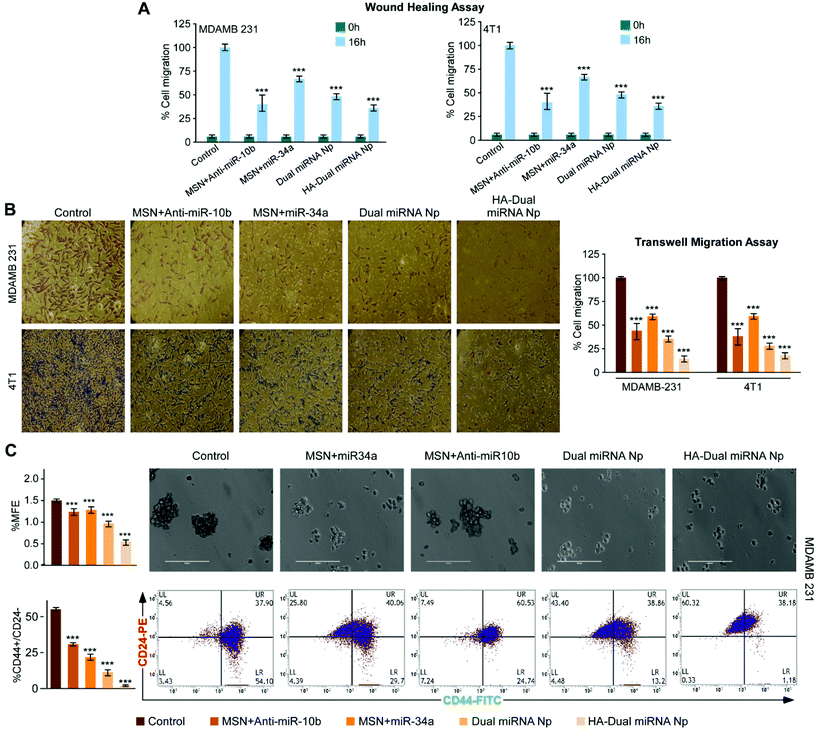
To further establish our findings in mouse mammary carcinoma, 4T1 cells were treated with different nanoparticles. Assessment of the percentage cell death showed a maximum cell death with HA-Dual miRNA NPs (Fig. 4A). The flow cytometer results indicated that the HA-Dual miRNA NPs could induce approximately 60% apoptosis in 4T1 cells at 24 h (Fig. 4B). Experiments were further extended in the presence and absence of NAC and there was an observable increase in ROS generation via the increase in DCFDA fluorescence after HA-Dual miRNA NPs treatment (IC50 dose) for 4T1 after 24 h (Fig. 4C). Next we investigated the changes in the mRNA expression level of Bax, Bcl-2, RhoC, and HOXD-10 using qRT-PCR analysis. Interestingly, we observed that the expressions of Bcl-2 and RhoC decreased, whereas the expression levels of Bax and HOXD10 increased upon treatment with HA-Dual miRNA NPs (Fig. 4D). These findings were further reinstated through western blot analysis (Fig. 4E), which also revealed similar observations.3.5 HA-Dual miRNA NPs retarded cell migration and reduced stemness in TNBC cells
A bidirectional wound healing assay in the human and murine TNBC cell lines, MDAMB-231 and 4T1, respectively, revealed that HA-Dual miRNA NPs attenuated cell migration more prominently compared to dual miRNA as well as single miRNA loaded sets (Fig. 5A and S7†). Transwell migration assay also validated the fact that cells treated with HA-Dual miRNA NPs were found to migrate and invade significantly less, thus proving the importance of targeted dual miRNA delivery through nanoparticles (Fig. 5B, left and right panels). Next, the number of lamellipodia formation in MDAMB-231 and 4T1 cells was monitored under SEM and was found to decrease after HA-Dual miRNA treatment with an observable structural deformation (Fig. S8†). Next, in order to examine the effect of our synthesized nanoparticles on stemness in MDAMB-231 cells, we calculated the MFE% upon treatment, which showed a substantial decline in the numbers of mammospheres (diameter ≥100 μm) formed in HA-Dual miRNA NPs sets (Fig. 5C, upper panel). Further, we performed flow cytometry to check its effect on the CD44+/24− breast cancer subpopulation. The results indicated that though the non-targeted miRNA-loaded MSN could decrease the CD44+/24− breast cancer subpopulation, the HA-Dual miRNA NPs could drastically reduce this percentage from 54.1% to 1.18% (Fig. 5C, lower panel). This may be an indication that cancer relapse due to the stem cell formation may also be controlled through these targeted dual miRNA loaded nanoparticles.3.6 Validation of the anti-cancer effect of HA-Dual miRNA NPs in an ex ovo model
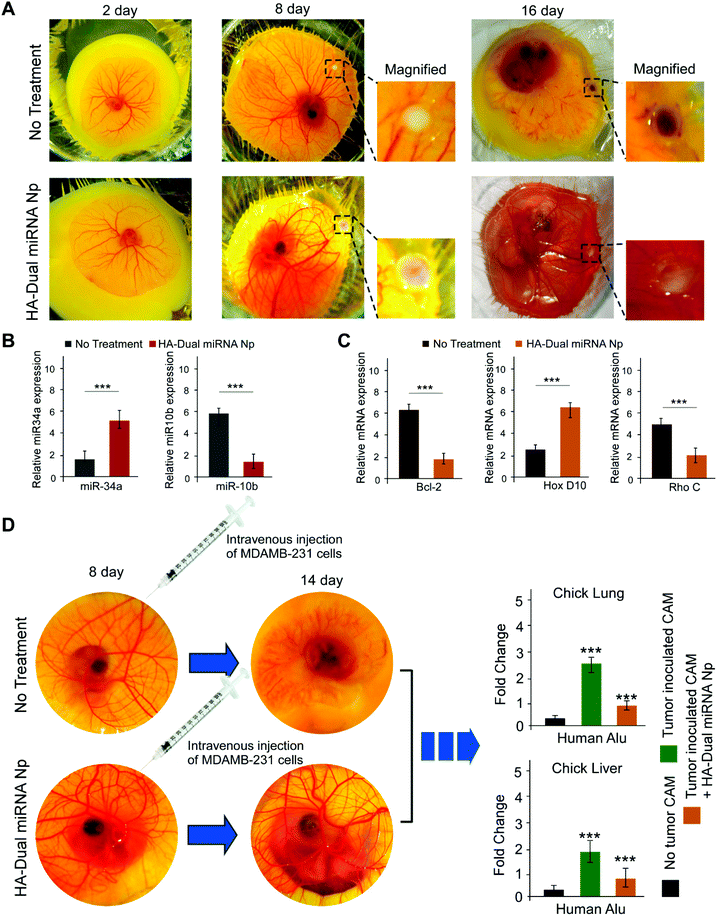
Chick embryo was used as an ex ovo xenograft model for studies of tumor regression as well as a metastasis assay. We inoculated MDAMB-231 (1 × 106) cells onto CAM in the 8-day-old chick embryos in order to carry out a study into tumor formation, for a developed tumor within 15 days. Along with this, a dose of 0.1 ml (15 μg mL−1) of HA-Dual miRNA NPs was given to the chick embryo. On day 16, we observed a formation of a whitish tumor, which indicated the presence of MDAMB-231 cells. Tumor regression in the case of the HA-Dual miRNA NPs-treated set was distinctly observable on day 16 in contrast to the untreated sets (Fig. 6A). To further validate our mechanism of action, we examined the expression of our desired microRNAs and mRNAs in the tumors with an ex ovo model and observed that the levels of miR-34a and HOXD10 were increased and those of miR-10b, Bcl-2, and Rho C were decreased in the HA-Dual miRNA NPs-treated sets as compared to the control sets (Fig. 6B and C).Additionally, to investigate the multi-organ metastasis, MDAMB-231 (1 × 106 cells) was intravenously injected in the 8-day-old chick embryos, followed by the harvesting of different organs, like the liver and lungs from the chick embryo on day 14. The untreated control chick embryos died at day 14, perhaps due to the toxicity imparted by TNBC cells upon tumor formation. The homogenized liver and lung were studied to detect the presence of MDAMB-231 cells through Alu PCR. Our study revealed that the HA-Dual miRNA NPs-treated sets had a significantly low expression of human Alu sequences as compared to a very high expression of Alu sequences in the untreated control sets, pointing sharply toward an anti-migratory role of HA-Dual miRNA NPs in TNBC cells in the ex ovo xenograft model (Fig. 6D).
3.7 In vivo sensitivity, biodistribution, and toxicity induced by HA-Dual miRNA NPs
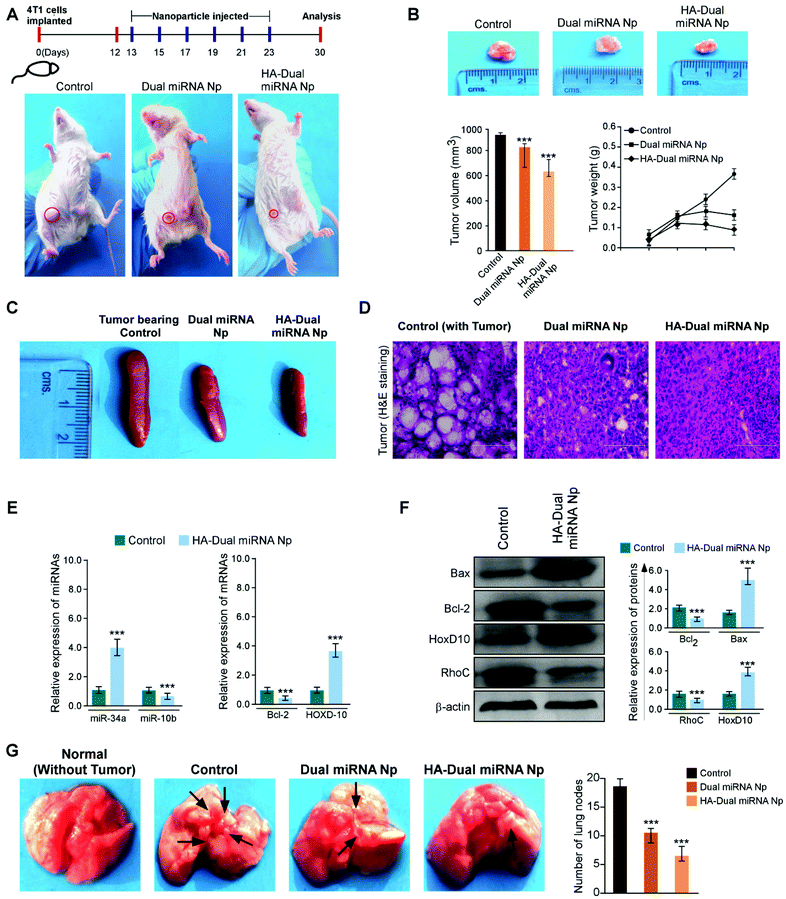
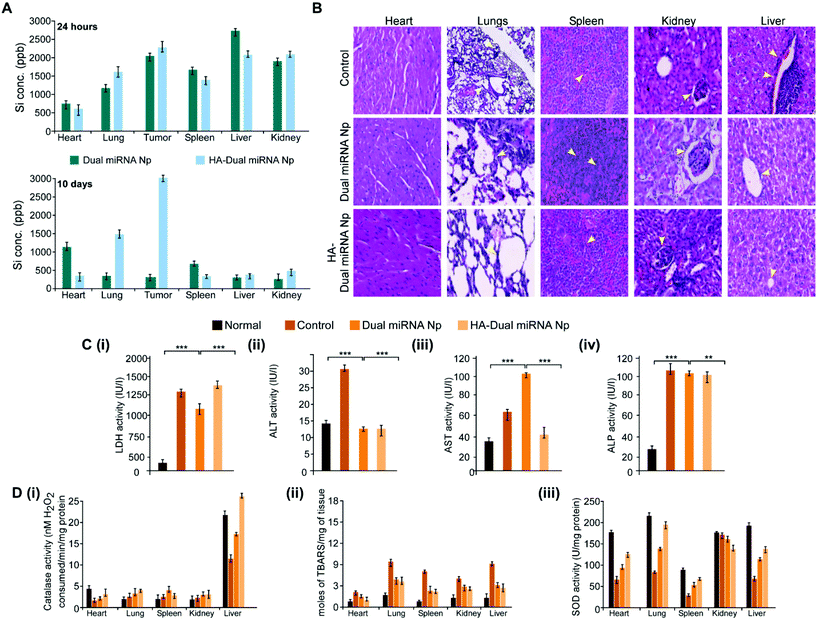
To investigate whether our desired miRNAs could suppress the tumor growth in an in vivo model, tumor-bearing BALB/c mice were treated with different nanoparticles in separate sets. As shown in Fig. 7A, the treatment with dual miRNA NPs did not show a significant inhibition of the tumor size, while HA-Dual miRNA NPs treatment in mice could significantly decrease the tumor size with respect to the control groups. At the end of the treatment, the tumors were excised from every mice, photographed, and their volumes and weights were measured, respectively (Fig. 7B). In the tumor-bearing BALB/c mice, the treatment with HA-Dual miRNA NPs showed tumor targeting and thus significantly hindered tumor progression over time. Splenomegaly was recognized in the spleen of the tumour-bearing control sets, which further restored its size after treatment with our nanoparticles (Fig. 7C). In addition, H&E staining data of tumor sections clearly indicated that the HA-Dual miRNA NPs showed no such higher infiltration and poorly differentiated structures in comparison to the control sets, thus causing no such damage (Fig. 7D). Next, through qRT-PCR and western blot analysis of homogenized tumors of different sets, we observed an increase in the expression levels of miR-34a and Bcl-2 and decreased expression levels of miR-10b and HOXD-10 as compared to the control group upon treatment with HA-Dual miRNA NPs (Fig. 7E and F). Also, the lungs were excised and photographed to study the presence of metastatic nodules. We observed that, as compared to the control groups, which showed significant nodule formation, the treatment sets showed less or no trace of nodules. Comparatively, the HA-Dual miRNA NPs-treated sets showed almost no presence of nodules as compared to a few nodules in the dual miRNA NPs-treated sets (Fig. 7G).Next, to check the biodistribution of our nanoparticles as silicon (as Si) in various organs, we performed ICP-AES. The tumor-bearing mice were injected with dual miRNA NPs and HA-Dual miRNA NPs and then the organs were extracted and digested to study the presence of silicon at two diverse times: (i) 24 h and (ii) 10 days. In the dual miRNA NPs-treated sets, the tumor, kidney, and spleen were found to show the presence of Si with the maximum accumulation in the liver at the 24 h time point, which remained till day 10 as well. However, the concentration of Si was less in the liver with an increasing concentration in other organs and the highest accumulation in the tumor in the case of the HA-Dual miRNA NPs-treated sets (Fig. 8A). Overall, these results indicate that the HA-Dual miRNA NPs could be expected to be a highly efficient delivery vehicle for achieving the targeted intracellular delivery of miRNAs. Additionally, the in vivo biodistribution highlighted that our HA-Dual miRNA NPs were able to show enhanced tumor targeting and could be retained within the system for up to 10 days. No significant problem related to renal clearance was found. These findings highlighted the potency of our nanoparticles to be used as a suitable delivery vehicle.
Furthermore, the toxicology parameters studies suggested a less significant toxicity of HA-Dual miRNA NPs as compared to the dual miRNA NPs and untreated sets. In addition to the antitumor activity, the safety profiles of the HA-Dual miRNA NPs were evaluated by studying the histology of the organs through H&E staining. As shown in Fig. 8B, the dual miRNA NPs group caused systemic toxicity as characterized by the vacuolization in cardiomyocytes, changes in the morphology of the bronchioles in the lungs and inflammatory cellular infiltration in the alveolar, and distorted hepato-architecture within the liver followed by the distortion of renal morphology (Fig. 8B), which might be related to the non-selected distribution of miRNAs in the case of dual miRNA NPs. However, no obvious organ toxicity was observed in the HA-Dual miRNA NPs-treatment groups in comparison with that in the control group. Further biochemical indices in liver function tests, such as aspartate aminotransferases (AST), alanine aminotransferases (ALT), and alkaline phosphatase (ALP) activity, showed changes in the serum as a result of membrane damage in the dual miRNA NPs-treated sets (Fig. 8C(i)–(iii)). These levels were somewhat similar to the control sets in the case of the HA-Dual miRNA NPs. Along with it, the oxidative stress parameters showed elevated amounts in different organs in the case of the dual miRNA NPs (Fig. 8D(i)–(iii)). These toxicities were decreased in the case of our targeted delivery. Combining these results, it can be inferred that the HA-Dual miRNA NPs were well-tolerated in vivo.
4. Conclusion
In this study, we reported a successful application of HA-Dual miRNA NPs as a dual nanocarrier system to simultaneously deliver anti-miR-10b and tumour suppressive miRNA-34a into triple-negative breast cancer cells for the improved efficacy of chemotherapy. We performed a comparative study, which demonstrated that among the different modifications of mesoporous silica nanoparticles, the cationic group showed the highest binding with the microRNAs. Furthermore after polymer coating, the release became more stable, which further enhanced the stability of our nanoparticles inside the cellular system. The furnished study shows how the modified nanoparticle gets internalized within the system, depicting a stable and sustained release of microRNAs, which further leads to cancer cell death and plays a role in the retardation of cell migration as well. The current research thus may provide ideas for addressing the dilemma regarding stability within the system. The systemic delivery of miRNA loaded nanoparticles in animal trials demonstrated high specificity toward the tumor and efficacy against its growth. This study provides an insight into the possible therapeutic application of these targeted nanoparticles in TNBC therapy.Conflicts of interest
Authors declare no conflict of interest.Acknowledgements
Authors would like to thank the Department of Science and Technology Nano Mission (grant no. SR/NM/NS-1185/2015(G)) and Government of India for financial support. Authors also thank CSIR (sanction no. 09/028(0989)/2016-EMR-I), Govt. of India for a scholarship to MA. We also thank the facility of Centre for Research in Nanoscience and Nanotechnology, University of Calcutta. We would like to acknowledge Mr Arpan Sarkar and Ms Arijita Basu for their help in some experiments. Our sincere thanks to Dr Ritesh Tiwari (CU-BD-CoE) for providing assistance and access to BD, Central facility. Thanks to Mr Samrat Kundu, Mr Pratyush Sengupta and Mrs Urmila Goswami for technical support to access FE-SEM, SEM and TEM respectively.References
- D. Austin, N. Hamilton, Y. Elshimali, R. Pietras, Y. Wu and J. Vadgama, Oncotarget, 2018, 9, 33912–33930 CrossRef PubMed.
- W. D. Foulkes, I. E. Smith and J. S. Reis-Filho, N. Engl. J. Med., 2010, 363, 1938–1948 CrossRef CAS PubMed.
- C. A. Hudis and L. Gianni, Oncologist, 2011, 16(Suppl 1), 1–11 CrossRef PubMed.
- S. L. Lin, H. Kim and S. Y. Ying, Front. Biosci., 2008, 13, 2216–2230 CrossRef CAS PubMed.
- A. Rodriguez, S. Griffiths-Jones, J. L. Ashurst and A. Bradley, Genome Res., 2004, 14, 1902–1910 CrossRef CAS PubMed.
- M. V. Iorio, M. Ferracin, C. G. Liu, A. Veronese, R. Spizzo, S. Sabbioni, E. Magri, M. Pedriali, M. Fabbri, M. Campiglio, S. Menard, J. P. Palazzo, A. Rosenberg, P. Musiani, S. Volinia, I. Nenci, G. A. Calin, P. Querzoli, M. Negrini and C. M. Croce, Cancer Res., 2005, 65, 7065–7070 CrossRef CAS PubMed.
- W. Zhang, J. E. Dahlberg and W. Tam, Am. J. Pathol., 2007, 171, 728–738 CrossRef CAS PubMed.
- S. Oliveto, M. Mancino, N. Manfrini and S. Biffo, World J. Biol. Chem., 2017, 8, 45–56 CrossRef PubMed.
- T. Frixa, S. Donzelli and G. Blandino, Cancers, 2015, 7, 2466–2485 CrossRef CAS PubMed.
- T. C. Chang, E. A. Wentzel, O. A. Kent, K. Ramachandran, M. Mullendore, K. H. Lee, G. Feldmann, M. Yamakuchi, M. Ferlito, C. J. Lowenstein, D. E. Arking, M. A. Beer, A. Maitra and J. T. Mendell, Mol. Cell, 2007, 26, 745–752 CrossRef CAS PubMed.
- L. Ma, F. Reinhardt, E. Pan, J. Soutschek, B. Bhat, E. G. Marcusson, J. Teruya-Feldstein, G. W. Bell and R. A. Weinberg, Nat. Biotechnol., 2010, 28, 341–347 CrossRef CAS PubMed.
- S. Yang, Y. Li, J. Gao, T. Zhang, S. Li, A. Luo, H. Chen, F. Ding, X. Wang and Z. Liu, Oncogene, 2013, 32, 4294–4303 CrossRef CAS PubMed.
- A. E. Labatut and G. Mattheolabakis, Eur. J. Pharm. Biopharm., 2018, 128, 82–90 CrossRef CAS PubMed.
- Z. Bai, J. Wei, C. Yu, X. Han, X. Qin, C. Zhang, W. Liao, L. Li and W. Huang, J. Mater. Chem. B, 2019, 7, 1209–1225 RSC.
- J. Tu, A. L. Boyle, H. Friedrich, P. H. H. Bomans, J. Bussmann, N. A. J. M. Sommerdijk, W. Jiskoot and A. Kros, ACS Appl. Mater. Interfaces, 2016, 8, 32211–32219 CrossRef CAS PubMed.
- X. Chen, H. Sun, J. Hu, X. Han, H. Liu and Y. Hu, Colloids Surf., B, 2017, 152, 77–84 CrossRef CAS PubMed.
- W. Cha, R. Fan, Y. Miao, Y. Zhou, C. Qin, X. Shan, X. Wan and J. Li, Molecules, 2017, 22, 782 CrossRef PubMed.
- X. Li, Y. Chen, M. Wang, Y. Ma, W. Xia and H. Gu, Biomaterials, 2013, 34, 1391–1401 CrossRef CAS PubMed.
- H. Meng, W. X. Mai, H. Zhang, M. Xue, T. Xia, S. Lin, X. Wang, Y. Zhao, Z. Ji, J. I. Zink and A. E. Nel, ACS Nano, 2013, 7, 994–1005 CrossRef CAS PubMed.
- J. Shen, H. C. Kim, H. Su, F. Wang, J. Wolfram, D. Kirui, J. Mai, C. Mu, L. N. Ji, Z. W. Mao and H. Shen, Theranostics, 2014, 4, 487–497 CrossRef CAS PubMed.
- D. Lin, Q. Cheng, Q. Jiang, Y. Huang, Z. Yang, S. Han, Y. Zhao, S. Guo, Z. Liang and A. Dong, Nanoscale, 2013, 5, 4291–4301 RSC.
- H. K. Makadia and S. J. Siegel, Polymers, 2011, 3, 1377–1397 CrossRef CAS PubMed.
- J. M. Lu, X. Wang, C. Marin-Muller, H. Wang, P. H. Lin, Q. Yao and C. Chen, Expert Rev. Mol. Diagn., 2009, 9, 325–341 CrossRef CAS PubMed.
- R. Devulapally, N. M. Sekar, T. V. Sekar, K. Foygel, T. F. Massoud, J. K. Willmann and R. Paulmurugan, ACS Nano, 2015, 9, 2290–2302 CrossRef CAS PubMed.
- S. Guo, Y. Huang, Q. Jiang, Y. Sun, L. Deng, Z. Liang, Q. Du, J. Xing, Y. Zhao, P. C. Wang, A. Dong and X.-J. Liang, ACS Nano, 2010, 4, 5505–5511 CrossRef CAS PubMed.
- A. M. Chen, M. Zhang, D. Wei, D. Stueber, O. Taratula, T. Minko and H. He, Small, 2009, 5, 2673–2677 CrossRef CAS PubMed.
- Y. Zhang, Z. Wang, W. Zhou, G. Min and M. Lang, Appl. Surf. Sci., 2013, 276, 769–775 CrossRef CAS.
- W. Ngamcherdtrakul, J. Morry, S. Gu, D. J. Castro, S. M. Goodyear, T. Sangvanich, M. M. Reda, R. Lee, S. A. Mihelic, B. L. Beckman, Z. Hu, J. W. Gray and W. Yantasee, Adv. Funct. Mater., 2015, 25, 2646–2659 CrossRef CAS PubMed.
- D. J. Gary, H. Lee, R. Sharma, J.-S. Lee, Y. Kim, Z. Y. Cui, D. Jia, V. D. Bowman, P. R. Chipman, L. Wan, Y. Zou, G. Mao, K. Park, B.-S. Herbert, S. F. Konieczny and Y.-Y. Won, ACS Nano, 2011, 5, 3493–3505 CrossRef CAS PubMed.
- I. A. Babar, C. J. Cheng, C. J. Booth, X. Liang, J. B. Weidhaas, W. M. Saltzman and F. J. Slack, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, E1695–E1704 CrossRef CAS PubMed.
- H. Jin, Y. Yu, W. B. Chrisler, Y. Xiong, D. Hu and C. Lei, Breast Cancer: Basic Clin. Res., 2012, 6, 9–19 Search PubMed.
- Y. Wu, M. Crawford, Y. Mao, R. J. Lee, I. C. Davis, T. S. Elton, L. J. Lee and S. P. Nana-Sinkam, Mol. Ther. – Nucleic Acids, 2013, 2, e84 CrossRef PubMed.
- A. Aruffo, I. Stamenkovic, M. Melnick, C. B. Underhill and B. Seed, Cell, 1990, 61, 1303–1313 CrossRef CAS PubMed.
- S. Song, F. Chen, H. Qi, F. Li, T. Xin, J. Xu, T. Ye, N. Sheng, X. Yang and W. Pan, Pharm. Res., 2014, 31, 1032–1045 CrossRef CAS PubMed.
- S. Bhattacharya, M. Ahir, P. Patra, S. Mukherjee, S. Ghosh, M. Mazumdar, S. Chattopadhyay, T. Das, D. Chattopadhyay and A. Adhikary, Biomaterials, 2015, 51, 91–107 CrossRef CAS PubMed.
- A. Roy, S. Sarker, P. Upadhyay, A. Pal, A. Adhikary, K. Jana and M. Ray, Biochem. Pharmacol., 2018, 156, 322–339 CrossRef CAS PubMed.
- A. Zijlstra, R. Mellor, G. Panzarella, R. T. Aimes, J. D. Hooper, N. D. Marchenko and J. P. Quigley, Cancer Res., 2002, 62, 7083–7092 CAS.
- T. D. Palmer, J. Lewis and A. Zijlstra, J. Visualized Exp., 2011, 2815, DOI:10.3791/2815.
- M. Ahir, S. Bhattacharya, S. Karmakar, A. Mukhopadhyay, S. Mukherjee, S. Ghosh, S. Chattopadhyay, P. Patra and A. Adhikary, Biomaterials, 2016, 76, 115–132 CrossRef CAS PubMed.
- S. L. Marklund, Mutat. Res., Fundam. Mol. Mech. Mutagen., 1985, 148, 129–134 CrossRef CAS PubMed.
- J. R. Wright, H. D. Colby and P. R. Miles, Arch. Biochem. Biophys., 1981, 206, 296–304 CrossRef CAS PubMed.
- N. S. Basakran, Saudi Med. J., 2015, 36, 273–279 CrossRef PubMed.
- C. Chen, S. Zhao, A. Karnad and J. W. Freeman, J. Hematol. Oncol., 2018, 11, 64–64 CrossRef PubMed.
- L. Ma, Breast Cancer Res., 2010, 12, 210–210 CrossRef PubMed.
- S. Imani, R.-C. Wu and J. Fu, J. Cancer, 2018, 9, 3765–3775 CrossRef CAS PubMed.
- L. Li, L. Yuan, J. Luo, J. Gao, J. Guo and X. Xie, Clin. Exp. Med., 2013, 13, 109–117 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Eight figures and four tables. It also consists of the synthesis protocol. See DOI: 10.1039/d0bm00015a |
| This journal is © The Royal Society of Chemistry 2020 |