 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Electrically conductive hydrogel-based micro-topographies for the development of organized cardiac tissues†
Ali Navaeia,
Nathan Moorea,
Ryan T. Sullivana,
Danh Truonga,
Raymond Q. Migrinobc and
Mehdi Nikkhah*a
aSchool of Biological and Health Systems Engineering (SBHSE), Arizona State University, 501 E Tyler Mall Building ECG, Suite 334, Tempe, Arizona 85287-9709, USA. E-mail: mnikkhah@asu.edu; Fax: +1-480-727-7624; Tel: +1-480-965-0339
bPhoenix Veterans Affairs Health Care System, Phoenix, AZ 85012, USA
cUniversity of Arizona College of Medicine, Phoenix, AZ 850004, USA
First published on 22nd November 2016
Abstract
The dense uniaxially aligned cardiac cytoarchitecture of the myocardium along with electrical and mechanical coupling between the cardiac cells are key factors in the synchronous contractility of the heart. Following myocardial infarction, the organized architecture of the myocardium is disrupted and non-functional scar tissues are produced. Engineered cardiac tissues, developed in hydrogel-based biomaterials, featuring biomimetic topographical cues have shown significant promise for regeneration and repair of injured myocardium. However, currently engineered tissues still do not exhibit an electrically conductive matrix integrated with highly oriented cellular constructs to promote tissue-level functionalities. To address this limitation, we utilized integrated micro- and nano-technologies to develop gelatin methacrylate (GelMA) hydrogel constructs comprised of surface micro-topographies (microgrooves with 50 μm width and depth) incorporated with electrically conductive gold nanorods (GNRs) to provide simultaneous electrical and topographical cues that mimic physiological relevant myocardium function. F-Actin stained images and fluorescent area coverage data revealed the formation of uniform, dense, and highly aligned cardiac tissues on GelMA–GNR hydrogels compared to discrete and disconnected cellular organization on pure GelMA constructs (control group). Immunostaining images of cardiac markers, specifically sarcomeric α-actinin and connexin 43 also showed higher cytoskeletal alignment and enhanced cellular connectivity on GelMA–GNR hydrogels. Tissues formed on both GelMA and GelMA–GNR constructs demonstrated spontaneous contractility from day 4 to 7 of culture. However, only electrically conductive GelMA–GNR cardiac tissues showed a consistent response in changing beat rate as a result of external stimulation. Overall, we demonstrate the enhanced formation of cardiac tissues with superior cellular organization, connectivity, and electrical properties with use of GNRs and microgrooved GelMA hydrogel that may have potential as a functional cardiac patch in the setting of infarcted myocardium.
1. Introduction
The native myocardium is a structurally orchestrated and adaptive tissue consisting of tightly packed uniaxial cytoarchitecture and electrically conductive Purkinje fibers, which provide electrical conduction through the heart.1–4 Such a unique cellular microenvironment contributes critical cues involved with initiation and development of the structure–function relationship. In particular, electrical and geometrical cues are two of the most impactful stimuli, influencing proper functionalities of the heart5 and efficient pumping of blood throughout the body.6 In vitro investigation of these signaling cues and their subsequent effects on cytoarchitecture can lead to better understanding of the tissue function and optimization of regenerative-based therapies for treatment of heart diseases (e.g. myocardial infarction).In the past two decades, tissue engineering has emerged as a powerful strategy to develop cardiac constructs with geometrical and electrical cues that mimic the native cardiac microenvironment. Numerous hydrogel-based scaffolds along with microscale techniques (i.e. micropatterning) have been integrated7–9 to develop biomimetic cardiac tissue substitutes that can be used in disease modeling and regeneration studies following injury. Micropatterned constructs fabricated by gelatin methacrylate (GelMA),10–12 methacrylated tropoelastin (MeTro),7 polyethylene glycol (PEG),13,14 collagen,15,16 poly(glycerol sebacate) (PGS),1 and chitosan16 are among the biomaterials that have been widely used for cardiac tissue engineering due to their excellent material and biological characteristics. For instance, in a pioneering study by Engelmayr et al.1 PGS accordion-like honeycombs with native-like anisotropic architecture were micro-fabricated to develop organized cardiac tissue constructs. Their findings indicated enhanced formation of aligned cardiac tissue constructs with anisotropic mechanical properties similar to native heart muscle. Photocrosslinkable hydrogels such as GelMA and MeTro have also shown promising results for cell-based studies and cardiac tissue engineering applications mainly due to their bioactivity and ease of precise fabrication.7,17,18 In particular, the integrin-based cell adhesion moieties in GelMA hydrogel provided a desirable microenvironment for cell growth and spreading, while methacrylated polymer chains enabled formation of micro-topographies.17,18 For example, Saini et al.10 fabricated cell-laden GelMA hydrogel constructs to develop 3D rectangular cardiac micro-tissues with variable geometrical features encapsulated with cardiomyocytes alone and with a co-culture with cardiac fibroblasts. Their results showed the necessity and critical impacts of cardiac fibroblasts and construct geometry on cell spreading, cytoskeleton alignment, tissue formation, and overall functionalities of the cardiac microtissues.
Despite these significant progresses, there remains critical shortcoming related to the electrical properties of the previously developed micropatterned hydrogel constructs that can be further improved for cardiac tissue engineering applications.12,19–22 Specifically, the electrically insulated macroporous matrix of hydrogel can interfere with cell–cell electrical coupling and signal propagation within the tissue.4,19 This phenomenon can impede full electrical integration of the tissue constructs with the native myocardium, which could potentially lead to undesired arrhythmias.3,23–25 To address this, incorporation of biocompatible nanomaterials with high electrical conductivity was demonstrated to significantly reduce the hydrogel matrix's resistivity and facilitate electrical signal propagation among the cardiac cells.2–4,20,22 For instance, in a recent work, Ramon-Azcon and Ahadian26 fabricated micropatterned GelMA hydrogel constructs incorporated with aligned carbon nanotubes via dielectrophoresis for skeletal muscle tissue engineering. The microgrooved hydrogel constructs demonstrated robust anisotropic electrical conductivity, leading to pronounced expression of skeletal muscle specific genes and proteins. However, there are numerous concerns regarding the utilization of carbon nanotubes for biomedical applications7 specifically with respect to biological toxicity,27–29 low solubility30,31 as well as high UV light absorption which limits development of three dimensional (3D) thick (i.e. millimeter scale) tissue constructs using photolithography.32 Alternatively, previous work of ours19 and others4,33–38 have exhibited the tremendous potential of gold nanomaterials (AuNMs) for myocardial tissue engineering. Particularly, cardiac tissue constructs incorporated with AuNMs (e.g. nanorods and nanowires) showed excellent viability and metabolic activity,19,34 enhanced cardiogenic differentiation,35,38,39 upregulated expressions of cardiac proteins and genes,4,40 and facilitated cell–cell electrical coupling.4,19,36,41 In addition, high biocompatibility,42,43 wide range of nanostructures,44,45 as well as ease of fabrication and biofunctionalization46,47 are among the other reported characteristics of AuNMs.
We previously developed gold nanorod-incorporated GelMA hybrid hydrogels (GelMA–GNR) to assess the applicability of the conductive material in prompting cardiac cell functions.19 Specifically, 5% (w/v) GelMA hydrogels with different concentrations of GNRs (0, 0.5, 1, and 1.5 mg mL−1) were formed using photolithography technique. Neonatal rat ventricular cardiomyocytes were seeded on the GelMA–GNR hydrogel constructs and subsequently cardiac cell phenotype (i.e. sarcomere organization, connexin 43) as well as tissue-level functionalities were assessed. Specifically, our results confirmed the formation of highly contractile tissues on the synthesized conductive hydrogels with 1 and 1.5 mg mL−1 GNRs, demonstrating synchronous cell–cell coupling and Ca2+ transients compared to pure GelMA hydrogel. However, the developed GelMA–GNR constructs did not exhibit biomimetic geometrical (i.e. topographical) cues to induce native-like anisotropic cellular organization. Therefore, in this study, we utilized an integrated micro- and nanoscale strategy by simultaneous incorporation of biomimetic geometrical (i.e. anisotropic microgrooved architecture) and electrical cues (i.e. GNRs) to develop highly oriented and contractile cardiac tissue constructs. We additionally optimized the concentration of GelMA hydrogel to 10% (w/v) incorporated with GNRs (1 mg mL−1), to maintain robust pattern fidelity. We performed material characterization to assess the electrical and mechanical properties of the new matrix. Neonatal rat cardiomyocytes were seeded on pristine GelMA and GelMA hydrogel incorporated with GNRs for 7 days, and cellular organization, cardiac specific markers expression as well as tissue contractility were further assessed. Our findings confirmed the formation of dense and highly organized cellular networks within GelMA–GNR microgrooved constructs. The electrically conductive and highly organized cardiac tissues exhibited continuous and synchronized contractions induced by external electrical stimuli, mimicking physiologic synchronous cardiac cell beatings. The proposed strategy demonstrates a unique approach, which can be used to develop biomimetic cardiac patches that could better integrate with native myocardium.3,23
2. Experimental section
2.1. Materials
Gelatin (type A), methacrylic anhydride, 3-(trimethoxysilyl)propyl methacrylate (TMSPMA), 2-hydroxy-1-(4-(hydroxyethoxy)phenyl)-2-methyl-1-propanone (photoinitiator), gold(III) chloride trihydrate (HAuCl4), sodium borohydride (NaBH4), silver nitrate (AgNO3), hexadecyltrimethylammonium bromide (CTAB), and L-ascorbic acid were purchased from Sigma-Aldrich and utilized with no further purifications.2.2. Fabrication of PDMS micromolds
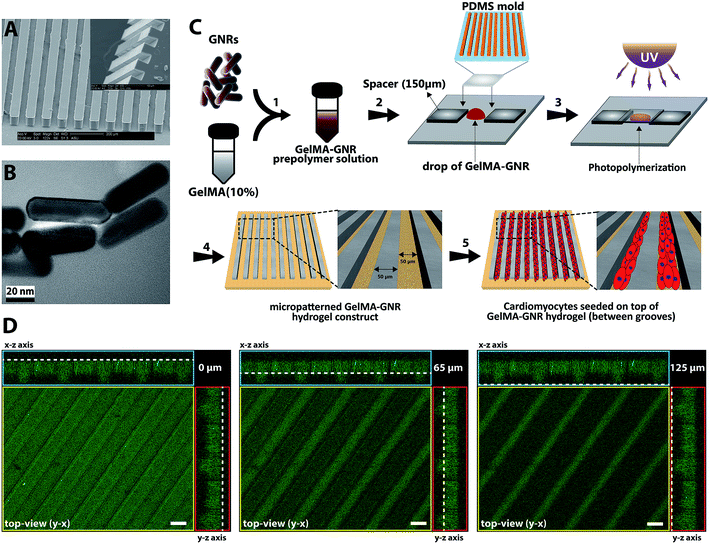
The micromolds were fabricated using standardized photo- and soft-lithography methods.48–50 First, the groove design (50 μm width and spacing) was created using a CAD (computer aided design) software. Next, a transparent mask was created and utilized in the SU-8 photolithography. SU8-2075 (MicroChem) was spun onto a silicon wafer until it reached a final height of 50 to 70 μm. The wafer was placed underneath the transparent mask with the microgrooved patterns and exposed to UV light. Subsequently, the wafer was transferred to a conventional oven for final crosslinking at 50 °C for 24 h. Finally, the wafers, containing of microgrooves, were developed at room temperature using SU-8 developer (MicroChem) and hard baked at 150 °C for 30 min. Wafers were salinized using trichloro(methyl)silane (TCMS, Sigma-Aldrich) for 20 min in a vacuum chamber. Next, polydimethylsiloxane (PDMS, Sylgard 184 Silicon Elastomer kit, Dow Corning) was casted over the SU-8 wafer and baked for 1 h at 80 °C. Afterwards, the PDMS micromolds (Fig. 1A) was peeled off and individual micromolds were cut and sterilized using 70% ethanol and UV.2.3. Fabrication of cell-seeded GelMA and GelMA–GNR microgrooved hydrogels
Gold nanorods (GNRs) and gelatin methacrylate (GelMA) prepolymer were synthesized based on previously reported protocols.10,19 Transmission electron microscopy (TEM) (Philips CM200-FEG, USA) operating at 200 kV accelerating voltage was used to view and confirm the formation of synthesized GNRs (Fig. 1B). Mean length, width, and aspect ratio of GNRs were calculated based on the TEM images (n = 10) using NIH ImageJ software. GelMA–GNR microgrooved hydrogels were fabricated using the soft lithography technique (Fig. 1C). A clear prepolymer solution of GelMA (10% w/v) in photoinitiator (0.5% w/v in DPBS) was prepared, and subsequently mixed with purified GNRs (1 mg mL−1 of GelMA prepolymer solution). GelMA–GNR solution was sonicated in water bath for 1 h to obtain a homogenous mixture. In the next step, PDMS micromolds (50 μm width, spacing, and depth) were treated by oxygen plasma (Harrick Plasma, USA) for 30 s to render the surface of the micromold more hydrophilic resulting in uniform penetration of hydrogel prepolymer solution within the microgrooves. The mold was placed on a droplet (15 μL) of GelMA–GNR hydrogel and the whole assembly was exposed to UV light (800 mW, 360–480 nm) for 25 seconds. To avoid any disruption in the structure, microgrooved hydrogels were soaked in the sterile DPBS for 10–15 min and then the PDMS mold was peeled off. GelMA microgrooved hydrogels (the control group) were fabricated also using the same procedure with 10 s UV light exposure. All microgrooved hydrogels were washed in an antibiotic solution (1% v/v penicillin/streptomycin in DPBS) twice with 10 min intervals, followed by a washing using cardiac culture media (containing Dulbecco's modified eagle medium, L-glutamine (1%), fetal bovine serum (FBS) (10%), and penicillin–streptomycin (100 units per mL)) in the same fashion. Afterwards, isolated 2 day old neonatal ventricular rat cardiomyocytes were seeded (1 × 106 cells/constructs) on GelMA–GNR microgrooved hydrogels (1 × 1 cm2 construct size) for 24 h, which were placed in 24 multiwell plates and incubated at 37 °C with 5% CO2. Unattached cells were washed off on the next day and 500 μL of fresh cardiac culture media were added to the samples. The media were exchanged every other day.2.4. Electrical, rheology, and morphological characterizations of GelMA and GelMA–GNR hydrogels
To evaluate the electrical conductivity of the GelMA and GelMA–GNR hydrogels, the constructs were placed between two conductive glass slides (indium tin oxide coated, 30–60 Ω sq−1, Sigma-Aldrich) and AC bias (Agilent 4284A LCR meter) was swept from 10 Hz to 1 MHz. Real portions of the impedance complex values were averaged (n = 4) and plotted versus the frequency. A rheometer (parallel plate rheometer, Anton Paar MCR-101) was utilized to measure the storage moduli (elasticity) of crosslinked GelMA and GelMA–GNR hydrogels. In this regard, hydrogels were placed between the parallel plates of the rheometer and storage moduli were measured as a function of strain sweep (0–100%). The values at 5% strain (n = 3) were considered the ultimate storage moduli of the hydrogels. Macroporous structure of GelMA and GelMA–GNR hydrogels were investigated using scanning electron microscopy (SEM). Hydrogels were fabricated and mounted onto the SEM sample holders, and sputter coated by Au/Pd. Matrix porosity of both hydrogels were calculated based on SEM images (n = 6) using ImageJ software.2.5. Viability, cellular alignment, and cardiac cells phenotype
Viability of cultured cardiac cells on GelMA and GelMA–GNR hydrogels were investigated using a standard Live/Dead assay kit (Thermo Fisher, USA). The samples were treated by the assay on day 1 and 7 of culture and fluorescent images were taken using an inverted microscope (Zeiss Observer Z1, USA). Cytoskeleton organization of microgrooved cardiac tissues was assessed based on F-actin fibers staining images. The hydrogel constructs were stained on day 1 and 7 of culture using Alexa Fluor® 488 Phalloidin (Thermos Fisher, USA) according to previously published protocols.51 Sequentially, samples were fixed in paraformaldehyde (4% v/v) for 30 min, permeabilized in Triton X-100 (1% v/v) for 45 min, and blocked in bovine serum albumin (1% v/v) for 1 h. Next, a mixture of Alexa Fluor® 488 Phalloidin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 dilution) and DAPI (40,6-diamidino-2-phenyl indole dihydrochloride, 1
40 dilution) and DAPI (40,6-diamidino-2-phenyl indole dihydrochloride, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution) in bovine serum albumin was added to the samples for 1 h, and z-stacked images were captured by a fluorescent microscope equipped with Apotome.2 (Zeiss, USA). The averaged area coverage and index of F-actin fibers were calculated (n > 15). The area coverage index specifically was defined as the area covered by cardiac cells delineated by the F-actin fibers inside the microgrooves. In addition, fiber alignment index (n > 10) was defined as the ratio of long axis to short axis of FFT (Fast Fourier Transform, built-in tool in ImageJ software) analysis images.10,52 To evaluate the maturation of the created cardiac tissues on the hydrogel microgrooves, cardiac-specific markers, sarcomeric α-actinin (SATN) and connexin 43 (Cx43) gap junctions were immunostained using primary antibodies (Abcam, USA).19 In brief, samples were fixed in paraformaldehyde (4% v/v) for 30 min, permeabilized in Triton X-100 (0.1% v/v) for 45 min, and blocked in goat serum (10% v/v) for 1 h. Afterwards, primary antibodies (1
1000 dilution) in bovine serum albumin was added to the samples for 1 h, and z-stacked images were captured by a fluorescent microscope equipped with Apotome.2 (Zeiss, USA). The averaged area coverage and index of F-actin fibers were calculated (n > 15). The area coverage index specifically was defined as the area covered by cardiac cells delineated by the F-actin fibers inside the microgrooves. In addition, fiber alignment index (n > 10) was defined as the ratio of long axis to short axis of FFT (Fast Fourier Transform, built-in tool in ImageJ software) analysis images.10,52 To evaluate the maturation of the created cardiac tissues on the hydrogel microgrooves, cardiac-specific markers, sarcomeric α-actinin (SATN) and connexin 43 (Cx43) gap junctions were immunostained using primary antibodies (Abcam, USA).19 In brief, samples were fixed in paraformaldehyde (4% v/v) for 30 min, permeabilized in Triton X-100 (0.1% v/v) for 45 min, and blocked in goat serum (10% v/v) for 1 h. Afterwards, primary antibodies (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 dilution in 10% goat serum) were loaded on the samples for 24 h at 4 °C, followed by staining using secondary antibodies (1
100 dilution in 10% goat serum) were loaded on the samples for 24 h at 4 °C, followed by staining using secondary antibodies (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 dilution in goat serum) (Thermo Fisher, USA) for 6 h. Finally, DAPI (1
200 dilution in goat serum) (Thermo Fisher, USA) for 6 h. Finally, DAPI (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution in DPBS) was added to stain the cells' nuclei. The area coverage of stained cardiac markers was averaged (n > 5) on day 7 of culture.
1000 dilution in DPBS) was added to stain the cells' nuclei. The area coverage of stained cardiac markers was averaged (n > 5) on day 7 of culture.
2.6. Spontaneous and stimulated beating behavior of microgrooved cardiac tissues
The spontaneous contractile behavior of GelMA and GelMA–GNR microgrooved cardiac constructs was monitored using an inverted microscope (Zeiss, USA) over 7 days, and presented as averaged beats per minute (n > 11). Also, a custom-made stimulation chamber was fabricated53 and utilized to induce electrical stimuli on the cardiac tissues on day 7. The cardiac microgrooved tissues were stimulated at 0.5, 1, 2 Hz (within physiologic range of human heart rate) and excitation thresholds (the minimum required voltage to induce synchronous cells' contraction) were recorded. To extract the beating signal from the videos, a custom-written Matlab (MathWorks, USA) code was used.19,542.7. Statistical analysis
All results were statistically analyzed with a Student's t-test (two-tailed) method using GraphPad Prism software. A p-value less than 0.05 (two-sided) was considered statistically significant difference within the samples.3. Results and discussion
3.1. Development and characterization of the electrically conductive hydrogel microgrooves
Fabricated PDMS micromolds (Fig. 1A) with microgrooved topographies were casted and used for creating GelMA and GelMA–GNR hydrogel constructs. Similar to our previously published work,19 the synthesized GNRs (Fig. 1B) exhibited nanorod-like structure with average length and width of ∼53 and ∼16 nm respectively (aspect ratio of ∼3.15). Purified GNRs were mixed and sonicated with GelMA prepolymer solution (10% w/v). A drop of GelMA–GNR mixture was then located on TMSPMA-coated glass slides, and PDMS mold was placed on top. Finally, the microgrooved hydrogel constructs were fabricated through UV photo-crosslinking, and submerged in sterile DPBS prior to biological studies (Fig. 1C). The microarchitecture of fabricated GelMA–GNR microgrooved hydrogels is demonstrated in Fig. 1D. As can be seen, the microgrooves with 60 μm depth (Fig. 1D, middle image) and 50 μm width were formed on the surface of hydrogel slab with 65 μm thickness. This result showed successful formation of surface microgrooves on GelMA–GNR hydrogel constructs.In our recent work,19 we performed extensive studies on the synthesis and characterization of GelMA (5% w/v) hydrogel embedded with variable concentrations of GNRs (0–1.5 mg mL−1) for the development of cardiac tissue constructs. However, our aim was to assess the role of matrix properties on behavior and function of cardiac cells. Our previously developed GelMA–GNR hydrogels did not exhibit biomimetic oriented topographical features to induce cardiac cells uniaxial alignment. In these studies, we optimized hydrogel concentration using 10% w/v GelMA incorporated with 1 mg mL−1 GNRs for fabrication of microgrooved features, due to its superior material and biological properties to conduct biological studies.19 In other words, the control condition in this study was the microgrooved GelMA hydrogel constructs without the presence of GNRs (GelMA vs. GelMA–GNR).
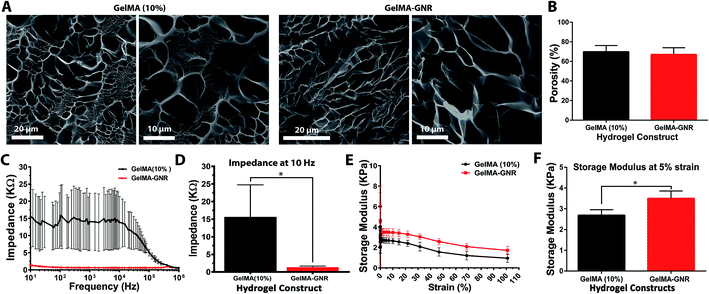
SEM micrographs (Fig. 2A) displayed the porous structure of GelMA and GelMA–GNR hydrogel matrices after 24 h hydration. As evident, both hydrogels showed similar porous architectures with no obvious aggregations of GNRs within the hydrogels' matrices. In addition, the architecture of GelMA and GelMA–GNR matrix were compared based on the measured porosity percentage (Fig. 2B) using SEM images. Both hydrogels demonstrated similar porosity with no statistically significant differences (GelMA 69 ± 6% and GelMA–GNR 67 ± 6%). The results showed that incorporation of GNRs within GelMA hydrogel did not disturb the macroporous architecture of the GelMA hydrogel matrix, consistent with our previously published work.19 Matrix porosity plays a critical role in cellular viability, spreading, and function especially in the case of cell encapsulation (cell-laden constructs) within hydrogel-based tissue constructs.55,56
Electrical conductivity of GelMA and GelMA–GNR hydrogels measured as the impedance of hydrogel matrix are shown in Fig. 2C. The results showed lower electrical impedance of GelMA–GNR hydrogel compared to pure GelMA within the frequency variations. Specifically, GNR-embedded hydrogels exhibited significantly less impedance (1.35 ± 0.36 kΩ) at the lowest tested frequency (10 Hz), which is relatively closer to the physiological range,40 compared to pure GelMA hydrogel (15.58 ± 9.18 kΩ) (Fig. 2D). Such characteristic can be correlated to the presence of GNRs within the GelMA matrix due to bridging of the electrically insulated hydrogel's pores. It has been shown that the electrically conductive hydrogel matrices provide an excellent microenvironment for the growth and functioning of cardiac cells.40,57
Mechanical properties of the hydrogels in terms of rheology (storage moduli) are shown in Fig. 2E. The storage moduli of GelMA and GelMA–GNRs hydrogels decreased as a function of strain. At 5% strain, both hydrogels exhibited their ultimate storage moduli, where it was statistically higher for GelMA–GNR hydrogel as shown in Fig. 2F. Improved mechanical characteristics have been consistently reported elsewhere for similar nanomaterial-incorporated hydrogels.19,22,58 Specifically, electrostatic interactions between GNRs (positively charged)59 and GelMA chemical structure (negatively charged carboxyl groups)17 along with the filler effect60 of incorporated GNRs are speculated to lead to higher mechanical stiffness (storage modulus) of GelMA–GNR hydrogel. However, such mechanical enhancements did not result in a several fold increase in GelMA–GNR (10% w/v) hydrogel storage modulus. It is speculated that as hydrogel prepolymer solution (GelMA) concentration increases (i.e. 10%), the influences in the mechanical properties caused by embedding of nanomaterials (GNRs) may be compromised. For example, in our previously published study, incorporation of 1 or 1.5 mg mL−1 GNRs within GelMA hydrogels with 5% w/v prepolymer concentration significantly increased the mechanical stiffness (Young's modulus) of the material (∼2 folds),19 measured by atomic force microscopy (AFM). However, it is important to note that although increasing GelMA prepolymer concentration enhances the mechanical properties of the hydrogel, it can have adverse influence by decreasing the electrical conductivity of the hydrogel.61,62 Nevertheless, nanoengineered hydrogels with enhanced mechanical properties have exhibited superior cell adhesion, spreading and tissue formation.3,63,64
3.2. Cellular viability and formation of organized cardiac tissues
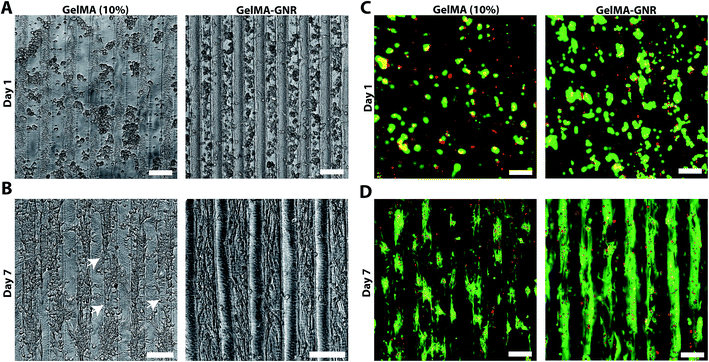
Fig. 3A illustrates phase contrast images of cardiomyocytes on the GelMA and GelMA–GNR hydrogel microgrooves on day 1 and day 7 of culture. Seeded cardiomyocytes formed discrete aggregated clusters inside the microgrooves on both substrates (GelMA vs. GelMA–GNR), on day 1. However, the cell adhesion pattern inside GelMA–GNR microgrooves was more homogenous. Over 7 days of culture, cardiomyocytes were spread and formed cellular networks within the microgrooves (Fig. 3B). Specifically, GelMA–GNR microgrooved cardiac tissues exhibited mostly concrete uniaxial cellular networks. In contrast, cell clusters exhibited discrete spreading within microgrooves on GelMA constructs. In some locations complete disconnections (Fig. 3B, white arrows) between the cellular clusters were evident. Cell retention comparing GelMA and GelMA–GNR on day 1 was not significantly different (data not shown). However, cardiomyocyte spreading and formation of organized tissues were profoundly improved on GelMA–GNR as compared to GelMA constructs (Fig. 3B, day 7). The dissimilarity in the cell adhesion and spreading patterns can be associated with the difference in elasticity (storage modulus) and surface characteristics between GelMA and GelMA–GNR hydrogels. As it has been demonstrated, the hydrogels with higher stiffness improved cell adhesion and spreading compared to the softer hydrogels, specifically in the case of two dimensional (2D) cell seeding on the constructs.19,22 Consistently, in our previous work, we confirmed the localization of GNRs within the micro-pores of the hydrogel constructs, which may subsequently lead to the formation of nanoscale local topographies, providing more anchoring points for cell adhesion and spreading. Higher surface roughness and nanoscale topographies were also observed in previous studies on the incorporation of carbon nanotubes within poly(N-isopropylacrylamide) (PNIPAAm) hydrogels.65 Regardless of hydrogel type (GelMA and GelMA–GNR), fluorescent Live/Dead images showed only few dead cells (red) on day 1 (Fig. 3C) and day 7 (Fig. 3D) of culture. In other words, purified GNRs (twice washed)43 incorporated within GelMA–GNR hydrogel did not induce major cytotoxicity over 7 days. Due to formation of dense cell clusters (day 1) and 3D topographical features of microgrooves (day 7), we were not able to quantify the percentage of viability.3.3. Cytoskeleton organization and cardiac cells phenotype
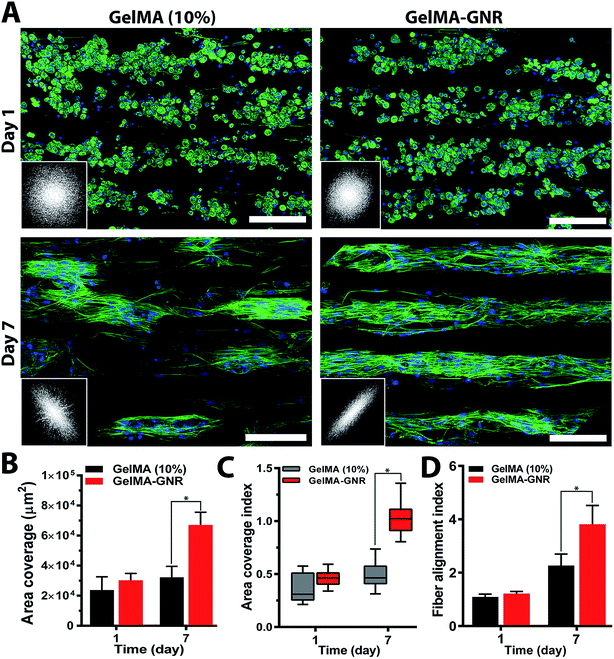
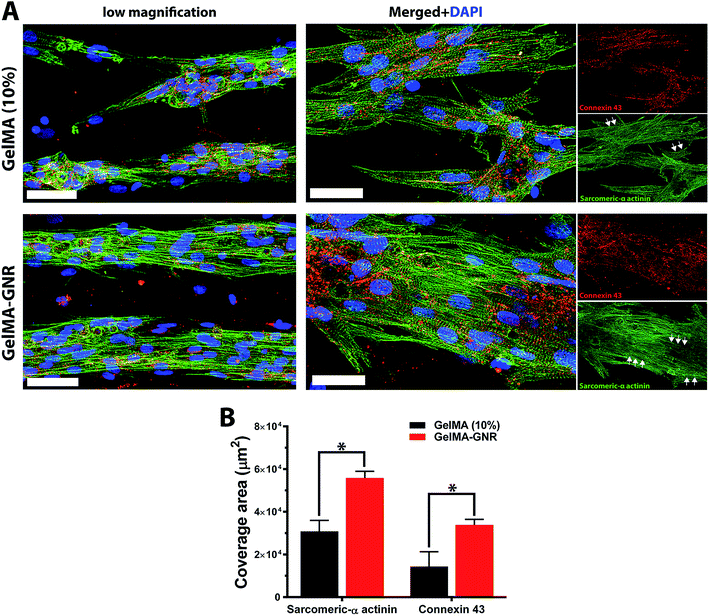
Cytoskeleton architecture of organized cardiac tissues on microgrooved GelMA and GelMA–GNR hydrogels were investigated through staining of F-actin fibers on day 1 (upon seeding) and day 7 of culture. As shown in Fig. 4A, cardiomyocytes were confined mostly within the microgrooves, and acquired round morphology on day 1 of culture within both hydrogel constructs. Quantified F-actin area coverage and index (Fig. 4B and C, day 1) also confirmed the same scenario as illustrated by immunostained images of round cells. After 7 days of culture, cardiac cells created compact and extended tissues (Fig. 4A, day 7) with significantly higher area coverage (Fig. 4B, day 7) on GelMA–GNR hydrogels as compared to disconnected cellular clusters on pure GelMA. Area coverage index (Fig. 4C) also confirmed considerable formation of organized tissues inside the GelMA–GNR microgrooves. Therefore, the major differences between GelMA and GelMA–GNR constructs were in terms of cellular connectivity, area coverage as well as formation of highly organized and compact tissues. Cytoskeleton alignment was further assessed using FFT analysis (inset images in Fig. 4A) on day 1 and 7. The outcomes indicated notably higher F-actin fiber alignment for cardiac tissues formed on GelMA–GNR microgrooves in comparison to GelMA constructs on day 7 (Fig. 4D). Despite the fact that GelMA–GNR hydrogels (∼3.5 kPa) provided stiffer matrices compared to GelMA (∼2.5 kPa), on day 1, cell adhesion coverage data (Fig. 4A and B) did not show a meaningful difference between both constructs. We speculate that GelMA hydrogel with 10% w/v prepolymer concentration provided a microenvironment with sufficient stiffness for early cell adhesion, but was not able to maintain cell spreading over time in comparison with GelMA–GNR hydrogel. Such behavior may be attributed to the excess cell anchoring points provided by embedded GNRs on the hydrogel matrix, as also demonstrated in our previous work,19 improving cellular spreading and tissue formation. Overall, the data in Fig. 4 were in agreement with our phase contrast images, confirming substantial formation of uniaxially aligned cardiac tissues directed within the microgrooves on GNR-incorporated GelMA hydrogels.The phenotype of cardiac cells on GelMA and GelMA–GNR microgrooved constructs was assessed using SATN and Cx43 gap junctions on day 7 of culture. As represented by immunostained images in Fig. 5A (separated channels), aligned SATN structures (white arrows) and well distributed Cx43 gap junctions are evident on both GelMA and GelMA–GNR hydrogels. However, similar to the cytoskeleton organization data (Fig. 4A) cellular disconnectivity and lack of tissue compactness were present on pure GelMA microgrooved constructs as compared to concrete and highly organized tissues on GelMA–GNR hydrogels. Fluorescent area coverage of SATN and Cx43 (Fig. 5B) further demonstrated significant differences in the formation of intact cardiac tissues comparing GelMA–GNR to GelMA (control) microgrooved constructs. These findings emphasize the critical role of hydrogel properties (e.g. stiffness) and the nanoscale cues provided by GNRs for the development of dense cardiac patches with native like organization and architecture. In addition, the electrically conductive matrix provided by GelMA–GNR hydrogel was involved in promoting cell–cell coupling.4,19,33,40
3.4. Spontaneous and stimulated contractile behavior of cardiac tissue microgrooves
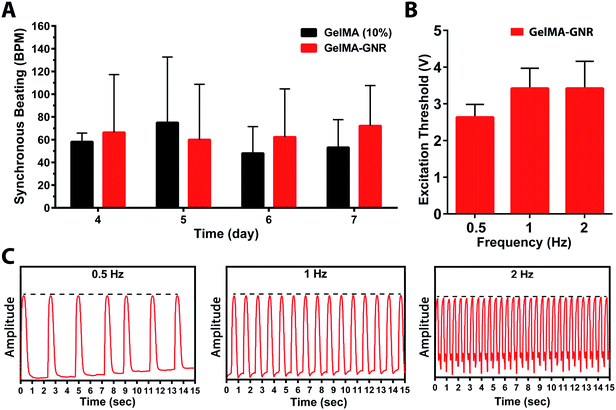
The functionality of aligned cardiac tissues in static culture condition and under external electrical stimulation on day 7 was further investigated using real time cellular contraction analysis (Fig. 6). As can be seen, cardiac tissues formed on GelMA and GelMA–GNR microgrooved constructs started beating at day 4 and continued until day 7 (ESI Movie SM1, SM2†). The results (Fig. 6A) exhibited no statistically significant differences in terms of spontaneous synchronous beats per minute (BPM) between GelMA and GelMA–GNR tissues. However, our previously published work19 on 1.5 mg mL−1 GNRs-incorporated 5% (w/v) GelMA hydrogel, without surface topographies (i.e. microgrooves), demonstrated significantly enhanced contractility (higher beats per minute, BPM) as compared to pure GelMA hydrogel. Despite the statistically higher mechanical stiffness of GelMA–GNR hydrogel (∼3.5 kPa) as compared to GelMA (∼2.5 kPa), we speculate that 10% (w/v) GelMA with high degree of methacrylation provided sufficient mechanical cues to initiate and maintain spontaneous contractility. Therefore, it is conjectured that incorporation of GNRs within GelMA hydrogel (10% w/v) and enhanced “electrical conductivity” (Fig. 2A and B) of the matrix would provide added benefits in terms of induced (controlled) electrical stimulation and contractility. Therefore, cardiac tissues were stimulated externally at 0.5, 1, and 2 Hz frequencies using an external pulse generator and custom made bioreactor setup.19,51 As the data shows (Fig. 6B), interestingly, it was not possible to induce stimulated contractions on tissues formed on pure GelMA constructs by electrical pulses up to 10 V amplitude. Conversely, highly organized tissues formed on GelMA–GNR hydrogel were successfully stimulated at all tested frequencies with relatively low excitation voltage thresholds (Fig. 6B). In addition, extracted beating signals (Fig. 6C and ESI Movie SM3†) demonstrated that cardiomyocytes were constantly contracted (no major peak-to-peak variation of amplitudes, dash black lines) with the same frequency provided by external stimulator. Therefore, addition of GNRs within the microgrooved GelMA hydrogel constructs not only induced higher cellular alignment, better connectivity and sarcomere organization but also provided a conductive matrix where the formed tissues could precisely follow external pulsed electrical stimulation regime. Therefore, it is envisioned that highly organized cardiac tissue formed on electrically conductive matrix would better integrate with the native myocardium following implantation.3 This is a critically important component of a viable cardiac patch for implantation as electrical signals in native cardiac tissue are often initiated and propagated by specialized cardiac cells (e.g. sinoatrial node and His-Purkinje cells) and then rapidly propagated throughout contractile cardiomyocyte tissue to effect synchronous contraction for optimum cardiac output. A highly organized tissue construct formed on microgrooved GelMA–GNR hydrogel that can be stimulated with low excitation threshold voltage would advance the ability to fully integrate with the synchronous contraction of native myocardium. Investigation of the functionalities of the proposed microengineered tissues in vivo, is the subject of our future work.4. Conclusions
In this study we developed and tested novel GNR-incorporated GelMA hydrogel microgrooved constructs (GelMA–GNR) for development of uniaxially aligned cardiac tissues. As a continuation of our previous work,19 we incorporated anisotropic topographical cues on electrically conductive GelMA–GNR hybrid hydrogels to provide simultaneous topographical and electrical signals to support cardiac cells functionalities in a biomimetic fashion. Our results showed improved mechanical elasticity (storage modulus) and electrical conductivity of GelMA–GNR hydrogels (lower impedance) compared to pure GelMA constructs. Phase-contrast images exhibited more uniform cell seeding fashion (day 1) on GelMA–GNR microgrooves compared to pure GelMA. Despite the resemblance in the cell coverage on day 1 for both constructs (i.e. with and without GNRs), cardiac tissues formed on GelMA–GNR hydrogels demonstrated highly dense and uniaxially aligned architectures, mainly inside the microgrooves. A similar scenario was observed for cardiac-markers demonstrating aligned cytoarchitecture. Specifically, cardiac cells on GelMA–GNR microgrooves revealed compact sarcomeric structures, while discrete pattern with considerably lower area coverage were detected on GelMA constructs without the presence of GNRs. Although both GelMA and GelMA–GNR microgrooved tissues demonstrated spontaneous contractility from day 4 to 7 of culture, only cardiac tissues formed on the electrically conductive GelMA–GNR microgrooves were able to be externally stimulated at different frequencies (0.5, 1, and 1 Hz) at low voltages. Overall, the data presented here demonstrate that the utility of integration of micro- and nanoscale strategy to developed highly organized and native-like cardiac tissues with superior functionalities for potential applications in regenerative medicine, disease modeling and drug screening. Finally, the use of human pluripotent stem cell-derived cardiomyocytes (e.g. hiPSCs) in regenerative medicine has dramatically increased in the past few years, due to their unique potential to model human myocardial pathophysiology and understand the mechanism of disease progression. In this regard, our future work will be focused on the use of hiPSC-derived cardiomyocytes to develop functional human cardiac patches for treatment of myocardial infarction.Acknowledgements
We acknowledge Ninad Chamele for the impedance characterization experiment, Amrita Pal for her contribution in the rheology assessment, and the LeRoy Eyring Center for Solid State Science at Arizona State University for SEM and TEM imaging.References
- G. C. Engelmayr Jr, M. Cheng, C. J. Bettinger, J. T. Borenstein, R. Langer and L. E. Freed, Nat. Mater., 2008, 7, 1003–1010 CrossRef PubMed.
- Z. Ling, S. Juan and H. Yuemei, Heart, 2011, 97, A67 CrossRef.
- J. Zhou, J. Chen, H. Sun, X. Qiu, Y. Mou, Z. Liu, Y. Zhao, X. Li, Y. Han, C. Duan, R. Tang, C. Wang, W. Zhong, J. Liu, Y. Luo, M. Mengqiu Xing and C. Wang, Sci. Rep., 2014, 4, 3733 Search PubMed.
- T. Dvir, B. P. Timko, M. D. Brigham, S. R. Naik, S. S. Karajanagi, O. Levy, H. Jin, K. K. Parker, R. Langer and D. S. Kohane, Nat. Nanotechnol., 2011, 6, 720–725 CrossRef CAS PubMed.
- D. H. Kim, E. A. Lipke, P. Kim, R. Cheong, S. Thompson, M. Delannoy, K. Y. Suh, L. Tung and A. Levchenko, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 565–570 CrossRef CAS PubMed.
- D. A. Pijnappels, S. Gregoire and S. M. Wu, Ann. N. Y. Acad. Sci., 2010, 1188, 7–14 CrossRef PubMed.
- N. Annabi, K. Tsang, S. M. Mithieux, M. Nikkhah, A. Ameri, A. Khademhosseini and A. S. Weiss, Adv. Funct. Mater., 2013, 23, 4950–4959 CrossRef CAS PubMed.
- N. Bursac, K. K. Parker, S. Iravanian and L. Tung, Circ. Res., 2002, 91, e45–54 CrossRef CAS PubMed.
- M. Radisic, H. Park, H. Shing, T. Consi, F. J. Schoen, R. Langer, L. E. Freed and G. Vunjak-Novakovic, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 18129–18134 CrossRef CAS PubMed.
- H. Saini, A. Navaei, A. Van Putten and M. Nikkhah, Adv. Healthcare Mater., 2015, 4, 1961–1971 CrossRef CAS PubMed.
- C. Cha, P. Soman, W. Zhu, M. Nikkhah, G. Camci-Unal, S. Chen and A. Khademhosseini, Biomater. Sci., 2014, 2, 703–709 RSC.
- M. Kharaziha, M. Nikkhah, S. R. Shin, N. Annabi, N. Masoumi, A. K. Gaharwar, G. Camci-Unal and A. Khademhosseini, Biomaterials, 2013, 34, 6355–6366 CrossRef CAS PubMed.
- Z. Ma, J. Wang, P. Loskill, N. Huebsch, S. Koo, F. L. Svedlund, N. C. Marks, E. W. Hua, C. P. Grigoropoulos, B. R. Conklin and K. E. Healy, Nat. Commun., 2015, 6, 7413 CrossRef CAS PubMed.
- Y. Yeo, W. Geng, T. Ito, D. S. Kohane, J. A. Burdick and M. Radisic, J. Biomed. Mater. Res., Part B, 2007, 81, 312–322 CrossRef PubMed.
- P. Camelliti, J. O. Gallagher, P. Kohl and A. D. McCulloch, Nat. Protoc., 2006, 1, 1379–1391 CrossRef CAS PubMed.
- L. L. Chiu, K. Janic and M. Radisic, Int. J. Artif. Organs, 2012, 35, 237–250 CrossRef CAS PubMed.
- J. W. Nichol, S. T. Koshy, H. Bae, C. M. Hwang, S. Yamanlar and A. Khademhosseini, Biomaterials, 2010, 31, 5536–5544 CrossRef CAS PubMed.
- B. J. Klotz, D. Gawlitta, A. J. Rosenberg, J. Malda and F. P. Melchels, Trends Biotechnol., 2016, 34, 394–407 CrossRef CAS PubMed.
- A. Navaei, H. Saini, W. Christenson, R. T. Sullivan, R. Ros and M. Nikkhah, Acta Biomater., 2016, 41, 133–146 CrossRef CAS PubMed.
- S. R. Shin, C. Zihlmann, M. Akbari, P. Assawes, L. Cheung, K. Zhang, V. Manoharan, Y. S. Zhang, M. Yuksekkaya, K. T. Wan, M. Nikkhah, M. R. Dokmeci, X. S. Tang and A. Khademhosseini, Small, 2016, 12, 3677–3689 CrossRef CAS PubMed.
- A. Paul, A. Hasan, H. A. Kindi, A. K. Gaharwar, V. T. Rao, M. Nikkhah, S. R. Shin, D. Krafft, M. R. Dokmeci, D. Shum-Tim and A. Khademhosseini, ACS Nano, 2014, 8, 8050–8062 CrossRef CAS PubMed.
- S. R. Shin, S. M. Jung, M. Zalabany, K. Kim, P. Zorlutuna, S. B. Kim, M. Nikkhah, M. Khabiry, M. Azize, J. Kong, K. T. Wan, T. Palacios, M. R. Dokmeci, H. Bae, X. S. Tang and A. Khademhosseini, ACS Nano, 2013, 7, 2369–2380 CrossRef CAS PubMed.
- W. H. Zimmermann, I. Melnychenko, G. Wasmeier, M. Didie, H. Naito, U. Nixdorff, A. Hess, L. Budinsky, K. Brune, B. Michaelis, S. Dhein, A. Schwoerer, H. Ehmke and T. Eschenhagen, Nat. Med., 2006, 12, 452–458 CrossRef CAS PubMed.
- A. Furuta, S. Miyoshi, Y. Itabashi, T. Shimizu, S. Kira, K. Hayakawa, N. Nishiyama, K. Tanimoto, Y. Hagiwara, T. Satoh, K. Fukuda, T. Okano and S. Ogawa, Circ. Res., 2006, 98, 705–712 CrossRef CAS PubMed.
- M. Rubart, K. B. Pasumarthi, H. Nakajima, M. H. Soonpaa, H. O. Nakajima and L. J. Field, Circ. Res., 2003, 92, 1217–1224 CrossRef CAS PubMed.
- S. Ahadian, J. Ramon-Azcon, M. Estili, X. Liang, S. Ostrovidov, H. Shiku, M. Ramalingam, K. Nakajima, Y. Sakka, H. Bae, T. Matsue and A. Khademhosseini, Sci. Rep., 2014, 4, 4271 Search PubMed.
- A. Simon, S. X. Maletz, H. Hollert, A. Schaffer and H. M. Maes, Nanoscale Res. Lett., 2014, 9, 396 CrossRef PubMed.
- Y. Zhu and W. X. Li, Sci. China, Ser. B: Chem., 2008, 51, 1021–1029 CrossRef CAS.
- C. P. Firme 3rd and P. R. Bandaru, Nanomedicine, 2010, 6, 245–256 Search PubMed.
- S. Vardharajula, S. Z. Ali, P. M. Tiwari, E. Eroglu, K. Vig, V. A. Dennis and S. R. Singh, Int. J. Nanomed., 2012, 7, 5361–5374 CAS.
- Y. Y. Huang and E. M. Terentjev, Polymers, 2012, 4, 275–295 CrossRef.
- S. R. Shin, H. Bae, J. M. Cha, J. Y. Mun, Y. C. Chen, H. Tekin, H. Shin, S. Farshchi, M. R. Dokmeci, S. Tang and A. Khademhosseini, ACS Nano, 2012, 6, 362–372 CrossRef CAS PubMed.
- S. Fleischer, M. Shevach, R. Feiner and T. Dvir, Nanoscale, 2014, 6, 9410–9414 RSC.
- P. Baei, S. Jalili-Firoozinezhad, S. Rajabi-Zeleti, M. Tafazzoli-Shadpour, H. Baharvand and N. Aghdami, Mater. Sci. Eng., C, 2016, 63, 131–141 CrossRef CAS PubMed.
- R. Ravichandran, R. Sridhar, J. R. Venugopal, S. Sundarrajan, S. Mukherjee and S. Ramakrishna, Macromol. Biosci., 2014, 14, 515–525 CrossRef CAS PubMed.
- M. Shevach, S. Fleischer, A. Shapira and T. Dvir, Nano Lett., 2014, 14, 5792–5796 CrossRef CAS PubMed.
- D. Jung, I. Minami, S. Patel, J. Lee, B. Jiang, Q. Yuan, L. Li, S. Kobayashi, Y. Chen, K. B. Lee and N. Nakatsuji, J. Nanobiotechnol., 2012, 10, 23 CrossRef CAS PubMed.
- T. J. Lee, S. Kang, G. J. Jeong, J. K. Yoon, S. H. Bhang, J. Oh and B. S. Kim, Tissue Eng., Part A, 2015, 21, 374–381 CrossRef CAS PubMed.
- S. Sridhar, J. R. Venugopal, R. Sridhar and S. Ramakrishna, Colloids Surf., B, 2015, 134, 346–354 CrossRef CAS PubMed.
- J. O. You, M. Rafat, G. J. Ye and D. T. Auguste, Nano Lett., 2011, 11, 3643–3648 CrossRef CAS PubMed.
- M. Shevach, B. M. Maoz, R. Feiner, A. Shapira and T. Dvir, J. Mater. Chem. B, 2013, 1, 5210–5217 RSC.
- R. Shukla, V. Bansal, M. Chaudhary, A. Basu, R. R. Bhonde and M. Sastry, Langmuir, 2005, 21, 10644–10654 CrossRef CAS PubMed.
- E. E. Connor, J. Mwamuka, A. Gole, C. J. Murphy and M. D. Wyatt, Small, 2005, 1, 325–327 CrossRef CAS PubMed.
- M. Hu, J. Chen, Z. Y. Li, L. Au, G. V. Hartland, X. Li, M. Marquez and Y. Xia, Chem. Soc. Rev., 2006, 35, 1084–1094 RSC.
- C. M. Cobley, J. Chen, E. C. Cho, L. V. Wang and Y. Xia, Chem. Soc. Rev., 2011, 40, 44–56 RSC.
- X. Huang, S. Neretina and M. A. El-Sayed, Adv. Mater., 2009, 21, 4880–4910 CrossRef CAS PubMed.
- P. Lloret, G. Longinotti, G. Ybarra, L. Socolovsky and C. Moina, Mater. Res. Bull., 2013, 48, 3671–3676 CrossRef CAS.
- D. Qin, Y. Xia and G. M. Whitesides, Nat. Protoc., 2010, 5, 491–502 CrossRef CAS PubMed.
- T. Betancourt and L. Brannon-Peppas, Int. J. Nanomed., 2006, 1, 483–495 CrossRef CAS PubMed.
- D. Truong, J. Pulel, A. Llave, G. Mouneimne, R. Kamm and M. Nikkhah, Sci. Rep., 2016, 6, 34094 CrossRef CAS PubMed.
- A. Navaei, D. Truong, J. Heffernan, J. Cutts, D. Brafman, R. W. Sirianni, B. Vernon and M. Nikkhah, Acta Biomater., 2016, 32, 10–23 CrossRef CAS PubMed.
- J. W. Nichol, G. C. Engelmayr Jr, M. Cheng and L. E. Freed, Biochem. Biophys. Res. Commun., 2008, 373, 360–365 CrossRef CAS PubMed.
- N. Tandon, C. Cannizzaro, P. H. Chao, R. Maidhof, A. Marsano, H. T. Au, M. Radisic and G. Vunjak-Novakovic, Nat. Protoc., 2009, 4, 155–173 CrossRef CAS PubMed.
- S. B. Kim, H. Bae, J. M. Cha, S. J. Moon, M. R. Dokmeci, D. M. Cropek and A. Khademhosseini, Lab Chip, 2011, 11, 1801–1807 RSC.
- Q. L. Loh and C. Choong, Tissue Eng., Part B, 2013, 19, 485–502 CrossRef CAS PubMed.
- N. Annabi, J. W. Nichol, X. Zhong, C. Ji, S. Koshy, A. Khademhosseini and F. Dehghani, Tissue Eng., Part B, 2010, 16, 371–383 CrossRef CAS PubMed.
- V. Martinelli, G. Cellot, F. M. Toma, C. S. Long, J. H. Caldwell, L. Zentilin, M. Giacca, A. Turco, M. Prato, L. Ballerini and L. Mestroni, Nano Lett., 2012, 12, 1831–1838 CrossRef CAS PubMed.
- A. K. Gaharwar, S. M. Mihaila, A. Swami, A. Patel, S. Sant, R. L. Reis, A. P. Marques, M. E. Gomes and A. Khademhosseini, Adv. Mater., 2013, 25, 3329–3336 CrossRef CAS PubMed.
- B. Nikoobakht and M. A. El-Sayed, Chem. Mater., 2003, 15, 1957–1962 CrossRef CAS.
- S. C. Tjong, Mater. Sci. Eng., R, 2006, 53, 73–197 CrossRef.
- B. Veleirinho, M. F. Rei and J. A. Lopes-da-Silva, J. Polym. Sci., Part B: Polym. Phys., 2008, 46, 460–471 CrossRef CAS.
- B. Tarus, N. Fadel, A. Al-Oufy and M. El-Messiry, Alexandria Eng. J., 2016, 55, 2975–2984 CrossRef.
- H.-B. Wang, M. Dembo and Y.-L. Wang, Am. J. Physiol.: Cell Physiol., 2000, 279, C1345–C1350 CAS.
- C. N. Grover, J. H. Gwynne, N. Pugh, S. Hamaia, R. W. Farndale, S. M. Best and R. E. Cameron, Acta Biomater., 2012, 8, 3080–3090 CrossRef CAS PubMed.
- H. Sun, S. Lü, X.-X. Jiang, X. Li, H. Li, Q. Lin, Y. Mou, Y. Zhao, Y. Han, J. Zhou and C. Wang, Biomaterials, 2015, 55, 84–95 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra26279a |
| This journal is © The Royal Society of Chemistry 2017 |