Open Access Article
Open Access ArticleNovel diagnostics for point-of-care bacterial detection and identification
Savannah Realia,
Elias Y. Najiba,
Krisztina E. Treuerné Balázsa,
Adeline Chern Hui Tana,
Linda Váradi b,
David E. Hibbs
b,
David E. Hibbs a and
Paul W. Groundwater
a and
Paul W. Groundwater *a
*a
aThe University of Sydney School of Pharmacy, Camperdown Campus, Sydney, NSW 2006, Australia
bCSIRO Manufacturing, Normanby Road, Clayton, VIC 3168, Australia. E-mail: paul.groundwater@sydney.edu.au
First published on 10th July 2019
Abstract
In addition to limiting the effectiveness of antimicrobial agents, antimicrobial resistance (AMR) is a significant global health concern as it is responsible for significant mortality/morbidity and increased economic burdens on healthcare systems. Diagnostic tests have been suggested as a means of prolonging the effectiveness of current antimicrobials; culture and other conventional diagnostics are hindered in their practicality as they are time- and labour intensive to perform. Point-of-care (POC) testing is performed near where the patient is being treated and can provide timely results that allow evidence based clinical interventions to be made. This review aims to outline the chemical principles behind some novel and emerging diagnostic techniques which have the required speed, simplicity, effectiveness and low-cost for incorporation into POC devices which can be used to inform and optimize antimicrobial use.
1. Introduction
1.1 The unmet need for rapid bacterial identification techniques
Antimicrobial resistance (AMR) is a significant global issue and has the potential to completely alter the landscape of modern healthcare. AMR already has devastating consequences on human health and the global economy as a result of associated increases in patient morbidity and mortality, increases in the length of hospital stays, risks associated with surgical procedures, and a significant economic burden due to losses in productivity from illness. It has been suggested that, at the current rate, AMR will be responsible for ten million deaths each year by 2050 (more than all cancers combined).1 During the period to 2050, the annual reduction in global gross domestic product (GDP) could be as large as the losses incurred during the 2008–2009 global financial crisis, with significantly greater implications, as the decrease in economic growth for low-income countries will be inordinately high, while the impacts will be felt during the entire period to 2050.2 Although a global problem, it is predicted that Asia and Africa will continue to share the greatest burden due to AMR, with 4.73 and 4.15 million deaths p.a., respectively, by 2050 (Fig. 1).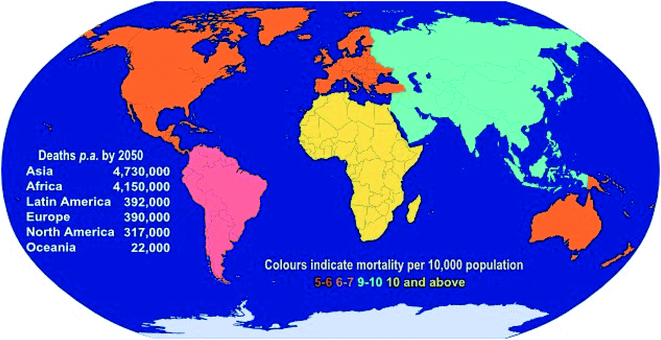 | ||
| Fig. 1 Predicted annual deaths (and mortality rates) due to AMR by 2050 (data taken from the 2016 O'Neill report1). | ||
Of particular concern to the growing trend of AMR are the ESKAPE bacteria (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter spp.). These multiple and pan drug resistant pathogens host a plethora of resistance mechanisms and contribute to the mortality of nosocomial infections.3 As the single leading cause of nosocomial infections, Clostridium difficile infections (CDIs)4–6 are also a major concern and are responsible for 500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 infections and 15
000 infections and 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths per year in the US.4
000 deaths per year in the US.4
In response to the urgent threat posed by AMR, the World Health Organization (WHO) initiated a global action plan calling for international collaboration in order to address the issue.7 The plan outlines five core objectives (Box 1), expected to be used by all countries during the development of their own national plans, including increased investment in new diagnostic tools and the optimization of antimicrobial use. In the last 20 years, there has been a 75% reduction in antimicrobial agents approved by the FDA8 and only two new classes have been developed with novel therapeutic actions.9 With few new antimicrobials in the pipeline, emphasis is now being placed on diagnostics to maintain the efficacy of current antimicrobials. Now, more than ever, in the face of a global crisis, rapid and accurate identification of infectious bacterial species and their susceptibility profile is vital so that the appropriate directed antibiotic therapy can be initiated, thus improving patient outcomes and helping to retard the further development of AMR. The WHO report suggests that antibiotic prescriptions are rarely based upon accurate/prompt diagnoses and that the standard of care should be evidence-based prescribing and dispensing, which could be informed by rapid, low-cost diagnostic tests, integrated into clinical, pharmacy, and veterinary practices.7 The 2016 O'Neill report on ‘Tackling Drug-Resistant Infections Globally’ suggests that by 2020 all clinicians should perform a rapid diagnostic test before prescribing antimicrobials.1 Particular emphasis is being placed on the development of point-of-care (POC) tests resulting in the rapid expansion of this diagnostic field, with the market volume estimated to grow to US $75.1 billion by 2020.10
A useful definition of a POC diagnostic test is one ‘that is performed near the patient or treatment facility, has a fast turnaround time, and may lead to a change in patient management’.11 Such tests, which do not require access to centralised laboratory facilities, should ideally be sufficiently rapid to allow clinically meaningful interventions (e.g. the initiation of directed, as opposed to empirical, antibacterial treatment) to be implemented at the place at which the patient is being treated. For example, it has been estimated that the use of rapid tests for three of the leading causes of death due to bacterial infections (community-acquired bacterial pneumonia (CAP) [Streptococcus pneumoniae, Haemophilus influenzae and Moraxella catarrhalis], antenatal syphilis [Treponema pallidum] and tuberculosis [Mycobacterium tuberculosis]) could help prevent more than one million deaths per annum in developing countries.12 Meanwhile the implementation of POC for S. pneumoniae and Legionella pneumophila in severe CAP cases has been shown to lower mortality rates by facilitating more accurate diagnoses, thus allowing more appropriate empirical treatment choices.13
Box 1. The five objectives outlined by the 2015 WHO global action plan on AMR7→ Utilize effective communication, education, and training to improve the awareness and understanding of AMR.→ Use surveillance and research to generate knowledge and then translate the evidence base into practice. → Utilize effective infection prevention (in conjunction with sanitation and hygiene measures) to help reduce the incidence of infection. → Optimize human (and animal) antimicrobial usage. → Develop a case for sustainable economic investment, taking into account the needs of all countries, in order to increase investment in the development of a range of interventions, including new medicines/vaccines and diagnostic tools. |
1.2 The current diagnostic landscape14
Phenotypic methods, such as culture and staining, have long been the gold standard for bacterial identification, but a major limitation is that results can take up to 48 hours (or more) to become available,15,16 and often do not provide sufficient information to inform antimicrobial prescription.14In the time taken to identification of the causative bacterium, the patient will probably receive empirical therapy, often with broad spectrum agents, the use of which is discouraged in antimicrobial stewardship programs (in favour of narrower spectrum, targeted agents) in order to minimize the use of less effective agents which may contribute to AMR.17 For severe infections such as sepsis, a delay in effective treatment can be severely detrimental, with the mortality rate increasing by 7.6% every hour targeted therapy is delayed.18 Furthermore, culturing may not lead to the specific identification of the causative bacterium in the presence of closely related species with similar phenotypic and metabolic properties; culturing must also be conducted by trained personnel.19 Molecular diagnostic methods, which rely upon the analysis of genomic markers (corresponding to nucleic acid sequences), can result in significantly shorter times to bacterial detection/identification but often require the use of specialized equipment and/or require specialist interpretation.14
In summary, conventionally used techniques are burdened by the length of time to diagnostic result, in some cases with the additional requirement of the isolation of the causative microorganism, and the need for trained personnel, and costly and highly specialised equipment.
While the definition of a POC device does not specify any particular technology, diagnostic techniques that are potentially low cost and can allow for rapid diagnostic results without the requirement for expensive specialized equipment have the potential to be implemented anywhere in the World, including well-resourced and resource-limited locations (such as developing countries in both Africa and Asia). Similar performance measures were also identified by, for example, the WHO Sexually Transmitted Diseases Diagnostic Initiative under the ASSURED criteria of ‘affordable, sensitive, specific, user-friendly, rapid and robust, equipment-free, and deliverable to end-users’.20
2. Diagnostic techniques suitable for the rapid detection and identification of bacteria in point-of-care (POC) devices
The need for rapid diagnosis has resulted in the development of novel diagnostic devices based upon the detection and quantification (e.g. colorimetric/fluorimetric or electrochemical21) of specific analytes by biorecognition elements that may be implemented in both community and primary care settings.22,23 Many of the novel detection technologies discussed in this review rely upon a combination of these approaches and we will discuss these different approaches in terms of the biorecognition elements employed (Fig. 2) providing examples of detection methods suitable for incorporation in POC devices.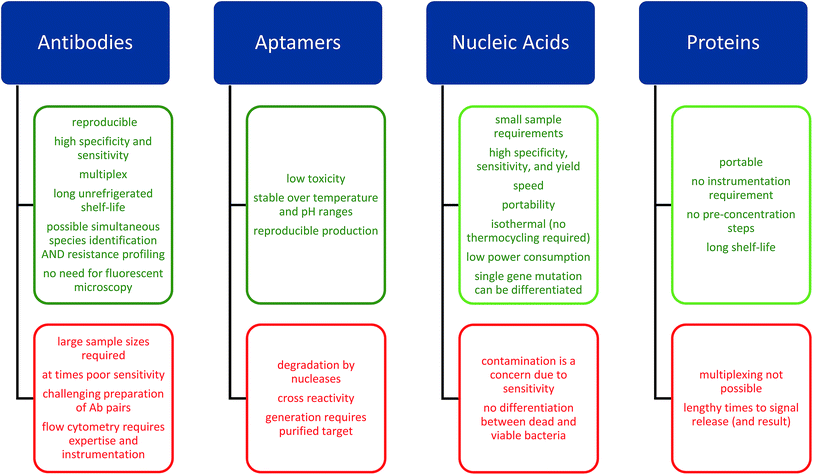 | ||
| Fig. 2 Advantages and disadvantages of biorecognition elements utilized in POC devices and discussed in this review. | ||
2.1 Antibodies
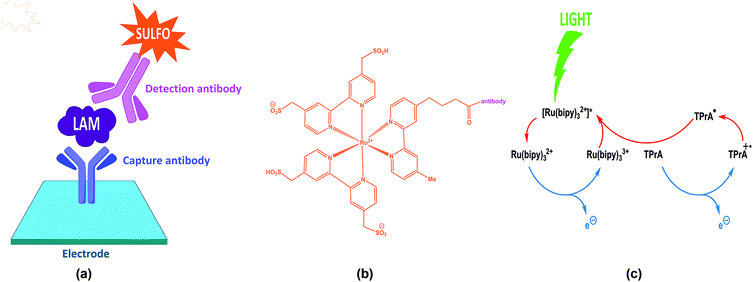 | ||
| Fig. 3 Schematic of the ECL-based detection of M. tuberculosis through an antibody sandwich complex for the 5-methythio-D-xylofuranose-lipoarabinomannan (MTX-LAM) epitope (adapted from https://www.mesoscale.com/en/technical_resources/our_technology/ecl).24 | ||
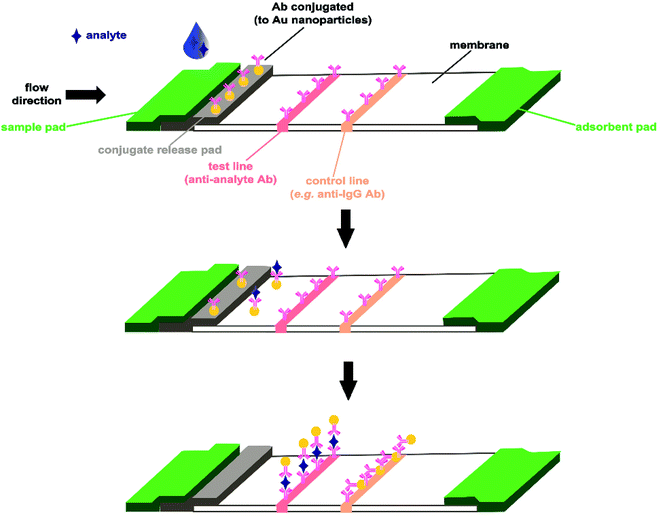 | ||
| Fig. 4 An example of a lateral flow immunoassay test platform (adapted from Koczula et al.25). The sample solution containing the analyte is deposited on the sample pad and migrates towards the antibody biorecognition element (in this case conjugated to gold nanoparticles). The analyte-conjugated antibodies are captured at the test line (positive outcome), while antibodies which are not bound to analyte are captured at the control line (providing test validation). | ||
Noble metal nanoparticles (NMNPs) have typically been used in LFIAs due to their potential of producing a diagnostic signal visible to the naked eye erasing the need for an external excitation source or emission sensor.28 Gold NPs (AuNPs) have typically been used as coloured conjugates due to their superior optical properties and ease of use,29 however their lack of sensitivity is an issue that needs to be addressed.
With the high levels of AMR among ESKAPE pathogens and other bacteria responsible for nosocomial infections, the combined ability to detect the presence of a bacterial species and identify resistant strains is highly advantageous. Immunochromatography has been effective in detecting the presence of carbapenemase resistance genes; Carba5 (NG Biotech, France) is a novel multiplex LFIA for the rapid detection of the five most prominent classes of carbapenemases, the NDM-, KPC-, IMP-, VIM-, and OXA-48-like types.30 This assay uses antibody pairs specific for these five carbapenemase classes. One of the antibodies is conjugated to AuNPs and the complementary antibody is immobilized on different test lines along the flow membrane surface. Carba5 exhibited 100% sensitivity and >90% specificity, therefore this robust, rapid assay has the potential for the identification of CPE in a POC setting.
Methods such as this, which incorporate LFIA, are capable of producing signals that can be detected by the naked eye within 15 minutes. They do not require large and expensive instrumentation, and so are relatively low in cost in comparison to other conventional molecular diagnostics, at an estimated one-quarter to one-third of the cost per test.30 As the colorimetric readout is determined by the number of bacteria present in a sample, these methods require large sample-sizes and inadequate sample volumes may give inaccurate results. The identification and preparation of unique antibody pairs are often time consuming, but with high stability and an unrefrigerated shelf life of 24 months, these systems can be produced and stored on a large-scale to minimize associated costs. With the primary issue of sensitivity having been addressed in newer LFIA systems, these techniques have demonstrated their suitability for inclusion in POC diagnostic methods for pathogenic bacteria. For example, a LFIA-based POC testing kit, aQcare Chlamydia TRF, for Chlamydia trachomatis (a leading cause of sexually transmitted disease), employs europium(III)-chelated NPs as fluorescent markers and had an overall sensitivity of 93.0% and specificity of 96.3%.31
A vertical flow immunoassay (VFI) demonstrated potential for use as a POC test for bio-threat agents targeting 1,3-linked 2-O-acetyl-6-dexoy-β-D-manno-heptopyanose, a capsular polysaccharide (CPS) from Burkholderia pseudomallei, the cause of melioidosis. The limit of detection for the CPS in spiked buffer solutions was 0.02 ng mL−1.32
2.2 Aptamers15,33
Aptamers are short, single-stranded DNA or RNA oligonucleotide biosensors that can interact with a wide variety of analyte targets with high affinity and specificity.15,34 Upon binding to their target analyte, aptamers fold into specific three-dimensional structures with many surface interactions for strong bonding,34 typically with a dissociation constant in the nano- or pico-molar level.35,36Aptamers are chemically synthesized in vitro by a process known as Systematic Evolution of Ligands by Exponential enrichment (SELEX), which involves three key steps: (1) the incubation of an oligonucleotide sequence library with the target analyte to assess which structures bind; (2) the elution of unbound oligonucleotides, separating them from those bound to the analyte; and (3) amplification of the remaining oligonucleotide sequences by PCR. These three steps can be repeated up to 20 times until a small collection of sequences with high affinity for the target are identified.34,35
When used as a biosensor, aptamers offer many advantages over antibodies; in contrast to antibodies, aptamers have low toxicity, are stable over a wide temperature and pH range, and are the products of simple and reproducible chemical syntheses.34,36 Although the determination of aptamer structure requires several steps, their in vitro synthesis is preferable to that of antibodies, which require synthesis in biological systems under highly specific conditions.33,37
Nucleic acid aptamers have been used with a variety of signal transduction instruments for development into POC tests. For example, for the detection of S. aureus, aptamer-based biosensors utilise colorimetric, mass spectrometry, or fluorescence-based detection. Of particular interest are electrochemical aptamer-based techniques due to their exceptional sensitivity, low cost of production, simplicity, and portability.36
An aptamer/graphene interlinked gold electrode has also been developed that utilizes a piezoelectric sensor;36,40 the aptamers are immobilized on a graphene surface and when conjugated with S. aureus there is a frequency shift of the piezoelectric quartz crystal, which is proportional to the bacterial concentration. The limit of detection of S. aureus is 41 CFU mL−1 and results are obtained within 60 minutes,40 making this rapid, simple, sensitive and label-free method suitable for use in a POC diagnostic test for S. aureus.
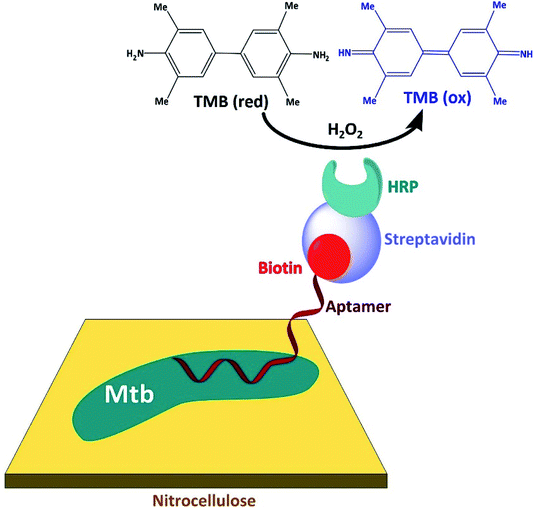 | ||
| Fig. 5 Outline of the LTBI diagnostic test which is based upon biotin-linked aptamers specific for the M. tuberculosis glycolipid, ManLAM (adapted from Li et al.41). | ||
2.3 Nucleic acids
Polymerase chain reaction (PCR)-based techniques form the basis of many diagnostic tests for bacterial infections,14 e.g. the use of PCR testing for nasal MRSA reduced the duration of empirical-MRSA therapy (vancomycin or linezolid) in patients with suspected pneumonia by ca. 2 days, without an increase in the number of adverse clinical outcomes.42 As it involves nucleic acid amplification (for example, by a factor of 1 million) from a small sample volume (e.g. a nasal swab), real time or quantitative PCR (qPCR) has been able to address some of the issues associated with culture-based methods (although some PCR-based methodologies require broth enrichment). Key developments in qPCR have resulted in significant diagnostic performance advantages over conventional PCR; for example, qPCR methods require small quantities of sample and provide rapid results,43,44 and also require minimal sample manipulation (thus further reducing the time required and the risk of contamination) due to the measurement of changes in the fluorescence signal from the start of the PCR process.On the other hand, as qPCR involves such high levels of amplification, contamination or the detection of nucleic acids which remain from a previously cleared infection is a genuine concern. Further, the discrimination between an asymptomatic colonization and a clinically relevant infection relies upon the development of standardized quantitative cut off cycle-threshold (Ct)† values.45 However, it is not clear that a relationship actually exists between the number of microorganisms present in an individual's specimen and the presence of healthy carriage or disease within the individual.
One means of overcoming the problematic amplification of non-target DNA is to utilize multiplex-touchdown PCR (MT-PCR), which combines the features of a multiplex PCR (incorporating primers for a number of DNA targets) with touchdown PCR (which involves a cycling program, in which the annealing temperature is gradually reduced from a value above the estimated melting temperature (Tm) of the primers‡ until it reaches the calculated annealing temperature [the touchdown temperature]), to increase the PCR specificity, sensitivity and yield.46
Such a MT-PCR method47 simultaneously detects the presence of the mecA gene (which encodes the penicillin binding protein 2a conferring MRSA resistance to methicillin48 by resulting in reduced binding affinity), and blaSHV, blaCTX-M, blaTEM and blaOXA49 (which encode extended spectrum β-lactamases [ESBLs]) genes in MRSA and extended spectrum β-lactamase (ESBL) positive blood cultures. This method resulted in 100% specificity for the detection of all genes, and an analytical sensitivity of 103 and 102 CFU mL−1 for mecA and other genes, respectively.
A proposed one-step multiplex PCR method targets the chromosomal class A β-lactamase genes blaSHV of Klebsiella pneumoniae, blaOKP of K. variicola and blaLEN of K. quasipneumoniae and their flanking gene (deoR) to create a rapid (less than 2.5 hours) and accurate method which can distinguish between these pathogens.50
The development of the above techniques has resulted in significant improvements in PCR, thus helping to satisfy the requirements of POC methods; devices employing PCR have become smaller, lower in energy consumption, more user-friendly, and provide accurate results more rapidly.
Recombinase polymerase amplification (RPA) amplifies target sequences in a sample by utilising an isothermal technique which operates at a relatively low constant temperature (between 37–42 °C) and, unlike the Taq polymerase used in the conventional PCR assay, does not require heating to 95 °C and thermocycling.19,53 RPA does not require thermal denaturing of the template; it employs recombinase–primer complexes which enable strand exchange at cognate sequences, single-stranded DNA binding proteins and a strand displacing DNA polymerase.54 This method had detection limits (of about 10 nucleic acid molecules) which are comparable to currently employed diagnostic devices, but its speed, portability, low cost and simplicity of operation make it suitable for use in any global setting. One drawback with this system is its inability to discriminate between live and dead bacteria, which may in future be overcome by detecting mRNA rather than DNA.
The cobas®Liat®Cdiff system is a completely automated approach to the diagnosis of C. difficile-associated diarrhoea by the direct detection of the toxin B (tcdB) gene in patient stool samples.55 This assay specifically detects this toxin gene to assess for a variety of toxigenic C. difficile strains (including a hypervirulent epidemic strain). The cobas®Liat®Cdiff system combines sample preparation, nucleic acid extraction, real-time PCR amplification and detection of target DNA sequences in a single sample analysis that has a turnaround time of 20 minutes, 95.1% specificity and 93.1% sensitivity, with limit of detection of 45–90 CFU per swab for toxigenic strains.55 A similar approach has also been used for the detection of MRSA using the cobas®MRSA/SA test.56
A magnetic barcode assay system was also the basis for a multiplexed detection platform for S. aureus and K. pneumoniae,57 which utilized both magnetic nanoparticles and fluorescent quantum dots (QDs) for the detection of the target bacteria (Fig. 6). Once again, as this technique is able to distinguish single-gene mutations, it is also able to detect drug-resistant strains; for example, it was able to differentiate between methicillin-sensitive (MSSA) and methicillin-resistant (MRSA) S. aureus. A target gene specific for each bacterium was selected for detection; fnbA for MSSA, mecA for MRSA and wcaG for K. pneumoniae.57
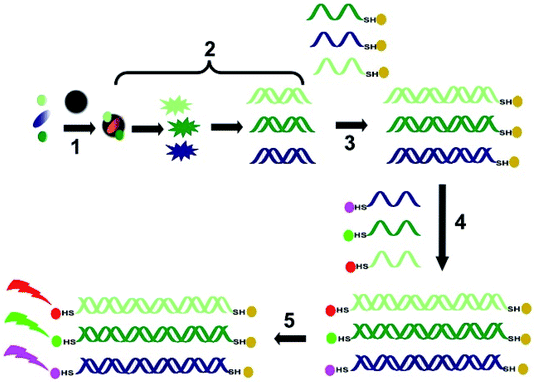 | ||
| Fig. 6 Schematic representation of the steps involved in bacterial identification using multiplex barcoding: (1) the bacteria are captured on the MNPs by a non-specific mechanism (presumably electrostatic attraction); (2) lysis of the bacterial cells on the MNP surface release the nucleic acids, which are amplified by multiplex PCR; (3) the amplicons are linked to gold-MNPs (Au-MNPs) labelled with complementary oligonucleotides, followed by magnetic purification; (4) QDs labelled with complementary oligonucleotide sequences to the fnbA, mecA and wcaG genes are added, followed by magnetic purification; (5) detection of fluorescence at different wavelengths (representative of the different QDs).57 This system is able to detect the presence of bacteria in concentrations as low as 100 CFU mL−1.58 MNPS demonstrate good thermal and chemical stability,58 are highly sensitive due to their high surface area to volume ratios, and have little interference from biological specimens. | ||
A magnetic barcode assay has been developed for the detection of Mycobacterium tuberculosis, and its clinical utility proven by the detection of M. tuberculosis in all M. tuberculosis-positive patient samples tested. Once more, this technique uses PCR to amplify the target mycobacterial genes (after off-chip DNA isolation from sputum) that are then captured by complementary nucleotide sequences attached to polymeric beads; the beads are then linked to magnetic nanoparticles (MNPs) through the opposite end of the amplicon.61 MNPs produce their own local magnetic field which can be detected by nuclear magnetic resonance spectroscopy (NMR) through its effect on the 1H NMR relaxation rate of the target DNA sequence. A disposable microfluidic device was developed which contained the on-chip components of this assay; PCR amplification, linkage of the amplicons to capture beads then MNPs, followed by NMR detection (through the incorporation of a microcoil NMR probe). Drug resistant strains could be detected from sputum samples within 2.5 hours, with estimated one-off costs for the DNA extraction device and reader containing the magnet, electronic circuit and thermocycler of $300 and $4000, respectively, and disposable costs of less than $3 for each assay.§
2.4 Proteins
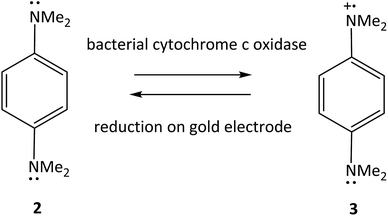 | ||
| Fig. 7 Oxidation of TMPD 2 by bacterial cytochrome c and reduction of the oxidized form 3 on a gold electrode.63 | ||
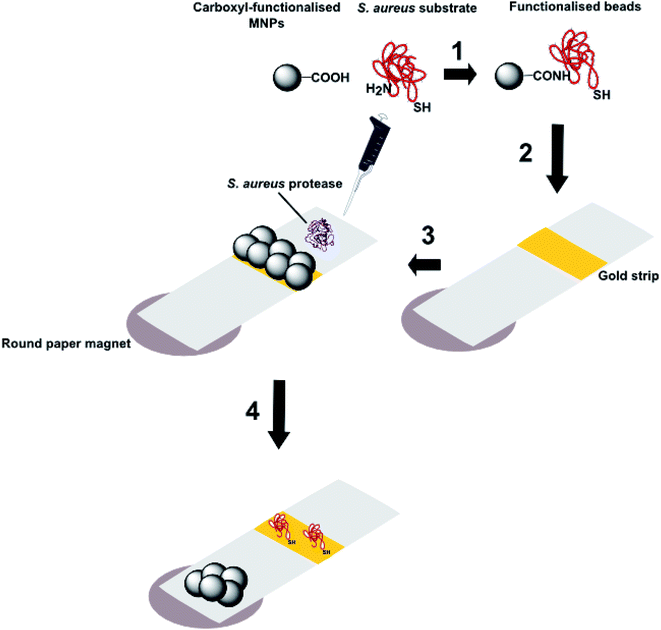 | ||
| Fig. 8 Schematic representation of the processes involved in a magnetic nanoparticle-peptide probe for the detection of Staphylococcus aureus (adapted from Suaifan et al.16): 1 Black coloured MNPs incorporating a terminal carboxyl group are conjugated to the N-terminus of a peptide substrate for S. aureus proteases and 2 are then immobilized on a gold sensor platform; 3 when the biosensor is exposed to S. aureus, enzymatic cleavage of the amide bond between the MNP and peptide substrate releases the nanoparticle; 4 the nanoparticles are attracted to external magnets located at the back of the sensor platform and thus expose the gold coloured surface. The colour change from black to gold occurs in 1 minute and can be detected with the naked eye. | ||
This system benefits from its simplicity in design and its lack of requirement for instrumentation, making it completely portable. The limit of detection can be as low as 100 CFU mL−1 for environmental samples of S. aureus, meaning that no sample pre-concentration or amplification steps are necessary. This method was also shown to be specific for S. aureus, as no colour change was observed when samples of P. aeruginosa, Escherichia coli, Listeria monocytogenesis or Candida albicans were tested. The test system was also stable upon storage for up to six months and does not require sample preparation or amplification prior to testing. S. aureus is a significant contributor to HAIs and this magnetic nanoparticle-peptide probe is a simple, inexpensive, rapid, equipment-free mode of detection that can be implemented in a wide range of settings. It is, however, limited in that there is no multiplexing capability, which is necessary for the timely diagnosis of severe HAIs, such as sepsis.18,65
3. Conclusions and future prospects
The discovery and development of antimicrobial agents have arguably been the most significant innovation in modern medicine. However, their overuse and misuse could result in society soon entering a ‘post-antibiotic’ era. Rapid POC techniques for the detection and identification of bacterial pathogens can complement antimicrobial prescribing to help reduce inappropriate use and extend the effectiveness of those agents which still retain efficacy. With the advances being made in the development of diagnostics that are suitable for POC tests, relying on empirical antimicrobial therapy is no longer justifiable; a rapid POC test for the diagnosis of bacterial infections should be routinely implemented prior to antimicrobial prescribing to enhance patient outcomes and limit AMR.The techniques discussed in this review have significant advantages over conventional diagnostics in their rapidity, low cost and ease of operation, as well as demonstrating specificity and sensitivity similar to more robust laboratory-based techniques. As evidenced by their low detection limits and time to detection and potential for portability, many of the techniques discussed in this review are suitable for use in a POC setting.
The role of emerging complimentary technologies to allow for the extended use of the discussed bacterial POC techniques in a variety of scenarios is undeniable. For example, the use of smartphones in POC bacterial diagnostics66 provides significant advantages due to their portability and their high-quality digital cameras, especially when incorporating complementary metal oxide-semiconductor (CMOS) sensors. The combination of these sensors, which enable detection and analysis through the capture of high-resolution images, and increasing processing power and memory storage, produces results which are readable and quantifiable.67 Smartphones can also be used in combination with external accessories, e.g. optics and light sources, or be integrated into complete analytical platforms, e.g. microfluidic lab-on-a-chip (LOC) platforms.68 Advances in the fabrication of materials also contributes to the accessibility of POC diagnostics; disposable microfluidic systems, e.g. screen-printed enzyme electrodes, are now available, increasing portability, affordability and opening up access to personalized devices. Other emerging technologies which enable microfluidic device fabrication, such as soft lithography69 and 3D printing,70 offer rapid and cost-effective production of high precision POC diagnostics.67
As evidenced by the examples given here, there have been significant technological developments, in terms of both materials- and device-design, as well as in the ever-improving performance and ease-of-use of POC technologies. However, the uptake and acceptance of POC tests as part of the clinical decision making process is burdened by a range of implementation issues, such as the lack of robust impact evaluations relating to patient outcomes and cost-effectiveness, and the development of effective guidelines or multi-level intervention strategies. In this respect, the European Joint Programming Initiative on Antimicrobial Resistance Translational Working Group ‘Antimicrobial Resistance – Rapid Diagnostic Tests’ (JPIAMR AMR-RDT) recommended that well-specified POCT guidelines and interventions should be targeted at individuals, communities, nations, and international networks.71 The JPIAMR also suggested that POCT is the ultimate tool in the combat against AMR, given that effective and efficient implementation strategies are in place as guidance and recommendation tailored to the largely differing needs and goals of healthcare providers, POCT innovators, and the general public. In the meantime, a sustainable future in terms of both AMR prevention and disease control should remain a primary focal point. As such responsibility will fall to biotechnology companies for the commercialization of these novel diagnostic tools, and to government and health strategists to ensure their implementation into clinical settings.
Conflicts of interest
None of the authors have any conflicts of interest to declare.References
- J. O'Neill, Tackling Drug-Resistant Infections Globally: Final Report and Recommendations, 2016, https://amr-review.org/, last accessed 16th April 2019 Search PubMed.
- O. O. Adeyi, E. Baris, O. B. Jonas, A. Irwin, F. C. J. Berthe, F. G. Le Gall, P. V. Marquez, I. A. Nikolic, C. A. Plante, M. Schneidman, D. E. Shriber and A. Thiebaud, Drug-resistant infections: a threat to our economic future, vol. 2, World Bank Group, Washington, D.C., 2017, http://documents.worldbank.org/curated/en/323311493396993758/final-report, last accessed 16th April 2019 Search PubMed.
- S. Santajit and N. Indrawattana, BioMed Res. Int., 2016, 2475067 Search PubMed.
- Centers for Disease Control and Prevention, https://www.cdc.gov/media/releases/2015/p0225-clostridium-difficile.html, last accessed 16th April 2019.
- F. Barbut, L. Surgers, C. Eckert, B. Visseaux, M. Cuingnet, C. Mesquita, N. Pradier, A. Thiriez, N. Ait-Ammar, A. Aifaoui, E. Grandsire and V. Lalande, Clin. Microbiol. Infect., 2014, 20, 136–144 CrossRef CAS.
- S. S. Magill, J. R. Edwards, W. Bamberg, Z. G. Beldavs, G. Dumyati, M. A. Kainer, R. Lynfield, M. Maloney, L. McAllister-Hollod, J. Nadle, S. M. Ray, D. L. Thompson, L. E. Wilson and S. K. Fridkin, N. Engl. J. Med., 2014, 370, 1198–1208 CrossRef CAS.
- World Health Organisation, Global action plan on antimicrobial resistance, https://www.who.int/antimicrobial-resistance/global-action-plan/en/, 2015, last accessed 16th April 2019 Search PubMed.
- H. W. Boucher, G. H. Talbot, J. S. Bradley, J. E. Edwards, D. Gilbert, L. B. Rice, M. Scheld, B. Spellberg and J. Bartlett, Clin. Infect. Dis., 2009, 48, 1–12 CrossRef.
- E. Tacconelli, E. Carrara, A. Savoldi, S. Harbarth, M. Mendelson, D. L. Monnet, C. Pulcini, G. Kahlmeter, J. Kluytmans, Y. Carmeli, M. Ouellette, K. Outterson, J. Patel, M. Cavaleri, E. M. Cox, C. R. Houchens, M. L. Grayson, P. Hansen, N. Singh, U. Theuretzbacher and N. Magrini, Lancet Infect. Dis., 2018, 18, 318–327 CrossRef.
- H. Inan, M. Poyraz, F. Inci, M. A. Lifson, M. Baday, B. T. Cunningham and U. Demirci, Chem. Soc. Rev., 2017, 46, 366–388 RSC.
- P. K. Drain, E. P. Hyle, F. Noubary, K. A. Freedberg, D. Wilson, W. R. Bishai, W. Rodriguez and I. V. Bassett, Lancet Infect. Dis., 2014, 14, 239–249 CrossRef.
- D. C. H. Burgess et al., Estimating the global health impact of improved diagnostic tools for the developing world, 2007, Rand Corporation, https://www.rand.org/pubs/research_briefs/RB9293.html, last accessed 16th April 2019 Search PubMed.
- G. Hansen, and E. J. Baron, Technical and Clinical Niches for Point-of-Care Molecular Devices, in: Advanced Techniques in Diagnostic Microbiology, ed. Y. W. Tang and C. Stratton, Springer, Cham, 2018, pp. 241–267 Search PubMed.
- L. Váradi, J. L. Luo, D. E. Hibbs, J. D. Perry, R. J. Anderson, S. Orenga and P. W. Groundwater, Chem. Soc. Rev., 2017, 46, 4818–4832 RSC.
- K. S. Park, Biosens. Bioelectron., 2018, 102, 179–188 CrossRef CAS PubMed.
- G. A. Suaifan, S. Alhogail and M. Zourob, Biosens. Bioelectron., 2017, 90, 230–237 CrossRef CAS.
- K. L. Buising, K. A. Thursky, M. B. Robertson, J. F. Black, A. C. Street, M. J. Richards and G. V. Brown, J. Antimicrob. Chemother., 2008, 62, 608–616 CrossRef CAS.
- J. Vila, M. D. Gómez, M. Salavert and J. Bosch, Enferm Infecc Microbiol Clin, 2017, 35, 41–46 CrossRef.
- L. D. Renner, J. Zan, L. I. Hu, M. Martinez, P. J. Resto, A. C. Siegel, C. Torres, S. B. Hall, T. R. Slezak, T. H. Nguyen and D. B. Weibel, Appl. Environ. Microbiol., 2017, 83, e02449-16 CrossRef.
- World Health Organization, Rapid Diagnostic Tests for Sexually Transmitted Infections, https://www.who.int/tdr/publications/journal-supplements/sti-way-forward/en/, last accessed 16th April 2019 Search PubMed.
- A. L. Furst and M. B. Francis, Chem. Rev., 2019, 119, 700–726 CrossRef CAS.
- M. A. Morales and J. M. Halpern, Bioconjugate Chem., 2018, 29, 3231–3239 CrossRef CAS PubMed.
- T. Y. Wei and C. M. Cheng, Cell Chem. Biol., 2016, 23, 1056–1066 CrossRef CAS.
- G. B. Sigal, A. Pinter, T. L. Lowary, M. Kawasaki, A. Li, A. Mathew, M. Tsionsky, R. B. Zheng, T. Plisova, K. Shen, K. Katsuragi, A. Choudhary, W. J. Honnen, P. Nahid, C. M. Denkinger and T. Broger, J. Clin. Microbiol., 2018, 56, e01338-18 CrossRef PubMed.
- K. M. Koczula and A. Gallotta, Essays Biochem., 2016, 60, 111–120 CrossRef.
- T. R. Kozel and A. R. Burnham-Marusich, J. Clin. Microbiol., 2017, 55, 2313–2320 CrossRef CAS.
- T. Tominaga, Food Sci. Biotechnol., 2017, 86, 566–570 CAS.
- C.-W. Yen, H. de Puig, J. Tam, J. Gómez-Márquez, I. Bosch, K. Hamad-Schifferli and L. Gehrke, Lab Chip, 2015, 15, 1638–1641 RSC.
- B. Amini, M. Kamali, M. Salouti and P. Yaghmaei, Biosens. Bioelectron., 2017, 92, 679–686 CrossRef CAS.
- H. Boutal, A. Vogel, S. Bernabeu, K. Devilliers, E. Creton, G. Cotellon, M. Plaisance, S. Oueslati, L. Dortet, A. Jousset, S. Simon, T. Naas and H. Volland, J. Antimicrob. Chemother., 2018, 73, 909–915 CrossRef CAS.
- J. Y. Ham, J. Jung, B.-G. Hwang, W.-J. Kim, Y.-S. Kim, E.-J. Kim, M.-Y. Cho, M.-S. Hwang, D. I. Won and J. S. Suh, Ann. Lab. Med., 2015, 35, 50–56 CrossRef PubMed.
- P. Chen, M. Gates-Hollingsworth, S. Pandit, A. Park, D. Montgomery, D. AuCoin, J. Gu and F. Zenhausern, Talanta, 2019, 191, 81–88 CrossRef CAS.
- A. Dhiman, P. Kalra, V. Bansal, J. G. Bruno and T. K. Sharma, Sens. Actuators, B, 2017, 246, 535–553 CrossRef CAS.
- N. Alizadeh, M. Y. Memar, S. R. Moaddab and H. S. Kafil, Biomed. Pharmacother., 2017, 93, 737–745 CrossRef CAS.
- M. Jarczewska, L. Gorski and E. Malinowska, Anal. Methods, 2016, 8, 3861–3877 RSC.
- M. Shahdordizadeh, S. M. Taghdisi, N. Ansari, F. Alebooye Langroodi, K. Abnous and M. Ramezani, Sens. Actuators, B, 2017, 241, 619–635 CrossRef CAS.
- V. Crivianu-Gaita and M. Thompson, Biosens. Bioelectron., 2016, 85, 32–45 CrossRef CAS.
- M. Rubab, H. M. Shahbaz, A. N. Olaimat and D.-H. Oh, Biosens. Bioelectron., 2018, 105, 49–57 CrossRef CAS.
- F. Jia, N. Duan, S. Wu, X. Ma, Y. Xia, Z. Wang and X. Wei, Microchim. Acta, 2014, 181, 967–974 CrossRef CAS.
- Y. Lian, F. He, H. Wang and F. Tong, Biosens. Bioelectron., 2015, 65, 314–319 CrossRef CAS.
- L. Li, Z. Liu, H. Zhang, W. Yue, C.-W. Li and C. Yi, Sens. Actuators, B, 2018, 254, 337–346 CrossRef CAS.
- N. Baby, A. C. Faust, T. Smith, L. A. Sheperd, L. Knoll and E. L. Goodman, Antimicrob. Agents Chemother., 2017, 61 Search PubMed.
- Y. Wang, L. Yu, X. Kong and L. Sun, Int. J. Nanomed., 2017, 12, 4789–4803 CrossRef CAS.
- S. Umesha and H. M. Manukumar, Crit. Rev. Food Sci. Nutr., 2018, 58, 84–104 CrossRef CAS.
- A. E. Zautner, U. Groß, M. F. Emele, R. M. Hagen and H. Frickmann, Front. Microbiol., 2017, 8, 1210 CrossRef PubMed.
- D. J. Korbie and J. S. Mattick, Nat. Protoc., 2008, 3, 1452–1456 CrossRef CAS.
- M. Y. Wang, J. L. Geng, Y. J. Chen, Y. Song, M. Sun, H. Z. Liu and C. J. Hu, Lett. Appl. Microbiol., 2017, 64, 138–143 CrossRef CAS.
- J. Fishovitz, J. A. Hermoso, M. Chang and S. Mobashery, IUBMB Life, 2014, 66, 572–577 CrossRef CAS.
- K. Bush and G. A. Jacoby, Antimicrob. Agents Chemother., 2010, 54, 969–976 CrossRef CAS.
- E. L. Fonseca, N. D. Ramos, B. G. Andrade, L. L. Morais, M. F. Marin and A. C. Vicente, Diagn. Microbiol. Infect. Dis., 2017, 87, 315–317 CrossRef CAS.
- B. Y. C. Ng, E. J. H. Wee, N. P. West and M. Trau, ACS Sens., 2016, 1, 173–178 CrossRef CAS.
- C. D. Chin, T. Laksanasopin, Y. K. Cheung, D. Steinmiller, V. Linder, H. Parsa, J. Wang, H. Moore, R. Rouse, G. Umviligihozo, E. Karita, L. Mwambarangwe, S. L. Braunstein, J. van de Wijgert, R. Sahabo, J. E. Justman, W. El-Sadr and S. K. Sia, Nat. Med., 2011, 17, 1015–1020 CrossRef CAS.
- T. N. T. Dao, E. Y. Lee, B. Koo, C. E. Jin, T. Y. Lee and Y. Shin, Anal. Biochem., 2018, 544, 87–92 CrossRef CAS.
- O. Piepenburg, C. H. Williams, D. L. Stemple and N. A. Armes, PLoS Biol., 2006, 4, e204 CrossRef.
- S. K. Garg, K. Lu, J. Duncan, L. R. Peterson and O. Liesenfeld, Eur. J. Microbiol. Immunol., 2017, 7, 310–318 CrossRef CAS.
- R. Moure, Á. Cañizares, M. Muíño, M. Lobato, A. Fernández, M. Rodríguez, M. J. Gude, M. Tomás and G. Bou, J. Microbiol. Methods, 2016, 120, 50–52 CrossRef CAS PubMed.
- K. Cihalova, D. Hegerova, A. M. Jimenez, V. Milosavljevic, J. Kudr, S. Skalickova, D. Hynek, P. Kopel, M. Vaculovicova and V. Adam, J. Pharm. Biomed. Anal., 2017, 134, 325–332 CrossRef CAS.
- K. Niemirowicz, K. H. Markiewicz, A. Z. Wilczewska and H. Car, Adv. Med. Sci., 2012, 57, 196–207 CrossRef CAS.
- S. M. Yoo and S. Y. Lee, Trends Biotechnol., 2016, 34, 7–25 CrossRef CAS.
- Z. Farka, T. Juriik, D. Kovaar, L. Trnkova and P. Sklaadal, Chem. Rev., 2017, 117, 9973–10042 CrossRef CAS.
- M. Liong, A. N. Hoang, J. Chung, N. Gural, C. B. Ford, C. Min, R. R. Shah, R. Ahmad, M. Fernandez-Suarez, S. M. Fortune, M. Toner, H. Lee and R. Weissleder, Nat. Commun., 2013, 4, 1752 CrossRef.
- S. Kuss, H. M. A. Amin and R. G. Compton, Chem.–Asian J., 2018, 13, 2758–2769 CrossRef CAS.
- S. Kuss, R. A. S. Couto, R. M. Evans, H. Lavender, C. C. Tang and R. G. Compton, Anal. Chem., 2019, 91, 4317–4322 CrossRef CAS.
- P. Damborský, J. Švitel and J. Katrlík, Essays Biochem., 2016, 60, 91–100 CrossRef.
- C. Dincer, R. Bruch, A. Kling, P. S. Dittrich and G. A. Urban, Trends Biotechnol., 2017, 35, 728–742 CrossRef CAS.
- S. C. B. Gopinath, T.-H. Tang, Y. Chen, M. Citartan and T. Lakshmipriya, Biosens. Bioelectron., 2014, 60, 332–342 CrossRef CAS.
- I. Hernandez-Neuta, F. Neumann, J. Brightmeyer, T. Ba Tis, N. Madaboosi, Q. Wei, A. Ozcan and M. Nilsson, J. Intern. Med., 2019, 285, 19–39 CrossRef CAS.
- X. Huang, D. Xu, J. Chen, J. Liu, Y. Li, J. Song, X. Ma and J. Guo, Analyst, 2018, 143, 5339–5351 RSC.
- M. Dou, D. C. Dominguez, X. Li, J. Sanchez and G. Scott, Anal. Chem., 2014, 86, 7978–7986 CrossRef CAS.
- E. Jue, N. G. Schoepp, D. Witters and R. F. Ismagilov, Lab Chip, 2016, 16, 1852–1860 RSC.
- J. P. Hays, K. Mitsakakis, S. Luz, A. van Belkum, K. Becker, A. van den Bruel, S. Harbarth, J. H. Rex, G. S. Simonsen, G. Werner, V. Di Gregori, G. Ludke, T. van Staa, J. Moran-Gilad and T. T. Bachmann, Eur. J. Clin. Microbiol. Infect. Dis., 2019, 38, 1015–1022 CrossRef.
Footnotes |
| † The cycle-threshold (Ct) value is the number of PCR cycles required for the fluorescence signal to cross the threshold (background level), with lower values suggesting high pathogenic bacterial loads. |
| ‡ Tm is melting temperature of the primer–template pair, the temperature at which half the molecules are single- and half are double-stranded. |
| § It is anticipated that these costs could be scaled down, from $4000 to $200 and <$3 to <$1. |
| This journal is © The Royal Society of Chemistry 2019 |