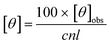Open Access Article
Open Access ArticleA non-zipper-like tetrameric coiled coil promotes membrane fusion†
Tingting
Zheng‡
a,
Monica
Bulacu‡
b,
Geert
Daudey
a,
Frank
Versluis
a,
Jens
Voskuhl
a,
Giuliana
Martelli
a,
Jan
Raap
a,
G. J. Agur
Sevink
c,
Alexander
Kros
 *a and
Aimee L.
Boyle
*a
*a and
Aimee L.
Boyle
*a
aSupramolecular and Biomaterials Chemistry, Leiden Institute of Chemistry, Leiden University, P.O. Box 9502, 2300RA, Leiden, The Netherlands. E-mail: a.l.boyle@chem.leidenuniv.nl; a.kros@chem.leidenuniv.nl
bCulgi BV, Galileiweg 8, 2333BD, Leiden, The Netherlands
cSolid State NMR, Leiden Institute of Chemistry, Leiden University, P.O. Box 9502, 2300RA, Leiden, The Netherlands
First published on 7th January 2016
Abstract
Two peptides, Coil-K and Coil-E, form a parallel heterodimeric coiled coil, CC-K/E, and have been shown to promote membrane fusion. This article examines the effects of reversing the sequence of Coil-E (to yield Coil-Er), on coiled-coil formation and membrane fusion. Coiled-coil assembly was studied using circular dichroism spectroscopy, paramagnetic proton NMR, fluorescence spectroscopy, analytical ultracentrifugation and computational simulations. Combined, the data show that Coil-K and Coil-Er combine in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio to form an antiparallel tetramer, reinforcing previous studies that show small changes to peptide sequences strongly affect the stoichiometry and orientation of the resulting assemblies. Cholesterol-modified Coil-K and Coil-Er variants were subsequently tested for their ability to promote membrane fusion and the results were compared to the CC-K/E model system. Surprisingly, no significant differences were found between the two systems, despite the Coil-K/Er complex being ‘non-zipper-like’.
1 ratio to form an antiparallel tetramer, reinforcing previous studies that show small changes to peptide sequences strongly affect the stoichiometry and orientation of the resulting assemblies. Cholesterol-modified Coil-K and Coil-Er variants were subsequently tested for their ability to promote membrane fusion and the results were compared to the CC-K/E model system. Surprisingly, no significant differences were found between the two systems, despite the Coil-K/Er complex being ‘non-zipper-like’.
Introduction
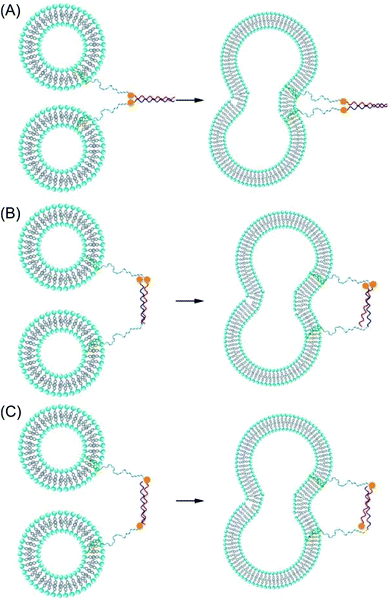
Membrane fusion occurs in all living cells where it regulates a variety of processes including fertilisation, viral infection, exocytosis, and vesicle trafficking.1–4 One of the most widely-studied fusion systems involves SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) proteins; these are a large family of proteins primarily responsible for vesicle trafficking and fusion.5–7 Whilst the exact mechanism of membrane fusion has yet to be determined, it is widely accepted that there are four steps: opposing membranes are brought into close proximity; local disruption of the membrane at the site of contact ensues; hemifusion, where outer membranes merge, occurs; and pore formation, facilitating content transfer between the two fused components, is the final result.8 SNARE proteins are located on opposing membranes and, in the first step of the fusion process, bind to form a tetrameric coiled-coil bringing the opposing membranes into close contact and triggering the fusion cascade.9 In an effort to mimic this system, and to increase the understanding of this complex process, numerous models for fusion have been created using a range of molecules including DNA, peptides, and organic small molecules as fusogens.10–18 One such model system, developed in our lab, takes inspiration from SNARE proteins and is based on the interaction between two coiled-coil forming peptides Coil-K ([KIAALKE]3) and Coil-E ([EIAALEK]3). These two peptides are designed to assemble into a parallel heterodimeric coiled coil (CC-K/E).19 Lipidated conjugates of Coil-K and Coil-E have been synthesised and incorporated into liposomes and, upon mixing, a parallel coiled coil is formed which forces the two opposing liposome membranes into close proximity resulting in fusion.20–23It has been postulated that, for fusion to occur, the fusogens need to be aligned in a parallel orientation allowing them to ‘zipper’ up from their distal N-termini to their membrane-proximal C-termini, generating an inward force that pulls the two membranes together and forces them to fuse.24,25 To probe this ‘zipper’ hypothesis, peptides that are anchored into the membrane at opposing termini,26 or peptides that have an antiparallel orientation can be used, Scheme 1. Previous studies have shown that both antiparallel SNARE derivatives and short antiparallel coiled coils cannot induce fusion.27,28 Intrigued by this, we endeavoured to discover whether non-zipper-like fusion was possible using antiparallel coiled coils incorporated into our membrane fusion system.
We hypothesized that, by reversing the amino acid sequence of either Coil-E or Coil-K we would generate an antiparallel coiled-coil. We were aware that changes to the core packing of coiled-coil structures can have large effects on the resulting assemblies: a well-known study of the GCN4 leucine zipper, an archetypal coiled coil, demonstrated that the oligomerisation state could be changed by altering the amino acids that comprise the hydrophobic core.29 A similar study reversed the entire GCN4 sequence, forming an antiparallel homotetramer; the native GCN4 is homodimeric.30 The reasons for these changes in oligomer state have been explained by the fact that different hydrophobic residues pack together in different manners at different positions within the coiled coil, meaning different oligomer states are necessary to facilitate the preferred packing of the different residues.31,32
With this in mind we decided to reverse the sequence of Coil-E, resulting in Coil-Er, and we analysed the resulting CC-K/Er complex using a variety of techniques including circular dichroism (CD) spectroscopy, paramagnetic proton nuclear magnetic resonance (NMR) spectroscopy, fluorescence spectroscopy, analytical ultracentrifugation, and computational simulations to determine the orientation and oligomer state of the assembly. Finally, the fusogenicity of cholesterol-modified variants of Coil-K and Coil-Er were compared to our CC-K/E model system to determine whether the CC-K/Er system was capable of effecting fusion and, if so, whether the rate and extent of fusion was affected by employment of this antiparallel, non-zipper-like coiled-coil.
Materials and methods
Materials
Fmoc-protected amino acids, rink amide resin, and HCTU (O-(1H-6-chlorobenzotriazole-1-yl)-1,1,2,2-tetramethyluronium hexafluorophosphate), were purchased from NovaBioChem. DIPEA (N,N-diisopropylethylamine), piperidine, acetic anhydride, NMP (N-methyl-2-pyrrolidone), DMF (N,N-dimethylformamide), MeCN (acetonitrile), and TFA (trifluoroacetic acid) were obtained from Biosolve. DCM (dichloromethane), diethyl ether, D2O (deuterium oxide), TIS (triisopropylsilane), EDT (1,2-ethanedithiol), cholesterol, PyBOP (benzotriazol-1-yl-oxytripyrrolidinophosphonium hexafluorophosphate), and sulforhodamine B were obtained from Sigma Aldrich. The Fmoc–NH–(PEG)12–COOH linker was purchased from Iris Biotech. MTSL (S-(2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate) was obtained from Toronto Research Chemicals Inc. DOPE (1,2-dioleyl-sn-glycero-3-phosphoethanolamine), DOPC (1,2-dioleyl-sn-glycero-phosphocholine), DOPE-LR (1,2-dioleyl-sn-glycero-3-phosphoethanolamine-N-lissamine rhodamine B sulfonyl), and DOPE-NBD (1,2-dioleyl-sn-glycero-3-phosphoethanolamine-N-7-nitro-2-1,3-benzoxadiazol-4-yl) were purchased from Avanti polar lipids.Cholesteryl-4-amino-4-oxobutanoic acid was synthesised according to a literature procedure.33
PBS (phosphate buffered saline) buffer contains 5 mM KH2PO4, 15 mM K2HPO4, 150 mM NaCl, pH 7.4.
Methods
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O
H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TIS (95
TIS (95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5
2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5 v/v) for 2 hours. Peptides with a Cys residue were cleaved from the resin with simultaneous side-chain deprotection using a mixture of TFA
2.5 v/v) for 2 hours. Peptides with a Cys residue were cleaved from the resin with simultaneous side-chain deprotection using a mixture of TFA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EDT
EDT![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O
H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TIS (92.5
TIS (92.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5
2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5
2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5 v/v) for 2 hours. The cleavage solution was added drop-wise into cold diethyl ether to precipitate the peptide, this was centrifuged and the liquid supernatant removed. All peptides were dried under vacuum, dissolved in deionised water and lyophilized yielding a white powder.
2.5 v/v) for 2 hours. The cleavage solution was added drop-wise into cold diethyl ether to precipitate the peptide, this was centrifuged and the liquid supernatant removed. All peptides were dried under vacuum, dissolved in deionised water and lyophilized yielding a white powder.
To form the lipidated peptide constructs CPE, CPEr, and CPK, the peptides were synthesised in the same way. After synthesis, the resin was washed with DMF. Fmoc–NH–(PEG)12–COOH (1.1 equivalents) was subsequently activated with HCTU (3 equivalents) and DIPEA (4 equivalents) in DMF and, after 5 minutes, added to the resin. The coupling reaction was allowed to proceed overnight. The linker was then Fmoc-deprotected by adding a solution of piperidine (20%) in DMF to the resin, and leaving for 15 minutes. This process was repeated three times before the resin was washed with DMF. Cholesteryl-4-amino-4-oxobutanoic acid (3 equivalents) was dissolved in 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v) DMF/DCM and activated with PyBOP (4 equivalents) and DIPEA (5 equivalents). After 5 minutes, this mixture was added to the resin and left overnight. The resin was then washed (DMF followed by DCM) and cleaved for 1 hour with a mixture of TFA
1 (v/v) DMF/DCM and activated with PyBOP (4 equivalents) and DIPEA (5 equivalents). After 5 minutes, this mixture was added to the resin and left overnight. The resin was then washed (DMF followed by DCM) and cleaved for 1 hour with a mixture of TFA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TIS (97.5
TIS (97.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5 v/v). The lipidated constructs were then precipitated in cold diethyl ether, collected by centrifugation and dried under vacuum.
2.5 v/v). The lipidated constructs were then precipitated in cold diethyl ether, collected by centrifugation and dried under vacuum.
where θobs is the observed ellipticity (mdeg), n is the number of peptide bonds, l is the path length of the cuvette (cm) and c is the peptide concentration (mM).
Spectra were recorded from 260 nm to 190 nm at 25 °C. Data points were collected with a 1 nm bandwidth at 1 nm intervals, using a scan speed of 1 nm s−1. Each spectrum was an average of 3 scans. For analysis, each spectrum had the background PBS spectrum subtracted. Thermal unfolding spectra were recorded at 222 nm, with a temperature gradient of 40 °C hour−1.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPE
DOPE![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cholesterol (50
cholesterol (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25
25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25 mol%) lipids in 1
25 mol%) lipids in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v) methanol
1 (v/v) methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) chloroform was prepared for all fusion experiments, except for the lipid mixing experiments. For these, a 1 mM stock solution of DOPC
chloroform was prepared for all fusion experiments, except for the lipid mixing experiments. For these, a 1 mM stock solution of DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPE
DOPE![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cholesterol
cholesterol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPE-LR
DOPE-LR![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPE-NBD (49.5
DOPE-NBD (49.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 24.75
24.75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 24.75
24.75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5
0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5, mol%) in 1
0.5, mol%) in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 methanol
1 methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) chloroform was made. Lipidated peptides were dissolved in a 1
chloroform was made. Lipidated peptides were dissolved in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v) choloroform
1 (v/v) choloroform![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol mixture, to a concentration of 50 uM. 1 mol% of these lipidated peptides were mixed with the appropriate liposome solution and the solvent was removed under a stream of air. For lipid mixing experiments, the dry lipid/peptide layer was rehydrated with PBS. For content mixing experiments, PBS buffer containing 20 mM sulphorhodamine B was added to the dried lipid/peptide mixtures containing Coil-E or Coil-Er. These solutions were subsequently sonicated for 5 minutes to yield ∼100 nm diameter liposomes. The solutions were used without further purification, with the exception of the sulforhodamine B containing liposomes, which were purified using a Sephadex column to remove any non-encapsulated sulforhodamine B.
methanol mixture, to a concentration of 50 uM. 1 mol% of these lipidated peptides were mixed with the appropriate liposome solution and the solvent was removed under a stream of air. For lipid mixing experiments, the dry lipid/peptide layer was rehydrated with PBS. For content mixing experiments, PBS buffer containing 20 mM sulphorhodamine B was added to the dried lipid/peptide mixtures containing Coil-E or Coil-Er. These solutions were subsequently sonicated for 5 minutes to yield ∼100 nm diameter liposomes. The solutions were used without further purification, with the exception of the sulforhodamine B containing liposomes, which were purified using a Sephadex column to remove any non-encapsulated sulforhodamine B.
| %F = (F(t) − F0)/(Fmax − F0) |
For the content mixing experiments, the sulphorhodamine B fluorescence intensity at 580 nm was monitored in a continuous fashion for 1 hour, after mixing non-fluorescent K-liposomes with sulforhodamine B loaded E- or Er-liposomes. F0 was obtained by measuring emission of sulforhodamine B-loaded E- or Er-liposomes, and Fmax was obtained by measuring the emission of plain liposomes loaded with 10 mM sulforhodamine B.
The FRET experiments were conducted using the same apparatus. Excitation and emission slits were set to 5 nm. Emission spectra were measured from 290 nm to 450 nm in 1 nm steps at a fixed excitation wavelength of 275 nm. The temperature was set at 25 °C. The spectra were corrected by subtraction of the background spectrum. The total peptide concentration was 50 μM.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–50
000–50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm) at 20 °C. The data were then fitted to a single-ideal species model using Ultrascan.35
000 rpm) at 20 °C. The data were then fitted to a single-ideal species model using Ultrascan.35
Results and discussion
Peptide design and synthesis
In this study, Coil-K retained its original sequence, with the addition of a Trp at the C-terminus to allow for quantification and analytical studies. A glycine (Gly, G) was introduced to act as a spacer between the chromophore and the coiled-coil forming portion of the sequence. This gave rise to a Coil-K variant with the sequence: (KIAALKE)3GW. Coil-E was redesigned to possess the reversed amino acid sequence, yielding Coil-Er. A Tyr residue was attached to the N-terminus, again to allow for quantification and analytical studies, whilst a Gly spacer was also included. This resulted in Coil-Er having the sequence: YG(EIAALEK)3.Additionally, the paramagnetic nitroxyl radical MTSL (S-(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate) was conjugated to Coil-K via a cysteine residue. Two MTSL-labelled derivatives were generated: attachment at the C-terminus yielded Coil-K*; whereas conjugation to the N-terminus produced Coil-*K. This label offered the opportunity to investigate the stoichiometry and relative peptide orientation of CC-K/Er by paramagnetic proton NMR and steady-state fluorescence measurements.43
Peptides were synthesised on Rink amide resin using standard Fmoc solid-phase synthesis techniques and purified by reversed-phase HPLC. Peptides were identified using LC-MS mass spectrometry, and analytical HPLC confirmed the purity of the peptides was >99% (ESI Fig. S1 and S2†).
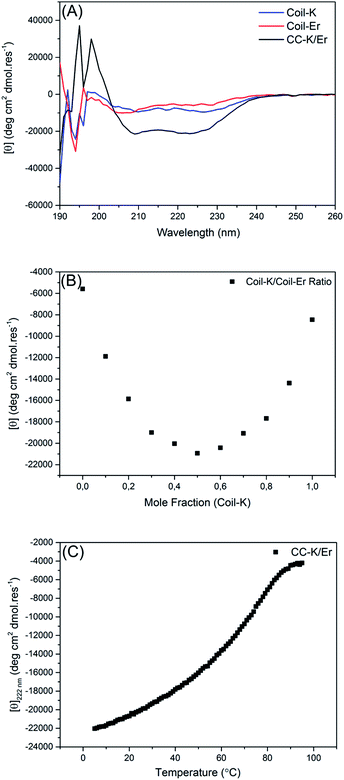
Circular dichroism spectroscopy
Circular dichroism (CD) spectroscopy was used to probe the secondary structure of the peptides. At room temperature, both Coil-K and Coil-Er are largely unfolded, exhibiting only small amounts of α-helical character, (Fig. 1A). The equimolar mixture, in contrast, is well folded as an α-helical assembly and exhibits typical coiled-coil characteristics with the [θ]222 nm/[θ]208 nm ellipticity ratio >1.44The binding stoichiometry of Coil-K to Coil-Er was subsequently determined. A Job-plot was created by changing the ratio of the mole fraction of Coil-K and Coil-Er, while keeping the total peptide concentration constant. A minimum molar ellipticity at [θ]222 nm was observed when Coil-K and Coil-Er were mixed in equimolar amounts, indicative of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding stoichiometry, (Fig. 1B).
1 binding stoichiometry, (Fig. 1B).
Finally, the thermal stability of CC-K/Er was investigated. A solution containing a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of Coil-K and Coil-Er was heated from 5 °C to 95 °C and a sigmoidal curve, indicative of a two-state transition from a folded to an unfolded complex was observed, (Fig. 1C). The melting temperature (Tm) was found to be 64 °C, similar to the Tm of 57 °C recorded for the CC-K/E complex.43
1 mixture of Coil-K and Coil-Er was heated from 5 °C to 95 °C and a sigmoidal curve, indicative of a two-state transition from a folded to an unfolded complex was observed, (Fig. 1C). The melting temperature (Tm) was found to be 64 °C, similar to the Tm of 57 °C recorded for the CC-K/E complex.43
Paramagnetic 1H-proton-NMR spectroscopy
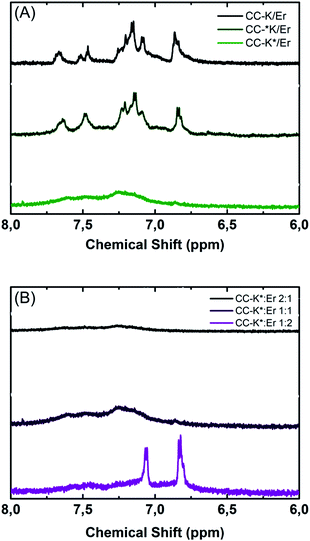
Paramagnetic NMR spectroscopy was used to confirm the stoichiometry of the CC-K/Er complex, and to explore the relative orientation of the peptides within the complex.Our previous work demonstrated that these properties can be facilely determined by examining whether suppression of the peptide-specific proton signals occurs due to the paramagnetic relaxation enhancement (PRE) effect.43,45,46 Full suppression of the proton signals occurs within a 10.5 Å radius of the paramagnetic center, and partial suppression occurs within 13 Å.47 To observe such an effect, peptides need to possess a functional group with a characteristic NMR signal, and one of the peptides is required to incorporate a signal suppressing paramagnetic spin label. Aromatic proton signals are well separated from other proton signals, therefore the signals from Trp and Tyr were monitored. In addition, the paramagnetic spin label MTSL was introduced onto either the N- or the C-terminus of Coil-K, yielding the aforementioned, Coil-*K and Coil-K*. To avoid spectral overlap of the aromatic protons with N–H amide protons, the latter were suppressed by H–D exchange.
The aromatic region of Trp exhibits four multiplets while Tyr shows two doublets, (ESI Fig. S3A & B†). Peptide Coil-K* shows full suppression of the Trp signals, (ESI Fig. S3C†), as the average distance between Trp and the MTSL label is 6.6 Å. In contrast, in peptide Coil-*K the average distance between the aromatic protons and the MTSL label is 36.7 Å, and therefore no PRE effect is observed, (ESI Fig. S3D†).
When Coil-K and Coil-Er are mixed in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio, all aromatic proton signals are observed, (Fig. 2A, black line). Similarly, when Coil-*K and Coil-Er are mixed, all aromatic proton signals are observed, hence there is no PRE effect (Fig. 2A, olive green line). When Coil-Er is combined with Coil-K* however, suppression of both Trp and Tyr signals are observed (Fig. 2A, lime green line), indicating that both residues are within 10.5 Å of the MTSL label. This clearly demonstrates that the peptides are assembled in an anti-parallel orientation, as the distance between Tyr and the MTSL label is >13.5 Å in the parallel orientation.
1 ratio, all aromatic proton signals are observed, (Fig. 2A, black line). Similarly, when Coil-*K and Coil-Er are mixed, all aromatic proton signals are observed, hence there is no PRE effect (Fig. 2A, olive green line). When Coil-Er is combined with Coil-K* however, suppression of both Trp and Tyr signals are observed (Fig. 2A, lime green line), indicating that both residues are within 10.5 Å of the MTSL label. This clearly demonstrates that the peptides are assembled in an anti-parallel orientation, as the distance between Tyr and the MTSL label is >13.5 Å in the parallel orientation.
As CD spectroscopy indicated that Coil-K and Coil-Er bind in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry, NMR experiments were performed to confirm this. When Coil-K* and Coil-Er were mixed in a 1
1 stoichiometry, NMR experiments were performed to confirm this. When Coil-K* and Coil-Er were mixed in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio (Fig. 2B, black line), the Tyr proton signals were fully suppressed. This was also observed for a 2
1 molar ratio (Fig. 2B, black line), the Tyr proton signals were fully suppressed. This was also observed for a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Coil-K*
1 (Coil-K*![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Coil-Er) mixture (Fig. 2B, purple line). However, when Coil-K* and Coil-Er were mixed in 1
Coil-Er) mixture (Fig. 2B, purple line). However, when Coil-K* and Coil-Er were mixed in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 molar ratio, Tyr proton signals were visible, (Fig. 2B, magenta line). This indicates there is an excess of Coil-Er which is not suppressed, and supports the conclusions drawn from the CD experiments that the two peptides bind in a 1
2 molar ratio, Tyr proton signals were visible, (Fig. 2B, magenta line). This indicates there is an excess of Coil-Er which is not suppressed, and supports the conclusions drawn from the CD experiments that the two peptides bind in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry.
1 stoichiometry.
Fluorescence spectroscopy
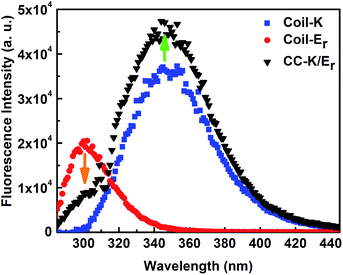
To confirm the relative peptide orientation, FRET (fluorescence resonance energy transfer) experiments were performed. FRET occurs when the donor and acceptor are within the Förster distance (10 Å).48 An equimolar mixture of Coil-K and Coil-Er was examined, and it revealed that the Coil-K fluorescence intensity increased while the Coil-Er fluorescence intensity decreased, Fig. 3. This is expected when Coil-Er and Coil-K form coiled coils with an antiparallel orientation, allowing FRET between the donor Tyr, and the acceptor Trp. This confirms the distance between the N-terminus of peptide Coil-Er and the C-terminus of peptide Coil-K is <10 Å, confirming an antiparallel orientation of CC-K/Er, which is consistent with the findings of the NMR experiments, and confirmed our hypothesis that reversing one of the peptide sequences would result in an antiparallel orientation of the resulting coiled coil.Analytical ultracentrifugation
To determine the oligomer state of the CC-K/Er species in solution, analytical ultracentrifugation (AUC) sedimentation equilibrium measurements were performed. Coil-K and Coil-Er were analysed in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio as both the CD and NMR data indicated the two peptides bind in a 1
1 ratio as both the CD and NMR data indicated the two peptides bind in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry. The collected datasets fitted well to a single-ideal species model, (ESI Fig. S6†), and the average mass of the species was determined to be 10
1 stoichiometry. The collected datasets fitted well to a single-ideal species model, (ESI Fig. S6†), and the average mass of the species was determined to be 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 620 Da, with 95% confidence limits (determined by Monte Carlo analysis) found to be +57 Da, −64 Da. Hence, weight-averaged molecular weights indicating a tetrameric species were observed.
620 Da, with 95% confidence limits (determined by Monte Carlo analysis) found to be +57 Da, −64 Da. Hence, weight-averaged molecular weights indicating a tetrameric species were observed.
The fact that Coil-K/Er forms a tetrameric species is unexpected, as previous studies have noted it forms a dimer, although no analytical studies were conducted to corroborate these claims.49 On reflection, the fact that a tetramer is formed is not surprising; this is in line with findings from previous studies of the GCN4 coiled coil which indicated that altering the residues that comprise the hydrophobic core also affects the oligomerisation state of the resulting assembly,29,30 although it should be noted such studies were conducted with homodimeric species; similar experiments with heteromeric sequences have not, to our knowledge, been conducted.
Molecular simulations
Collectively, the experimental data strongly indicate that Coil-K and Coil-Er associate in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio to form an antiparallel tetramer. However, the exact arrangement of the peptides within this tetramer cannot be determined without obtaining a crystal structure. Coarse-grained molecular dynamics (CGMD) simulations were performed to obtain additional information about the preferred orientations of the peptides within the tetramer. In addition, simulations of 6 peptides in solution, (results not shown), i.e. containing 3 copies of both Coil-K and Coil-Er, showed the formation of a dimer and a tetramer indicating the tetramer is indeed the preferred structure. Formation of a trimer was not observed during the simulations.
1 ratio to form an antiparallel tetramer. However, the exact arrangement of the peptides within this tetramer cannot be determined without obtaining a crystal structure. Coarse-grained molecular dynamics (CGMD) simulations were performed to obtain additional information about the preferred orientations of the peptides within the tetramer. In addition, simulations of 6 peptides in solution, (results not shown), i.e. containing 3 copies of both Coil-K and Coil-Er, showed the formation of a dimer and a tetramer indicating the tetramer is indeed the preferred structure. Formation of a trimer was not observed during the simulations.
Focussing on the tetramer simulations, two copies of both Coil-K and Coil-Er were initially placed in an 11 nm3 simulation box that also contained explicit solvent (water and Na+/Cl− ions) to mimic experimental conditions. The simulations were run for 40 μs, after which time the formation pathways and resulting assemblies were analysed.
In all simulations, the randomly distributed peptides spontaneously aggregate into supramolecular structures. The four peptides first come into contact, and then orientate parallel to each other, forming a 4-helix-bundle with a hydrophobic core. This aggregation process is relatively fast, approximately 0.5 microseconds, while 20 to 40 microseconds are needed for further equilibration; during this stage misaligned peptides rotates around their own axis or flip to form an antiparallel structure and optimize the hydrophobic core. After equilibration, each peptide forms two longitudinal interfaces with two neighbouring peptides, and the diameter of the cylindrical assembly is ∼2.5 nm.
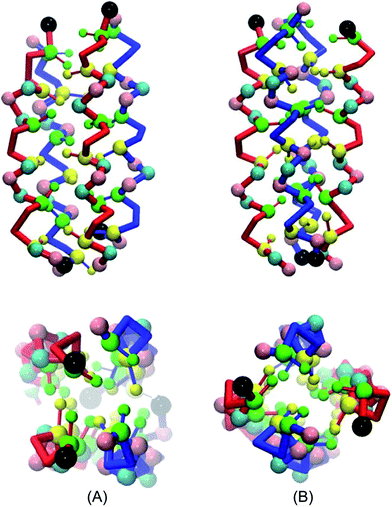
All possible tetrameric structures can be divided into two major classes: those with four K–Er interfaces; or assemblies with two K–Er interfaces, together with one K–K and one Er–Er interface. Accounting for parallel or antiparallel peptide orientations, as well as symmetry, 11 distinct peptide arrangements within the tetramer are possible.
Our 40 μs simulations provide the means to discriminate between these potential arrangements. From the 20 independent simulations that were performed, we identified only two stable structures, (Fig. 4). The first structure has two K–Er interfaces, along with one K–K and one Er–Er interface. The helices at the K–Er interfaces are antiparallel, whilst the helices at the K–K and Er–Er interfaces are aligned in a parallel orientation, (Fig. 4A). The second structure possesses four K–Er interfaces, with antiparallel orientations of the helices at these interfaces but parallel orientations of the two Coil-K and two Coil-E helices (Fig. 4B).
Our computation setup, which relies on extensive pathway sampling for the various starting conditions, gives us confidence that the most probable binding conformations are indeed sampled by the CGMD simulations. This is further supported by observations of a complete reorientation of the peptides, even after association, an event that is generally considered to be rare on the timescale of molecular simulations. In the tetramer simulations no dimers were observed in the simulation time, and the only two stable structures observed has an antiparallel orientation, supporting the conclusion from the experimental evidence that an antiparallel tetrameric assembly is formed by the association of these peptides.
Fusion studies
A variety of analytical and computational techniques have shown that Coil-K and Coil-Er form an antiparallel tetramer, whereas previous studies showed that Coil-K and Coil-E form a parallel dimer.CC-K/E is known to induce efficient and targeted membrane fusion when both peptides are modified with a lipid anchor and incorporated into liposome membranes.20,22,23,26 These fusogenic constructs comprise three distinct segments: the peptide segment, which serves as the recognition unit; a hydrophobic cholesterol anchor, which enables the construct to be secured in a liposome membrane, and; a hydrophilic linker, which is thought to be crucial for allowing force transduction and therefore helping to facilitate fusion in natural SNARE systems.6,50,51
In addition to the linking region being crucial for fusion, it is postulated that the peptides need to be in a parallel orientation, as their ‘zippering’ from one terminus to the other as they fold provides the force that draws the two membranes close together and induces them to fuse.24,25 Non-zipper-like coiled coils, that is, those with an antiparallel orientation should not induce fusion if it is indeed the ‘zipper-like’ folding of the fusogens that induces fusion.
To determine whether CC-K/Er, which assembles in a non-zipper-like manner, is able to induce fusion, cholesterol-anchored constructs were generated with the peptide and cholesterol being separated by a polyethyleneglycol (PEG)12 spacer. The resulting lipidated peptides were denoted CPK and CPEr. CPE was also synthesised so comparisons regarding the rate and efficiency of fusion of CC-K/E and CC-K/Er could be made. Peptide-decorated liposomes were prepared, and fusion experiments were performed by combining equimolar amounts of Coil-K liposomes with either Coil-E- or Coil-Er-containing liposomes.
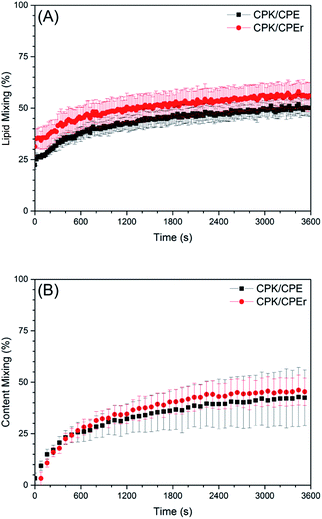
For these experiments, Coil-E or Coil-Er decorated liposomes were loaded with sulforhodamine B at a self-quenching concentration of 20 mM. If full fusion between these liposomes and liposomes decorated with Coil-K occurs, the sulforhodamine B concentration decreases, resulting in relief of self-quenching and an increase in fluorescence intensity, which is indeed what was observed for both systems, (Fig. 5B). As with the lipid mixing experiments, no significant differences in the rates of content mixing were observed, and after one hour both systems demonstrated a similar extent (45%) of content mixing.
Taken together these results indicate that, for this peptide-mediated fusion system, both lipid- and content-mixing can be facilitated by zipper-like and non-zipper-like fusogens. This suggests that the way in which the peptides fold is not important, what is important is that energy is generated upon folding and that this can be transduced, presumably via the linking region, to the liposomes themselves resulting in fusion events.
Conclusions
Reversing the peptide sequence of Coil-E yields Coil-Er which, in the presence of equimolar amounts of Coil-K, results in the formation of an antiparallel tetrameric coiled coil and not an antiparallel dimeric coiled coil as previously reported.27 This shows that, in line with previous studies on proteins and peptides, reversing the peptide sequence significantly effects the orientation and oligomer state of the resulting assemblies.19,42,45 The generation of an antiparallel coiled coil offered a new ‘non-zipper-like’ peptide candidate for our membrane fusion model system. Cholesterol modified variants of Coil-K and Coil-Er were produced and were subsequently demonstrated to induce liposome fusion to an extent comparable to, and in the case of lipid-mixing surpassing that of, CC-K/E induced fusion. This shows that, for this peptide-mediated membrane fusion system, both zipper-like and non-zipper-like coiled-coils are capable of inducing efficient liposome fusion. Future work will focus on elucidating which elements are crucial for fusion if it is indeed not the way in which the peptides fold. As previous studies have implicated the linking region between the fusogen and the membrane anchor as being crucial for efficient fusion,6,50,51 this would be an intriguing candidate for future studies.Acknowledgements
T. Z. acknowledges the support of the CSC. A. L. B. acknowledges the support of the FOM. M. B. and G. J. A. S. acknowledge the NCF (Netherlands Computing Facilities). A. K. acknowledges the support of the European Research Council (ERC) via an ERC starting grant (240394) which funded F. V. and G. M, and the support of the Netherlands Organisation for Scientific Research (NWO) via a ChemThem grant that funded G. D. We are grateful to Dr Hana Robson Marsden for productive discussions, and to Dr Thomas Sharp for critical reading of this manuscript.References
- J. E. Gerst, Cell. Mol. Life Sci., 1999, 55, 707–734 CrossRef CAS PubMed.
- M. E. Hemler, Annu. Rev. Cell Dev. Biol., 2003, 19, 397–422 CrossRef CAS PubMed.
- R. Jahn and R. H. Scheller, Nat. Rev. Mol. Cell Biol., 2006, 7, 631–643 CrossRef CAS PubMed.
- M. Kielian and F. A. Rey, Nat. Rev. Microbiol., 2006, 4, 67–76 CrossRef CAS PubMed.
- A. Maximov, J. Tang, X. F. Yang, Z. P. P. Pang and T. C. Sudhof, Science, 2009, 323, 516–521 CrossRef CAS PubMed.
- T. C. Sudhof and J. E. Rothman, Science, 2009, 323, 474–477 CrossRef PubMed.
- C. Hu, M. Ahmed, T. J. Melia, T. H. Sollner, T. Mayer and J. E. Rothman, Science, 2003, 300, 1745–1749 CrossRef CAS PubMed.
- T. Weber, B. V. Zemelman, J. A. McNew, B. Westermann, M. Gmachl, F. Parlati, T. H. Sollner and J. E. Rothman, Cell, 1998, 92, 759–772 CrossRef CAS PubMed.
- R. Jahn, T. Lang and T. C. Sudhof, Cell, 2003, 112, 519–533 CrossRef CAS PubMed.
- Y. H. Chan, B. van Lengerich and S. G. Boxer, Biointerphases, 2008, 3, FA17 CrossRef CAS PubMed.
- Y. Gong, Y. Luo and D. Bong, J. Am. Chem. Soc., 2006, 128, 14430–14431 CrossRef CAS PubMed.
- A. Kashiwada, K. Matsuda, T. Mizuno and T. Tanaka, Chemistry, 2008, 14, 7343–7350 CrossRef CAS PubMed.
- A. Kashiwada, M. Tsuboi, N. Takamura, E. Brandenburg, K. Matsuda and B. Koksch, Chem.–Eur. J., 2011, 17, 6179–6186 CrossRef CAS PubMed.
- A. S. Lygina, K. Meyenberg, R. Jahn and U. Diederichsen, Angew. Chem., Int. Ed., 2011, 50, 8597–8601 CrossRef CAS PubMed.
- M. M. Ma and D. Bong, Acc. Chem. Res., 2013, 46, 2988–2997 CrossRef CAS PubMed.
- V. Marchi-Artzner, T. Gulik-Krzywicki, M. A. Guedeau-Boudeville, C. Gosse, J. M. Sanderson, J. C. Dedieu and J. M. Lehn, ChemPhysChem, 2001, 2, 367–376 CrossRef CAS PubMed.
- G. Stengel, L. Simonsson, R. A. Campbell and F. Hook, J. Phys. Chem. B, 2008, 112, 8264–8274 CrossRef CAS PubMed.
- J. Voskuhl and B. J. Ravoo, Chem. Soc. Rev., 2009, 38, 495–505 RSC.
- J. R. Litowski and R. S. Hodges, J. Biol. Chem., 2002, 277, 37272–37279 CrossRef CAS PubMed.
- H. R. Marsden, N. A. Elbers, P. H. H. Bomans, N. Sommerdijk and A. Kros, Angew. Chem., Int. Ed., 2009, 48, 2330–2333 CrossRef PubMed.
- H. R. Marsden, A. V. Korobko, T. T. Zheng, J. Voskuhl and A. Kros, Biomater. Sci., 2013, 1, 1046–1054 RSC.
- F. Versluis, J. Voskuhl, B. van Kolck, H. Zope, M. Bremmer, T. Albregtse and A. Kros, J. Am. Chem. Soc., 2013, 135, 8057–8062 CrossRef CAS PubMed.
- T. T. Zheng, J. Voskuhl, F. Versluis, H. R. Zope, I. Tomatsu, H. R. Marsden and A. Kros, Chem. Commun., 2013, 49, 3649–3651 RSC.
- Y. A. Chen and R. H. Scheller, Nat. Rev. Mol. Cell Biol., 2001, 2, 98–106 CrossRef CAS PubMed.
- R. C. Lin and R. H. Scheller, Neuron, 1997, 19, 1087–1094 CrossRef CAS PubMed.
- F. Versluis, J. Dominguez, J. Voskuhl and A. Kros, Faraday Discuss., 2013, 166, 349–359 RSC.
- G. Pahler, C. Panse, U. Diederichsen and A. Janshoff, Biophys. J., 2012, 103, 2295–2303 CrossRef PubMed.
- K. Weninger, M. E. Bowen, S. Chu and A. T. Brunger, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 14800–14805 CrossRef CAS PubMed.
- P. B. Harbury, T. Zhang, P. S. Kim and T. Alber, Science, 1993, 262, 1401–1407 CAS.
- P. R. Mittl, C. Deillon, D. Sargent, N. Liu, S. Klauser, R. M. Thomas, B. Gutte and M. G. Grutter, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 2562–2566 CrossRef CAS.
- F. H. C. Crick, Acta Crystallogr., 1953, 6, 689–697 CrossRef CAS.
- D. N. Woolfson, Adv. Protein Chem., 2005, 70, 79–112 CrossRef CAS PubMed.
- Q. Sun, S. Cai and B. R. Peterson, Org. Lett., 2009, 11, 567–570 CrossRef CAS PubMed.
- J. R. Litowski and R. S. Hodges, J. Pept. Res., 2001, 58, 477–492 CrossRef CAS PubMed.
- B. Demeler, in Modern Analytical Ultracentrifugation: Techniques and Methods, ed. D. J. Scott, S. E. Harding and A. J. Rowe, Royal Society of Chemistry, Cambridge, 2005, vol. 11, pp. 210–229 Search PubMed.
- B. Hess, C. Kutzner, D. van der Spoel and E. Lindahl, J. Chem. Theory Comput., 2008, 4, 435–447 CrossRef CAS PubMed.
- S. J. Marrink, H. J. Risselada, S. Yefimov, D. P. Tieleman and A. H. de Vries, J. Phys. Chem. B, 2007, 111, 7812–7824 CrossRef CAS PubMed.
- L. Monticelli, S. K. Kandasamy, X. Periole, R. G. Larson, D. P. Tieleman and S. J. Marrink, J. Chem. Theory Comput., 2008, 4, 819–834 CrossRef CAS PubMed.
- D. Sengupta and S. J. Marrink, Phys. Chem. Chem. Phys., 2010, 12, 12987–12996 RSC.
- L. V. Schafer, D. H. de Jong, A. Holt, A. J. Rzepiela, A. H. de Vries, B. Poolman, J. A. Killian and S. J. Marrink, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 1343–1348 CrossRef CAS PubMed.
- X. Periole, A. M. Knepp, T. P. Sakmar, S. J. Marrink and T. Huber, J. Am. Chem. Soc., 2012, 134, 10959–10965 CrossRef CAS PubMed.
- D. A. Lindhout, J. R. Litowski, P. Mercier, R. S. Hodges and B. D. Sykes, Biopolymers, 2004, 75, 367–375 CrossRef CAS PubMed.
- T. T. Zheng, A. Boyle, H. R. Marsden, D. Valdink, G. Martelli, J. Raap and A. Kros, Org. Biomol. Chem., 2015, 13, 1159–1168 CAS.
- N. E. Zhou, C. M. Kay and R. S. Hodges, J. Biol. Chem., 1992, 267, 2664–2670 CAS.
- B. Apostolovic and H. A. Klok, Biomacromolecules, 2008, 9, 3173–3180 CrossRef CAS PubMed.
- T. Gruene, M. K. Cho, I. Karyagina, H. Y. Kim, C. Grosse, K. Giller, M. Zweckstetter and S. Becker, J. Biomol. NMR, 2011, 49, 111–119 CrossRef CAS PubMed.
- C. Peggion, M. Jost, W. M. de Borggraeve, M. Crisma, F. Formaggio and C. Toniolo, Chem. Biodiversity, 2007, 4, 1256–1268 CAS.
- J. Eisinger, B. Feuer and A. A. Lamola, Biochemistry, 1969, 8, 3908–3915 CrossRef CAS PubMed.
- K. Meyenberg, A. S. Lygina, G. van den Bogaart, R. Jahn and U. Diederichsen, Chem. Commun., 2011, 47, 9405–9407 RSC.
- J. A. McNew, T. Weber, D. M. Engelman, T. H. Sollner and J. E. Rothman, Mol. Cell, 1999, 4, 415–421 CrossRef CAS PubMed.
- J. A. McNew, T. Weber, F. Parlati, R. J. Johnston, T. J. Melia, T. H. Sollner and J. E. Rothman, J. Cell Biol., 2000, 150, 105–117 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5ra26175a |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2016 |