Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Roll-to-roll fabrication of integrated PDMS–paper microfluidics for nucleic acid amplification†
Jussi
Hiltunen
 *a,
Christina
Liedert
a,
Marianne
Hiltunen
a,
Olli-Heikki
Huttunen
a,
Johanna
Hiitola-Keinänen
a,
Sanna
Aikio
a,
Mikko
Harjanne
a,
Marika
Kurkinen
a,
Leena
Hakalahti
a and
Luke P.
Lee
*bcde
*a,
Christina
Liedert
a,
Marianne
Hiltunen
a,
Olli-Heikki
Huttunen
a,
Johanna
Hiitola-Keinänen
a,
Sanna
Aikio
a,
Mikko
Harjanne
a,
Marika
Kurkinen
a,
Leena
Hakalahti
a and
Luke P.
Lee
*bcde
aVTT-Technical Research Centre of Finland, Kaitoväylä 1, FIN-90590 Oulu, Finland. E-mail: jussi.hiltunen@vtt.fi
bDepartment of Bioengineering, University of California Berkeley, CA 94720, USA. E-mail: lplee@berkeley.edu
cBerkeley Sensor and Actuator Center, University of California Berkeley, CA 94720, USA
dDepartment of Electrical Engineering and Computer Science, University of California Berkeley, CA 94720, USA
eBiophysics Graduate Program, University of California Berkeley, CA 94720, USA
First published on 30th April 2018
Abstract
Microfluidic-based integrated molecular diagnostic systems, which are automated, sensitive, specific, user-friendly, robust, rapid, easy-to-use, and portable, can revolutionize future medicine. Current research and development largely relies on polydimethylsiloxane (PDMS) to fabricate microfluidic devices. Since the transition from the proof-of-principle phase to clinical studies requires a vast number of integrated microfluidic devices, there is a need for a high-volume manufacturing method of silicone-based microfluidics. Here we present the first roll-to-roll (R2R) thermal imprinting method to fabricate integrated PDMS–paper microfluidics for molecular diagnostics, which allows production of tens of thousands of replicates in an hour. In order to validate the replicated molecular diagnostic platforms, on-chip amplification of viral ribonucleic acid (RNA) with loop-mediated isothermal amplification (LAMP) was demonstrated. These low-cost, rapid and accurate molecular diagnostic platforms will generate a wide range of applications in preventive personalized medicine, global healthcare, agriculture, food, environment, water monitoring, and global biosecurity.
Introduction
Microfluidics is a multidisciplinary field which includes chemistry, physics, life sciences, and engineering to create platforms for precision control and manipulation of fluids at a submillimeter scale, enabling rapid and parallel analysis with a small sample volume. The main driver for the huge research and development activities in academia and industry has been its foreseen potential to produce scientific breakthroughs and practical devices in life sciences, medical, pharmaceutical, and chemical industries, environmental monitoring, and biosecurity applications.1–9 Over the years, extensive varieties of theoretical investigations and experimental studies have been realized to prove the various advantages of microfluidic devices over conventional analytical procedures. The core of microfluidics is its intrinsic ability to manipulate very small volumes of fluids in a wide variety of integrated ways including fast sample processing, accurate control of fluids and delivery of results with a fast turnaround time. In particular, the huge potential of microfluidics technology is expected to advance automated point-of-care (POC) systems since low cost per analysis, portability and/or wear ability is an important requirement.2Originally, microfluidic devices were mostly fabricated using photolithographic methods as microelectromechanical systems (MEMS) in silicon or glass.1 To avoid the risk of sample cross-contamination, most applications require that chips be disposed of after usage, setting strict demands for fabrication costs. To complement conventional MEMS processing in the fabrication of disposable microfluidic devices, other fabrication approaches have been developed including printing, embossing and molding polymers.3 The vast usage of polymeric materials in commercial applications is largely due to the fact that versatile, low-cost, high-volume fabrication methods are available. Therefore, the possibility of replacing inorganic materials and associated fabrication methods with polymeric material-compatible production technologies in microfluidics has attracted research interest.3 Most of the micro- and nanoscale polymer structuring methods are based on the replication of a master structure into the polymeric replica. In particular, during the last 10 years, these replication methods have contributed to the large progress in micro- and nanofabrication fields.2,10,11
The elastomer poly(dimethylsiloxane) (PDMS) has become one of the most widely used materials in the development of microfluidic devices in academic environments.1,3,8,12 The popularity of PDMS materials originates from a number of factors, most importantly, the relatively cheap and easy prototyping of small numbers of devices with soft lithography using master molds for structure replication.13 PDMS surface properties can be chemically tuned so that the microfluidic devices can easily bond with glass and PDMS itself.6 These factors have enabled rapid prototyping of devices at a lower cost than what is feasible using silicon technology.1 Despite of all the benefits, a number of limitations are affiliated with PDMS. It has been found that PDMS can release uncrosslinked oligomers from the curing process, and, on the other hand, it has been shown to absorb small molecules affecting the functionality such as biological assays.14 In addition, PDMS is highly permeable to gases and thus evaporation at micro- and nanolitre aqueous volumes can occur rapidly at increased temperature, e.g. during polymerase chain reaction for nucleic acid amplification. To overcome these material-related limitations, methods such as parylene coating of the microchannel surface can be implemented. Still, probably the biggest factor in limiting the usage of PDMS outside laboratory settings relates to the lack of high-volume fabrication methods to produce PDMS-based microfluidics.6 The common industrial scale polymer processes, such as injection molding, favor polymers other than PDMS.8 Additionally, academic researchers could benefit greatly from the high-volume chip fabrication if the number of samples does not limiting the assessment of the designs and associated test protocols. Clinical tests typically require a vast number of samples to obtain adequate statistical relevance. In this article, a large-volume fabrication method for PDMS-based microfluidics is presented to aid the transition of prototypes into commercial devices.
In 2008, Ahn and Guo showed that PDMS is roll-to-roll (R2R)-processable by fabricating sub-μm test structures into PDMS on a flexible polyethylene terephthalate (PET) substrate.15 The objective of our work was to show that functional PDMS-based microfluidics for biomedical usage can be processed by the R2R method and produced in high numbers. Furthermore, an important part of our work was to investigate the usage of paper as a carrier/substrate for R2R-produced PDMS microfluidics. The use of paper as a carrier for microfluidics has been studied with the aim of producing robust and low-cost analytical systems. In the case of typical down-conversion fluorescent detection, strict requirements are set for the paper quality due to the strong autofluorescence of paper additives significantly limiting the number of suitable paper materials.9 Therefore, other detection methods, such as colorimetry, have been used in paper-based microfluidics.19–21 To achieve the benefits of fluorescence detection, with the most notably low limit of detection, fluorescent upconversion sensing has been developed for paper-based microfluidics.9 For medical applications, paper is an attractive material as it is inexpensive and provides an integrated flammable material for chip disposal. The protocol for the disposal of analytical accessories is generally strictly regulated when dealing with potentially harmful substances, such as blood samples. Thus, sample disposal should be considered as an integral part of the envisioned test protocol.
For analytes present at relatively low concentrations in biological samples, either target or signal amplification is usually required.8 For nucleic acid detection, powerful target amplification methods have been developed and adopted for routine usage. Polymerase chain reaction (PCR) is the most notable among them.16 In PCR, temperature cycling is utilized to multiply specific sequences of nucleic acids allowing the detection of even a single target. Isothermal amplification chemistries, on the other hand, are well suited for POC devices since the nucleic acid detection times are short, and the amplification occurs at constant temperature, needing only simple instrumentation with reduced power requirements. For example, loop-mediated amplification (LAMP) is an efficient and highly selective reaction able to identify a few copies of DNA by employing four to six primers, which recognize six distinct sequences of the target DNA. The amplification occurs at temperature 68–72 °C and the amplification threshold is reached in 8–20 minutes.17 In this work, we demonstrate rapid LAMP of viral RNA using R2R-processed PDMS microfluidics on paper.
Methods
Mold preparation
The mold for roll-to-roll imprinting was patterned with photolithography on a single-side polished 150 μm thick nickel plate (ESI† Fig. S1). The first planarization layer of EpoCore_50 (Micro resist technology) was spin-coated (40 s, 2000 rpm) on a 10 cm × 10 cm size nickel plate and UV-cured with 210 mJ cm−2 (KarlSuss MA6) exposure dose for improving the adhesion of the lithography mold on the surface. The edges of the nickel plates were left unexposed in order to weld the plates on the nickel shim. The planarization layer was developed with Mr-Dev600 (Micro resist technology) for 40 seconds, followed by isopropanol flushing. After the planarization layer was hard baked for one hour at 120 °C, the thick EpoCore_50 layer was spin-coated (step 1: 5 s/500 rpm/200 ramp, step 2: 60 s/800 rpm/100 ramp), followed by soft-mode photolithography patterning through a mask with i-line filtered light. The used exposure dose was 700 mJ cm−2. The patterned EpoCore layer was post-baked (5 min@50 °C, 10 min@85 °C) and left for one day to relax before development with Mr-Dev600 and isopropanol flushing. The total development time was ten minutes. The molds were hard-baked for 1.5 hour at 120 °C. Three identically processed epoxy-based molds on Ni-plates were welded on the 150 μm thick nickel plate. The plate was cut into precise length according to an imprinting cylinder diameter and rolled to form a sleeve for the imprinting cylinder. The cylinder was then dip-coated in a Teflon AF solution (diluted in a weight ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 with an FC-40 solvent) and cured for 60 min at 200 °C.
3 with an FC-40 solvent) and cured for 60 min at 200 °C.
Roll-to-roll replication
R2R processing is illustrated in Fig. 1. First, a 100 mm wide aluminized paper substrate (Zanders Chromolux Alu Magic Chrome, 80 g m−2) was treated with atmospheric corona (Vetaphone Corona-Plus Type TF-415, 1 kW) and gravure coated in line with an adhesion promoter (Wacker Primer G790, Anilox, 80 lines per cm, 10 ml m−2, speed of 6 m min−1). During PDMS replication, the Al-coated paper substrate was again treated with corona (1 kW) and the premixed PDMS (Wacker Elastosil RT604) was deposited on the running substrate by spiral bar application with a thickness of 270 μm (RK K-control coater bar for a nominal wet thickness of 300 ml m−2, speed of 1.5 m min−1) and roll-to-roll imprinted in line. The imprinting cylinder was heated to a temperature of 130 °C to cure the PDMS.Replication accuracy
Scanning electron microscopy images (Jeol Neoscope JCM-5000) were taken from the mold and PDMS replica with 10 kV accelerating voltage in a high-vacuum mode. Prior to the imaging, the samples were coated with a thin layer of gold by a sputtering method. Dimensional analysis of the mold and reaction chambers was performed with a white light interferometer (Wyko NT3300) using vertical scanning interferometry (VSI) mode, 2.5× objective, 1× field of view and 3× speed. The modulation threshold in mold and chamber analysis was 0.5% and 2%, respectively.Fluorescence set-up
The schematic of the optical fluorescence set-up is shown in ESI† Fig. S2 and the calculus in ESI† Fig. S3. A mounted light emitting diode (LED) (Thorlabs M470L3) was used as a light source, followed by a condenser lens with a diffuser (Thorlabs ACL2520U-DG6-A) and an excitation filter (Chroma Technology AT480/30x). The chip was illuminated using a slanted light beam. Fluorescent light emitted by the sample was filtered using an emission filter (Chroma Technology AT535/40m) positioned in front of the camera objective (Schneider-Kreuznach Xenon 0.95/17). Images were captured using a camera (IDS Imaging Development System UI-3240CP-NIR-GL) connected to a computer. An optical power meter (Ophir Nova II) was used to monitor the LED power when studying the suppression of paper autofluorescence and the enhancement of the fluorescence signal by a highly reflective coating.LAMP and primer dispense
LAMP assays were performed using a commercial kit from Lucigen Corporation. The control reaction mixture was prepared according to Lucigen's OmniAmp RNA & DNA LAMP kit manual containing (final concentration): 1× DNA polymerase buffer, 800 μM dNTPs, 8 mM MgSO4, 0.15 M betaine, 0.4× EvaGreen fluorescent dye (Biotium Inc), 1× LAMP RNA control I primer mix,22 1× OmniAmp DNA polymerase, and 8% (V/V) MS2 phage RNA, and brought to a volume of 25 μl with DNase–RNase-free water. Reagents with or without RNA and primers were premixed prior to application to the microfluidic chip. A roll-to-roll compatible dispenser (Ginolis Delilah) was used to dispense 20 nl of primers per chamber (ESI† Fig. S4). Dispensed primer solution dried instantly at room temperature. In the amplification experiments, the samples were positioned on a Peltier element with a mounted thermocouple. The temperature was controlled with a temperature controller (Thorlabs TED200C).By using a separate two-channel thermometer (Yokogawa 2455) with one thermoelement positioned directly on the Peltier element and the other thermoelement on the PDMS-coated paper, it was concluded that the reaction chambers reached the temperature of about 70 °C when the Peltier element was heated to a temperature of 73 °C.
Results and discussion
Roll-to-roll imprinting of integrated PDMS microfluidics on Al-coated paper
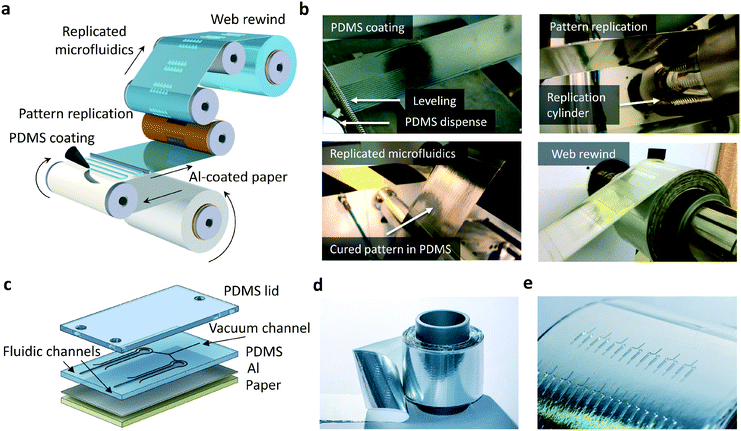
PDMS-based microfluidics were fabricated by R2R thermal imprinting (Fig. 1a and b). In principle, any solid support, for example a transparent polyester that withstands the R2R processing conditions required for thermal curing of PDMS, can be exploited as a carrier material. In this work, aluminum (Al)-coated paper was used as a solid support in R2R processing. Prior to the fabrication of PDMS structures, the paper carrier was treated with an R2R atmospheric corona unit and R2R gravure coated with an adhesion promoter. Uncured PDMS was dispensed on the running web and the meandering dispense pattern was leveled with a spiral bar to form a continuous layer of uncured PDMS.The paper web was subsequently brought into contact with the rotating cylinder. In a simultaneous step, PDMS was cured and lithographically patterned features on the cylinder (ESI† Fig. S1) were replicated into the PDMS layer during the contact with the heated cylinder. The contact time of a 270 μm thick PDMS layer with the heated cylinder was 8 s. PDMS-based devices on metal-coated paper (Fig. 1c) contain fluidic channels for sample transport and amplification, as well as parallel vacuum channels enabling bubble-free filling of fluidic channels and dead-end chambers discussed below. A roll of PDMS-based microfluidics on Al-coated paper and a line of microfluidic devices are presented in Fig. 1d and e, respectively.
R2R replication accuracy and device operation
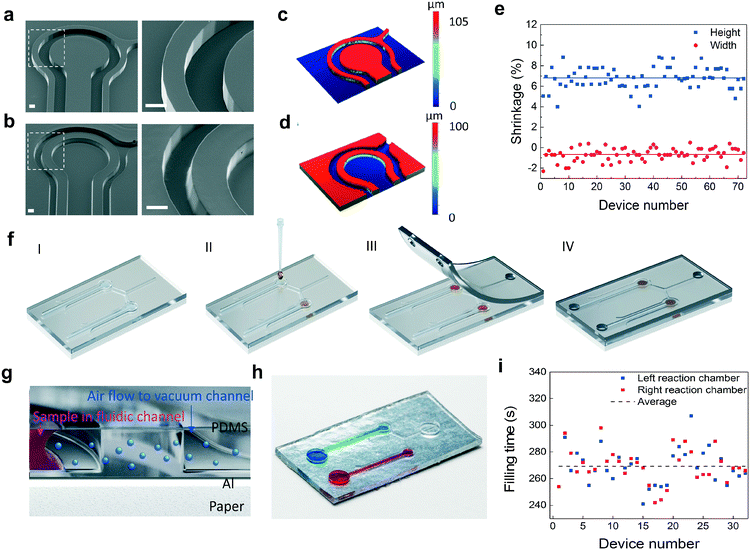
Scanning electron microscopy (SEM) images from the imprinting tool and PDMS replica (Fig. 2a and b) reveal that the structures in the mold were transferred with high accuracy. The results also show that steep sidewalls with only slight beveling can be obtained by using the R2R method. The reaction chamber is separated from the vacuum channels by a wall having a thickness of 200 μm. Dimensional variation between the mold and PDMS replica was investigated by measuring the vertical and lateral dimensions of the surface patterns with white light interferometry (Fig. 2c and d). The curing of PDMS results in a dimensional shrinkage of about 6.5% in the vertical direction. In the lateral direction, the dimensions were slightly increased with an average value below 1% (Fig. 2e). The average diameter of the reaction chambers, the width of the fluidic channels and the width of the vacuum channels are 1 mm, 408 μm and 109 μm, respectively. The average depth of the structures is 97 μm. Following PDMS replication, the primers for LAMP were dispensed into the reaction chambers and coverlids were assembled (Fig. 2f, ESI† Fig. S5). The coverlids were R2R-processed onto a PET carrier utilizing an imprinting cylinder without surface structures resulting in an unstructured PDMS layer that was easily removable from the carrier (ESI† Fig. S5). Vias for fluidic and vacuum connections were manually punched and lids were then bonded onto the fluidic layer with a plasma treatment method. The operation principle of this configuration is explained in detail in ref. 18. In brief, PDMS is a highly air permeable material. When there is a pressure difference between two adjacent channels that are not directly connected, air flow is induced from the higher pressure channel towards the lower pressure channel through gas permeable PDMS (Fig. 2g, ESI† Fig. S6). When the vacuum channels of the chip are connected to a vacuum pump, the fluid injected into the inlet of the fluidic channels fills the reaction chambers without trapped air bubbles. Fig. 2h illustrates a single device containing two microfluidic channel-connected reaction chambers that are filled with blue and red colored water. An average filling time of 270 s with an RMS deviation of 13 s was determined for the produced devices (Fig. 2i).Enhancement of the fluorescence signal and suppression of background fluorescence emission
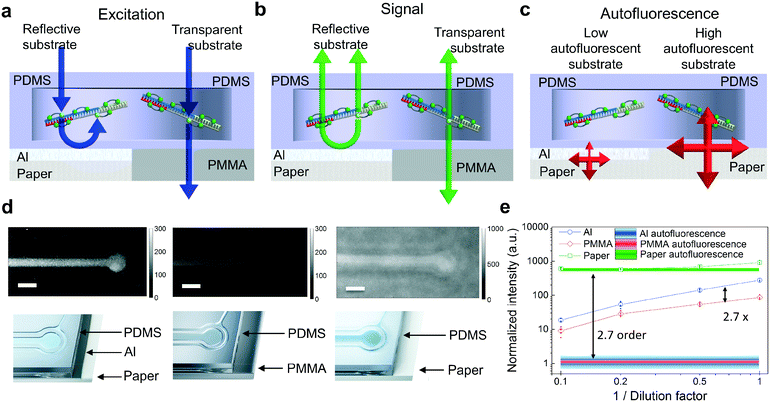
The packaging industry has developed ultra-high throughput thin film evaporation processes of metals, typically aluminum, on paper to improve the barrier properties of paper against the penetration of vapor, water and aromas. Since the Al coating isolates paper optically from the PDMS layer, commercially available Al-coated paper (Chromolux Alu Magic) was utilized as a carrier in this work. It is also expected that other paper materials with different metal coatings can be used, providing that strongly autofluorescent paper is optically isolated. The advantages of using produced PDMS devices on metal are illustrated in Fig. 3a–c. First, the Al coating reflects the excitation light that has not been absorbed by the fluorophores. This results in the enhancement of the excitation light intensity in the reaction chambers (Fig. 3a). The enhancement effect is much weaker if the PDMS devices are fabricated on highly transparent substrates. Secondly, the generated fluorescence signal inside the fluidic reaction chamber is directed upwards due to the metal coating with high reflectivity (Fig. 3b). Thirdly, the thin metal layer suppresses the paper substrate-originated autofluorescence by partially reflecting and absorbing the excitation light. This results in the reduced intensity of the excitation field reaching the paper. In addition, the autofluorescence generated by the paper must also penetrate through the reflecting and absorbing Al coating to reach the detector (Fig. 3c).The fluorescence properties of the devices were investigated experimentally by fabricating the reference PDMS-based devices on poly(methyl methacrylate) (PMMA) polymer and paper substrates and comparing their operation against the R2R-processed devices on Al-coated paper. PMMA was chosen as a substrate material with low reflectivity and low autofluorescence. In the experiments, the reaction chambers were filled with LAMP reagents and a green fluorophore (EvaGreen). The mixture is fluorescent at ambient temperature. The mixture was sequentially diluted by 10-fold with water to evaluate the substrate effects on the fluorescence properties. Fig. 3d illustrates how the fluorescence signal is stronger in reaction chambers on Al-coated paper compared to the one on PMMA. Also, though white paper with high reflectivity enhances the fluorescence signal of interest, it is strongly concealed by the paper autofluorescence. Shaded lines (Fig. 3e) represent the background fluorescence levels for Al-coated paper, PMMA and paper obtained by measuring the intensity at the blank area of the samples. By comparing the autofluorescence values between the Al-coated and non-coated sides of the paper, it was found that the Al-coating suppresses the autofluorescence by an order of about 2.7. When comparing the fluorescence signal values between devices on the highly reflective Al film and transparent PMMA, the metal layer exhibited enhancement of the excitation field and upwards directed fluorescent emission resulting in an overall signal enhancement factor of about 2.7.
Imprinted LAMP chip for viral RNA detection
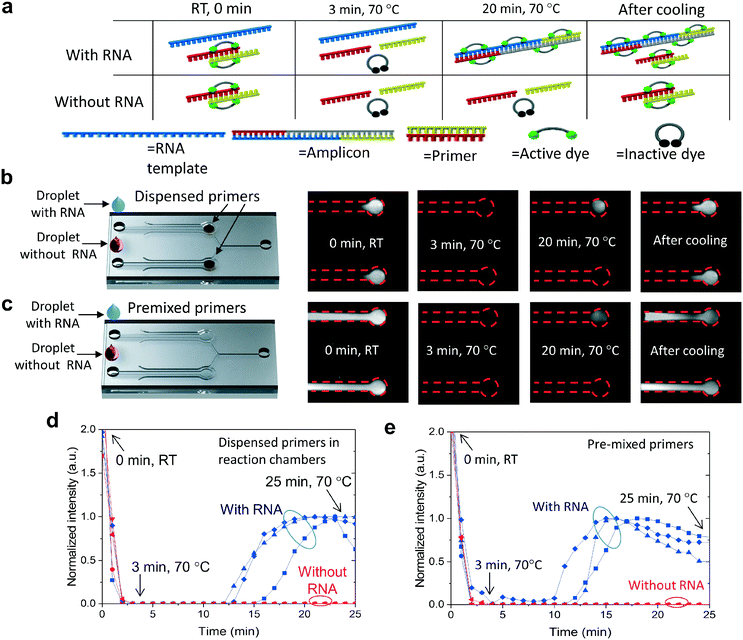
The functionality of the R2R-processed fluidics was demonstrated by performing LAMP of viral RNA. The reaction volume is fluorescent prior to the amplification at room temperature (RT) and the fluorescence can be observed from both the positive and negative channels (Fig. 4a). We assume that the binding of EvaGreen dye to the primers causes the background fluorescence, as there is no autofluorescence when the primers are omitted. When the temperature is increased from RT to 70 °C, the fluorescence intensity diminishes rapidly due to the assumed dissociation of the primers and dyes. The fluorescence level remained very low until LAMP produced double-stranded DNA, turning the intercalating dye fluorescently active due to binding with DNA. Without RNA templates, no fluorescence signal is generated at 70 °C. After cooling, fluorescence becomes clearly observable in both the negative control and positive reactions due to label binding with the primers and the amplified DNA. Two protocols were used in the experiments, where LAMP primers were either dispensed into the microfluidic chambers (Fig. 4b) or, in the control experiment reactions, they were premixed with the other reagents (Fig. 4c). Comparative experiments were performed to see how dried primers affect the amplification. Amplification of the negative control (no RNA) and positive control (RNA) was monitored in real-time by taking fluorescence images with an interval of 1 min (Fig. 4b with dispensed primers and Fig. 4c with premixed primers). In both protocols, vacuum channels were first connected to a vacuum pump and the sample was pipetted into the inlets of the fluidic channels (ESI† Fig. S6). One vacuum channel was utilized to fill both reaction chambers, one for the negative and one for the positive sample. LAMP was triggered by increasing the chip temperature from room temperature (RT) to 70 °C within a time period of about 3 min. The onset of amplification with dispensed primers (Fig. 4d) occurred at a time period of about 13–16 min which was a few minutes delayed compared to amplification with pre-mixed primers (Fig. 4e). When comparing the amplification between dispensed (Fig. 4b) and premixed primers (Fig. 4c), a significant difference can be observed between the fluorescent areas at room temperature before and after the amplification. When the primers were premixed with the sample, fluorescence was observed across the entire fluidic channel, whereas, in the dispensed channels, the fluorescence was localized. The strong fluorescence after cooling indicated that the fluidics were dried during the amplification and the dispensed primers did not diffuse considerably during the assay.Conclusions
This is the first report describing a roll-to-roll replication method for PDMS microfluidic fabrication in large volumes. The novel method bridges the gap between academic research and industrial usage when the same material and device designs can be used in upscale fabrication processes.The functionality of the R2R-produced devices was demonstrated by performing isothermal amplification of viral RNA on chip. Paper was used as a carrier material in the R2R processing of PDMS microfluidics. The strong autofluorescence of paper was found to be effectively suppressed by applying a thin metal layer isolating optically the paper carrier and fluidics. The highly reflective metal layer was found to enhance the fluorescence signal generated during amplification. Therefore, other (bio)analyses that use fluorescence detection would also benefit from the PDMS–aluminium-coated paper microfluidics. The accurately replicated microfluidic structures with dead-end chambers inhibit contamination and enable volume control. The amplification occurred in less than 20 minutes, making the devices attractive for POC applications. By dispensing different sets of primers on chip, it would be possible to customize each chamber for different targets, thus enabling multiplexed molecular diagnostic platforms, which can create a wide range of applications in preventive personalized medicine, global healthcare, agriculture, food, environment, water monitoring, and global biosecurity.
Author contributions
L. P. L conceived the study and together with J. H. led the work. O.-H. H., J. H.-K., L. H. and M. K. developed the R2R PDMS replication process. C. L. and Ma. H. designed the microfluidic devices and performed the chip post-processing, RNA amplification and channel filling experiments. M. H. analysed the chamber replication quality. J. H. and S. A. designed and implemented the fluorescence set-up. J. H., Ma. H., Mi. H., and S. A. C. L. interpreted the data and prepared the data presentation. All the authors contributed to the manuscript text preparation.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge financial support by a Fulbright Scholarship for Luke P. Lee's proposal on “Intelligent paper opto-electro-microfluidic systems (iPOEMs) for preventive personalized medicine and global healthcare,” and the Academy of Finland grant No. 284907. Jaakko Pennanen, Ulla Sarajärvi, Pekka Ontero and Jenni Tomperi at VTT are acknowledged for their assistance in R2R fabrication. Janne Aikio and Antti Veijola at VTT-Technical Research Centre of Finland are acknowledged for filming and taking the photographs of the PDMS samples. We thank Topi Hintsala, Ginolis Ltd, for his assistance with primer dispensing.Notes and references
- G. M. Whitesides, Nature, 2006, 442, 368–373 CrossRef CAS PubMed.
- H. Becker and C. Gärtner, Anal. Bioanal. Chem., 2008, 390, 89–111 CrossRef CAS PubMed.
- L. Gervais, N. de Rooij and E. Delamarche, Adv. Mater., 2011, 23, H151–H176 CrossRef CAS PubMed.
- Y. Temiz, R. D. Lovchik, G. V. Kaigala and E. Delamarche, Microelectron. Eng., 2015, 132, 156–175 CrossRef CAS.
- T. M. Squires and S. R. Quake, Rev. Mod. Phys., 2005, 77, 977–1025 CrossRef CAS.
- E. K. Sackmann, A. L. Fulton and D. J. Beebe, Nature, 2014, 507, 181–189 CrossRef CAS PubMed.
- D. Mark, S. Haeberle, G. Roth, F. von Stettenzab and R. Zengerlez, Chem. Soc. Rev., 2010, 39, 1153–1182 RSC.
- C. D. Chin, V. Linder and S. K. Sia, Lab Chip, 2012, 12, 2118–2134 RSC.
- M. He and Z. Liu, Anal. Chem., 2013, 85, 11691–11694 CrossRef CAS PubMed.
- N. Kooy, K. Mohamed, L. T. Pin and O. S. Guan, Nanoscale Res. Lett., 2014, 9, 320 CrossRef PubMed.
- L. Peng, Y. Deng, P. Yi and X. Lai, J. Micromech. Microeng., 2014, 24, 013001 CrossRef.
- X. Hou, Y. S. Zhang, G. Trujilo-de Santiago, M. M. Alvarez, J. Ribas, S. J. Jonas, P. S. Weiss, A. M. Andrews, J. Aizenberg and A. Khademhosseini, Nat. Rev. Mater., 2017, 2, 17016 CrossRef CAS.
- Y. Xia and G. M. Whitesides, Annu. Rev. Mater. Sci., 1998, 28, 153–184 CrossRef CAS.
- K. J. Regehr, M. Domenech, J. T. Koepsel, K. C. Carver, S. J. Ellison-Zelski, W. L. Murphy, L. A. Schuler, E. T. Alaridc and D. J. Beebea, Lab Chip, 2009, 9, 2132–2139 RSC.
- S. H. Ahn and L. J. Guo, Adv. Mater., 2008, 20, 2044–2049 CrossRef CAS.
- Y. Zhang and P. Ozdemir, Anal. Chim. Acta, 2009, 13, 115–125 CrossRef PubMed.
- T. Notomi, H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino and T. Hase, Nucleic Acids Res., 2000, 28, e63 CrossRef CAS PubMed.
- L. Xu, H. Lee, D. Jetta and K. W. Oh, Lab Chip, 2015, 15, 3962–3979 RSC.
- X. Li, D. R. Ballerini and W. A. Shen, Biomicrofluidics, 2012, 6, 11301 CrossRef PubMed.
- A. K. Yetisen, M. S. Akram and C. R. Lowe, Lab Chip, 2013, 13, 2210–2251 RSC.
- C. Parolo and A. Merkoci, Chem. Soc. Rev., 2013, 42, 450–457 RSC.
- Y. Chander, J. Koelbl, J. Puckett, M. J. Moser, A. J. Klingele, M. R. Liles, A. Carrias, D. A. Mead and T. W. Schoenfeld, Front. Microbiol., 2014, 5, 395 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8lc00269j |
| This journal is © The Royal Society of Chemistry 2018 |