Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Sortase-mediated labelling of lipid nanodiscs for cellular tracing†
A. Ivona
Petrache
,
Darren C.
Machin
,
Daniel J.
Williamson
,
Michael E.
Webb
* and
Paul A.
Beales
*
School of Chemistry and Astbury Centre for Structural Molecular Biology, University of Leeds, Leeds, UK. E-mail: m.e.webb@leeds.ac.uk; p.a.beales@leeds.ac.uk
First published on 6th April 2016
Abstract
Lipid nanodiscs have broad applications in membrane protein assays, biotechnology and materials science. Chemical modification of the nanodiscs to expand their functional attributes is generally desirable for all of these uses. We present a method for site-selective labelling of the N-terminus of the nanodisc's membrane scaffold protein (MSP) using the Sortase A protein. Labelling of the MSP was achieved when assembled within the lipid nanodisc architecture, demonstrating that this method can be used as a retrofit approach to modification of preformed nanodiscs before or during application. We label the MSP with a fluorescent fluorescein moiety and use them to image nanodisc uptake into HeLa cells. The Sortase A labelling method could be employed as a general approach to labelling nanodiscs with application-specific functionalities.
Lipoprotein and lipoprotein-mimetic particles have recently attracted significant interest in the field of biotechnology.1,2 This is due to their potential for applications in areas such as the stabilisation and handling of membrane proteins3 and formulation of active pharmaceutical ingredients in nanomedical delivery technologies.4–11 These nanoscale particles, which we will refer to as lipid nanodiscs, consist of a discotic segment of lipid bilayer membrane where the hydrophobic line tension at the disc perimeter is stabilised by an amphipathic protein or polymer, e.g. natural apolipoproteins,12 synthetic apolipoprotein derivatives,13,14 or styrene maleic acid copolymers.15,16
Chemical functionalisation of lipid nanodiscs for biotechnology applications is highly desirable as this facilities the incorporation of new, additional functionalities within these particles. Examples include the addition of image contrast agents,17 targeting ligands,18 or environmental sensors. These functionalities could be attached to the lipids in the bilayer, for example DNA-functionalised lipids have been used to adapt nanodiscs for directed assembly into quasi-1D columnar stacks, which were used as templates to direct gold nanoparticle assembly.19,20 Similarly, the scaffold proteins (or polymers) are desirable substrates for functionalisation. The scaffold proteins of lipid nanodiscs have previously been functionalised with tetramethylrhodamine dyes by engineering a cysteine into the protein for site-specific labelling.17 However, labelling strategies for these proteins that can be performed on the original construct, without the need to reengineer and express a batch of new protein, would be highly attractive.
Here we present a method for specifically labelling the N-terminus of the MSP1D1 scaffold proteins of lipid nanodiscs. MSP1D1 (MSP hereafter) is a truncated derivative of apolipoprotein A-1, developed in the laboratory of Stephen Sligar.21 To achieve site specific labelling, we use the Sortase A (SrtA)-mediated labelling and the optimised depsipeptide substrates previously reported by Williamson et al.22 SrtA is a transpeptidase required for the attachment of proteins to the cell wall of Gram-positive bacteria but can also be used for linkage of peptides and proteins using C-terminal or internal LPXTG motifs in one substrate together with an N-terminal glycine in the second substrate. Replacement of the LPXTG peptide sequence with LPXToG depsipeptide leaves the labelling reaction irreversible and can be used for quantitative labelling.23 For SrtA-mediated labelling to be effective, the target protein must possess an N-terminal glycine residue. The expressed MSP scaffold protein has a His-tag, used for affinity purification, linked to the scaffold protein by a Tobacco Etch Virus (TEV) protease recognition site (ENLYFQG). We therefore used the TEV protease to cleave the His-tag from the scaffold protein, which hydrolyses the amide bond between the QG amino acid residues. This conveniently results in a glycine residue at the N-terminus of MSP, as required.
As a proof-of-principle for SrtA-mediated labelling of lipid nanodiscs, we functionalised MSP with a fluorescent fluorescein-depsipeptide. The MSP1D1 scaffold protein was overexpressed in E. coli BL21 (DE3) from the expression plasmid reported by Sligar and coworkers24 and purified by sequential Ni–NTA affinity chromatography and size-exclusion chromatography. Our initial attempts to directly label monomeric MSP using SrtA were unsuccessful due to protein precipitation during both the TEV protease cleavage and SrtA labelling steps. We therefore chose to directly label MSP in pre-formed lipid nanodiscs.
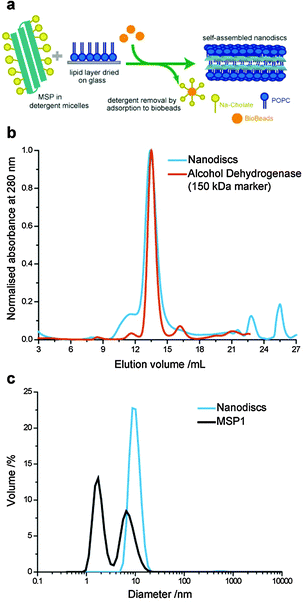
Lipid nanodiscs were formed using a slightly modified method compared to previously reported protocols (Fig. 1a).24 Previous protocols have used two-step formation of lipid–detergent micelles followed by addition of the scaffold proteins. In our optimised protocol, the detergent-solubilised MSP solution was added directed to the dried lipid (POPC) film in a MSP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) lipid
lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cholate ratio of 1
cholate ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 65
65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 and incubated with gentle mixing at 4 °C for 2.5 h. The sodium cholate detergent is then removed by incubation with 0.3 g ml−1 of wet Bio-beads at 4 °C for 4 to 5 h. We found that storage of MSP in detergent-containing buffer (40 mM sodium cholate) improved the stability of the protein against aggregation, which can hinder nanodisc formation once protein aggregates in stock solutions become large.
100 and incubated with gentle mixing at 4 °C for 2.5 h. The sodium cholate detergent is then removed by incubation with 0.3 g ml−1 of wet Bio-beads at 4 °C for 4 to 5 h. We found that storage of MSP in detergent-containing buffer (40 mM sodium cholate) improved the stability of the protein against aggregation, which can hinder nanodisc formation once protein aggregates in stock solutions become large.
The expected size distribution of MSP lipid nanodiscs was confirmed by size exclusion chromatography (SEC) and dynamic light scattering (DLS) (Fig. 1b and c). Nanodiscs eluted at a similar volume to a 150 kDa alcohol dehydrogenase molecular weight marker in the SEC with a narrow size distribution, as has previously been reported for MSP nanodiscs.21 The average hydrodynamic diameter of nanodiscs was measured to be 10.1 nm by DLS, close to the 9.7 nm diameter previously reported by SAXS measurements.21 Furthermore, we analysed the MSP stock solution by DLS, which showed a bimodal size distribution with peaks at 1.7 nm and 6.5 nm hydrodynamic diameters. This is consistent with the presence of monomeric and small oligomeric protein populations; the presence of the cholate detergent inhibited the formation of larger MSP aggregates, which inhibit efficient nanodisc formation.
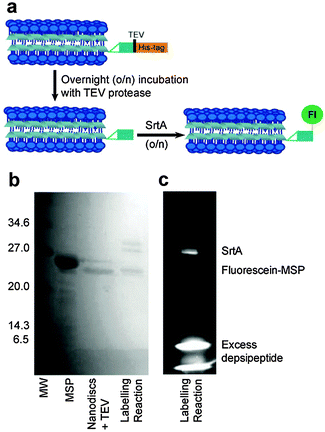
Two approaches were taken to fluorescence labelling of the nanodiscs; either labelling MSP before or after nanodisc formation. In practice, labelling MSP prior to nanodisc formation was impractical due to the prolonged incubation times required for both TEV protease and sortase-mediated labelling as detailed below. The final SrtA-mediated nanodisc labelling protocol is schematically outlined in Fig. 2a. Following nanodisc formation, the His-tag of MSP was cleaved with TEV protease by overnight incubation at room temperature. Complete cleavage was confirmed by mass spectrometry following disassembly of the nanodiscs (Fig. S1, ESI†). The size change of the nanodiscs following His-tag cleavage was also resolvable by SEC: nanodiscs with cleaved His-tags elute slightly later from the column than nanodiscs with the His-tags intact (Fig. S2, ESI†). The cleaved MSP (two proteins per nanodisc25) was then modified via SrtA-mediated ligation using a fluorescein-depsipeptide synthesized as previously described26 for labelling. The nanodiscs were incubated overnight at room temperature with an estimated 10 equivalents of fluorescein-depsipeptide and 0.2 equivalents of SrtA (based on complete conversion of MSP to nanodiscs). An excess of depsipeptide label was added to the nanodiscs to ensure a high protein modification yield and reduce the effect of sortase-mediated hydrolysis during the prolonged incubation period required for labelling of the very low effective concentration of nanodiscs. Successful labelling of MSP was confirmed by shift in position and fluorescence of the MSP band in the SDS-PAGE (Fig. 2b) and mass spectrometry (Fig. S3, ESI†). A fluorescent band corresponding to the molecular weight of MSP1D1 was observed by SDS-PAGE and the measured molecular weight of 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 959 Da by mass spectrometry is consistent with the theoretical value of 22
959 Da by mass spectrometry is consistent with the theoretical value of 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 957 Da. SrtA, which contains a His-tag for purification, was then removed from the nanodiscs by overnight incubation of the sample with Ni resin followed by separation of the nanodiscs from the resin by centrifugation.
957 Da. SrtA, which contains a His-tag for purification, was then removed from the nanodiscs by overnight incubation of the sample with Ni resin followed by separation of the nanodiscs from the resin by centrifugation.
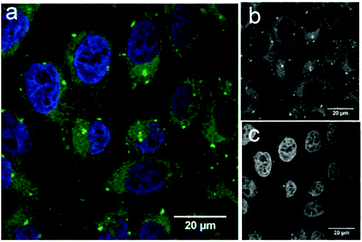
To demonstrate the potential application of fluorescein-functionalised nanodiscs, we investigated their traceable delivery into mammalian cells. Labelled nanodiscs were incubated with HeLa cells for 4 h before paraformaldehyde (4%) fixation, washing and imaging by confocal microscopy (Fig. 3). Bright, punctate spots of fluorescence inside these cells are clearly visible above the background autofluorescence in the green (fluorescein) channel that are not apparent in control images of these cell lines with the addition of unlabeled nanodiscs (Fig. S4, ESI†). These spots mark the location of the MSP scaffold protein, but not necessarily the location of the nanodisc lipids. Previous work by Carney et al. showing the delivery of nanodiscs containing Gd-labeled lipids to HeLa cells indicates that these lipids are dispersed throughout the cell membranes (implying dynamic exchange of lipids between the nanodiscs and the HeLa cell), while the MSP is localised as punctate spots inside the cell, in agreement with our observations.17 The brightness of these spots in Fig. 3 is indicative of colocalisation of several nanodiscs within HeLa cells: this is suggestive of entrapment within intracellular vesicles, but a detailed study of the pathway of nanodisc trafficking by these cells is not an aim of this initial study.
Our data demonstrates the uptake of lipid nanodiscs by HeLa cells, showing their potential as carriers for intracellular delivery. These constructs will allow future detailed mechanistic studies of nanodisc uptake in different cell lines using organelle-specific immunostaining. In particular it will allow investigation of how the scaffold proteins are specifically trafficked by these cells.
Conclusions
We have successfully demonstrated a method for SrtA-mediated site-specifically labelling of the N-terminus of MSP1D1 scaffold proteins of lipid nanodiscs. Nanodiscs were labelled with a fluorescent fluorescein probe and their application for traceable delivery was demonstrated in HeLa cells. The SrtA approach to labelling the scaffold protein is an alternative method to inserting a site-specific mutation (e.g. a cysteine) within the scaffold protein for chemical labelling.17 Furthermore this can be used as a retrofit approach to chemically functionalise scaffold proteins within preformed lipid nanodiscs. This general approach to site-specific labelling of MSP1D1 nanodiscs by SrtA has the potential to be used to introduce a wide variety of desirable chemical functionalisations for wide-ranging nanodisc applications by conjugation to alternative depsipeptide substrates.We acknowledge funding from European Union Marie Curie Career Integration Grant BioNanoMuTT (No. PCIG09-GA-2011-293643).
Notes and references
- D. A. Bricarello, J. T. Smilowitz, A. M. Zivkovic, J. B. German and A. N. Parikh, ACS Nano, 2011, 5, 42–57 CrossRef CAS PubMed.
- A. Nath, W. M. Atkins and S. G. Sligar, Biochemistry, 2007, 46, 2059–2069 CrossRef CAS PubMed.
- T. H. Bayburt and S. G. Sligar, FEBS Lett., 2010, 584, 1721–1727 CrossRef CAS PubMed.
- G. Almer, H. Mangge, A. Zimmer and R. Prassl, Curr. Med. Chem., 2015, 22, 3631–3651 CrossRef CAS PubMed.
- C. Chen, H. Hu, M. Qiao, X. Zhao, Y. Wang, K. Chen, X. Guo and D. Chen, Int. J. Pharm., 2015, 480, 116–127 CrossRef CAS PubMed.
- S. K. Kim, M. B. Foote and L. Huang, Biomaterials, 2012, 33, 3959–3966 CrossRef CAS PubMed.
- A. J. Luthi, H. Zhang, D. Kim, D. A. Giljohann, C. A. Mirkin and C. S. Thaxton, ACS Nano, 2012, 6, 276–285 CrossRef CAS PubMed.
- K. M. McMahon, R. K. Mutharasan, S. Tripathy, D. Veliceasa, M. Bobeica, D. K. Shumaker, A. J. Luthi, B. T. Helfand, H. Ardehali, C. A. Mirkin, O. Volpert and C. S. Thaxton, Nano Lett., 2011, 11, 1208–1214 CrossRef CAS PubMed.
- K. G. Nelson, J. V. Bishop, R. O. Ryan and R. Titus, Antimicrob. Agents Chemother., 2006, 50, 1238–1244 CrossRef CAS PubMed.
- K. K. Ng, J. F. Lovell and G. Zheng, Acc. Chem. Res., 2011, 44, 1105–1113 CrossRef CAS PubMed.
- K. A. Redmond, T.-S. Nguyen and R. O. Ryan, Int. J. Pharm., 2007, 339, 246–250 CrossRef CAS PubMed.
- P. G. Frank and Y. L. Marcel, J. Lipid Res., 2000, 41, 853–872 CAS.
- T. H. Bayburt, Y. V. Grinkova and S. G. Sligar, Nano Lett., 2002, 2, 853–856 CrossRef CAS.
- I. G. Denisov, M. A. McLean, A. W. Shaw, Y. V. Grinkova and S. G. Sligar, J. Phys. Chem. B, 2005, 109, 15580–15588 CrossRef CAS PubMed.
- T. J. Knowles, R. Finka, C. Smith, Y.-P. Lin, T. Dafforn and M. Overduin, J. Am. Chem. Soc., 2009, 131, 7484–7485 CrossRef CAS PubMed.
- V. Postis, S. Rawson, J. K. Mitchell, S. C. Lee, R. A. Parslow, T. R. Dafforn, S. A. Baldwin and S. P. Muench, Biochim. Biophys. Acta, Biomembr., 2015, 1848, 496–501 CrossRef CAS PubMed.
- C. E. Carney, I. L. Lenov, C. J. Baker, K. W. MacRenaris, A. L. Eckermann, S. G. Sligar and T. J. Meade, Bioconjugate Chem., 2015, 26, 899–905 CrossRef CAS PubMed.
- G. Zheng, J. Chen, H. Li and J. D. Glickson, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 17757–17762 CrossRef CAS PubMed.
- P. A. Beales, N. Geerts, K. K. Inampudi, H. Shigematsu, C. J. Wilson and T. K. Vanderlick, J. Am. Chem. Soc., 2013, 135, 3335–3338 CrossRef CAS PubMed.
- N. Geerts, C. F. Schreck, P. A. Beales, H. Shigematsu, C. S. O'Hern and T. K. Vanderlick, Langmuir, 2013, 29, 13089–13094 CrossRef CAS PubMed.
- I. G. Denisov, Y. V. Grinkova, A. A. Lazarides and S. G. Sligar, J. Am. Chem. Soc., 2004, 126, 3477–3487 CrossRef CAS PubMed.
- D. J. Williamson, M. A. Fascione, M. E. Webb and W. B. Turnbull, Angew. Chem., Int. Ed., 2012, 51, 9377–9380 CrossRef CAS PubMed.
- D. J. Williamson, M. E. Webb and W. B. Turnbull, Nat. Protoc., 2014, 9, 253–262 CrossRef CAS PubMed.
- T. K. Ritchie, Y. V. Grinkova, T. H. Bayburt, I. G. Denisov, J. K. Zolnerciks, W. M. Atkins and S. G. Sligar, in Methods in Enzymology; Liposomes, Pt F, ed. N. Duzgunes, 2009, vol. 464, pp. 211–231 Search PubMed.
- M. T. Marty, H. Zhang, W. Cui, R. E. Blankenship, M. L. Gross and S. G. Sligar, Anal. Chem., 2012, 84, 8957–8960 CrossRef CAS PubMed.
- P. Morrison, M. Balmforth, S. Ness, D. Williamson, M. Rugen, B. Turnbull and M. E. Webb, ChemBioChem, 2016 DOI:10.1002/cbic.201500547.
Footnote |
| † Electronic supplementary information (ESI) available: Detailed materials and methods and supporting figures. See DOI: 10.1039/c6mb00126b |
| This journal is © The Royal Society of Chemistry 2016 |