Performance of optimized noncanonical amino acid mutagenesis systems in the absence of release factor 1†
Abstract
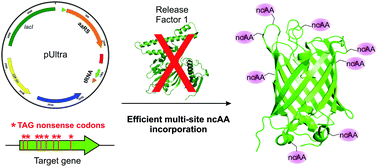
Site-specific incorporation of noncanonical amino acids (ncAAs) into proteins expressed in E. coli using UAG-suppression competes with termination mediated by release factor 1 (RF1). Recently, unconditional deletion of RF1 was achieved in a genomically recoded E. coli (C321), devoid of all endogenous UAG stop codons. Here we evaluate the efficiency of ncAA incorporation in this strain using optimized suppression vectors. Even though the absence of RF1 does not benefit the suppression efficiency of a single UAG codon, multi-site incorporation of a series of chemically distinct ncAAs was significantly improved.
- This article is part of the themed collections: Chemical Biology in Molecular BioSystems and Protein Labelling

 Please wait while we load your content...
Please wait while we load your content...