Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Immobilization of an artificial imine reductase within silica nanoparticles improves its performance†
Martina
Hestericová
a,
M. Rita
Correro
b,
Markus
Lenz
c,
Philippe F.-X.
Corvini
c,
Patrick
Shahgaldian
*b and
Thomas R.
Ward
*a
aDepartment of Chemistry, University of Basel, Spitalstrasse 51, CH-4056 Basel, Switzerland. E-mail: thomas.ward@unibas.ch
bInstitute of Chemistry and Bioanalytics, School of Life Sciences, University of Applied Sciences and Arts Northwestern Switzerland, Gründenstrasse 40, CH-4132 Muttenz, Switzerland
cInstitute for Ecopreneurship, School of Life Sciences, University of Applied Sciences and Arts Northwestern Switzerland, Gründenstrasse 40, 4132 Muttenz, Switzerland
First published on 6th July 2016
Abstract
Silica nanoparticles equipped with an artificial imine reductase display remarkable activity towards cyclic imine- and NAD+ reduction. The method, based on immobilization and protection of streptavidin on silica nanoparticles, shields the biotinylated metal cofactor against deactivation yielding over 46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 turnovers in pure samples and 4000 turnovers in crude cellular extracts.
000 turnovers in pure samples and 4000 turnovers in crude cellular extracts.
Catalysis plays a crucial role in a variety of scientific fields including synthetic organic chemistry. Given this importance, a wide range of synthetic systems, including heterogeneous solids, organocatalysts, metal complexes and enzymes, have been developed in the past century.1 Transition metal catalysts and enzymes possess complementary properties, which can be exploited in the synthesis of various enantiopure compounds.2
The industrial use of enzymes is partially limited by their operational instability and, in some cases, high price.3 Therefore, significant efforts have been invested in developing immobilization strategies that may contribute to stabilize enzymes, while allowing their recycling.4
An artificial metalloenzyme (ArM) results from the incorporation of a synthetic metal complex cofactor within a host protein.5 The biotin–streptavidin technology has been widely exploited to ensure the localization of a biotinylated cofactor within the host protein.6 As ArMs often bear a soft metal, cellular metabolites, including organosulfur compounds present in the E. coli cellular debris, frequently deactivate the metal.7 Such hybrid catalysts can be viewed as complementary to both homogeneous catalysts and enzymes.2,8–14
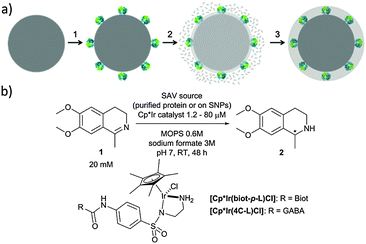
d6-piano stool complexes have shown great potential for in vivo catalysis as they display promising stability in biological environments.15 With the aim of improving its catalytic performance, we set out to shield an artificial transfer hydrogenase (ATHase) in a protective organosilica layer on silica nanoparticles (SNPs) and evaluate its catalytic performance towards the reduction of imines and the regeneration of NAD+.16
Working on the development of nanomaterials endowed with virus recognition properties,17,18 we have designed a chemical strategy that allows embedding enzymes in a biomimetic and soft organosilica layer.19,20
In the present work, we have expanded this shielding strategy to artificial metalloenzymes and tested the resulting nanocatalysts for the reduction of cyclic imine 1 in the presence of various cellular extracts. As shown previously with soluble ArMs, embedding the biotinylated pianostool iridium complex [Cp*Ir(biot-p-L)Cl] within various streptavidin mutants (Sav hereafter) allows to access either (R)- and (S)-salsolidine 2 (Fig. 1), depending on the Sav mutant (Table 1). This observation ensures that the biotinylated cofactor [Cp*Ir(biot-p-L)Cl] is indeed incorporated within Sav, as the bare cofactor yields (rac)-salsolidine 2.
| Entry | Sav mutant | eea,b (%) | Conv.a (%) | TON |
|---|---|---|---|---|
| Reactions were performed in 200 μl of reaction mixture.a Enantiomeric excess and conversion were determined by HPLC analysis on a Chiralpak-IC column.b Positive ee values correspond to (R)-salsolidine and negative ee values correspond to (S)-salsolidine. | ||||
| 1 | [Cp*Ir(biot-p-L)Cl]·Sav WT | 38 | 70 | 140 |
| 2 | [Cp*Ir(biot-p-L)Cl]·S112A | 87 | 89 | 178 |
| 3 | [Cp*Ir(biot-p-L)Cl]·S112K | −70 | 86 | 173 |
| 4 | [Cp*Ir(biot-p-L)Cl]·K121A | −35 | 99 | 198 |
| 5 | [Cp*Ir(biot-p-L)Cl]·S112A-K121A | 79 | 98 | 196 |
| 6 | [Cp*Ir(biot-p-L)Cl]·R84A-S112A-K121A | 68 | 99 | 198 |
Single mutants [Cp*Ir(biot-p-L)Cl]·S112A Sav and [Cp*Ir(biot-p-L)Cl]·S112K Sav were selected for immobilization and protection. These ArMs afford opposite enantiomers of amine 2. Additionally, the double mutant [Cp*Ir(biot-p-L)Cl]·S112A-K121A Sav was selected for immobilization in view of its remarkable activity as purified ArM.21
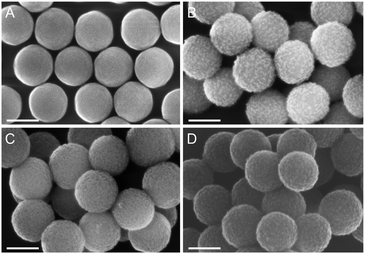
Native SNPs (240 nm in diameter) were prepared by the Stöber method.22 Next, in order to covalently anchor Sav on the surface of the nanoparticles, the SNPs were amino-modified by incubation with (3-aminopropyl)-triethoxysilane (APTES) and further reacted with glutaraldehyde. Sav isoforms preincubated with the iridium cofactor, [Cp*Ir(biot-p-L)Cl]·S112A Sav, [Cp*Ir(biot-p-L)Cl]·S112K Sav and [Cp*Ir(biot-p-L)Cl]·S112A-K121A Sav, were immobilized at the surface of SNPs by reacting with the free aldehyde functions present at the surface of the SNPs. The synthesis of a protective organosilica layer was achieved by incubating the SNPs with tetraethyl orthosilicate and APTES for one- and four hours respectively, yielding protected SNPs (prot-SNPs, Fig. 2).
SNPs were imaged using a field emission scanning electron microscope (FESEM). The micrographs revealed monodisperse and well-defined nanoparticles with an average diameter of 260 nm. As illustrated in Fig. 2, the SNPs equipped with the additional protective silica layer (prot-SNPs) display a homogeneous size distribution with a diameter increase of approx. 10 nm upon protection.
Gratifyingly, immobilization and additional protection of the ATHase on the SNPs to yield [Cp*Ir(biot-p-L)Cl]·Sav@SNP and [Cp*Ir(biot-p-L)Cl]·Sav@prot-SNP respectively only led to a modest variation of the enantioselectivity of the hybrid catalyst towards the reduction of imine 1 (Table 1, entries 6 and 10). The activity (i.e. total turnover number, TON) of the SNPs was established based on the iridium concentration. This latter was determined by inductively coupled plasma mass spectrometry (ICP-MS) for each SNP batch. Dried SNPs contain approximately 0.04 ng Ir per mg SNPs. Salsolidine reduction using [Cp*Ir(biot-p-L)Cl]·S112A Sav SNPs yielded total TON of 4294 after 2 days (Table 2, entry 1).
| Entry | Sav sourcea | Ir concentrationb (μM) | eec,d (%) | Conv.c (%) | TON |
|---|---|---|---|---|---|
| a Reactions performed in a reaction buffer using 1 mg of SNPs in 100 μl reaction mixture with 20 mM substrate concentration at RT for 48 hours. b Determined by means of ICP-MS. c The enantiomeric excess and conversion were determined by HPLC analysis on a Chiralpak-IC column (see ESI for details). d Positive ee values correspond to (R)-salsolidine 2 and negative ee values correspond to (S)-salsolidine 2. e Reactions performed in a reaction buffer using 1 mg of SNPs in 100 μl reaction mixture with 200 mM substrate concentration for 48 hours. f Reactions were performed with free purified enzymes with the same iridium concentration as used with SNPs. g Reactions performed without the presence of Sav using the iridium ligand 10 [Cp*Ir(4C-L)Cl] in reaction buffer. | |||||
| 1 | [Cp*Ir(biot-p-L)Cl]·S112A@prot-SNP | 2.6 | 90 | 56 | 4294 |
| 2 | [Cp*Ir(biot-p-L)Cl]·S112A@prot-SNP 1st recycling | 2.6 | 83 | 28 | 2172 |
| 3 | [Cp*Ir(biot-p-L)Cl]·S112A@prot-SNP 2nd recycling | 2.6 | 80 | 24 | 1863 |
| 4 | [Cp*Ir(biot-p-L)Cl]·S112A@prot-SNP 200 mM substratee | 2.6 | 88 | 22 | 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 990 990 |
| 5 | [Cp*Ir(biot-p-L)Cl]·S112A Sav purified proteinf | 2.6 | 79 | 15 | 1154 |
| 6 | [Cp*Ir(biot-p-L)Cl]·S112K@prot-SNP | 1.2 | −56 | 22 | 3775 |
| 7 | [Cp*Ir(biot-p-L)Cl]·S112K@prot-SNP 1st recycling | 1.2 | −53 | 12 | 2000 |
| 8 | [Cp*Ir(biot-p-L)Cl]·S112K@prot-SNP 2nd recycling | 1.2 | −58 | 9 | 1580 |
| 9 | [Cp*Ir(biot-p-L)Cl]·S112K@prot-SNP 200 mM substratee | 1.2 | −48 | 10 | 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 667 667 |
| 10 | [Cp*Ir(biot-p-L)Cl]·S112K Sav purified proteinf | 1.2 | −66 | 5 | 833 |
| 11 | [Cp*Ir(biot-p-L)Cl]·S112A-K121A@prot-SNP | 1.5 | 70 | 97 | 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 885 885 |
| 12 | [Cp*Ir(biot-p-L)Cl]·S112A-K121A@prot-SNP 1st recycling | 1.5 | 68 | 56 | 7294 |
| 13 | [Cp*Ir(biot-p-L)Cl]·S112A-K121A@prot-SNP 2nd recycling | 1.5 | 67 | 50 | 6570 |
| 14 | [Cp*Ir(biot-p-L)Cl]·S112A-K121A@prot-SNP 200 mM substratee | 1.5 | 76 | 36 | 46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 747 747 |
| 15 | [Cp*Ir(biot-p-L)Cl]·S112A-K121A Sav purified proteinf | 1.5 | 89 | 19 | 2513 |
| 16 | Native SNPs | 0 | 0 | 0 | 0 |
| 17 | Empty prot-SNPs | 0 | 0 | 0 | 0 |
| 18 | [Cp*Ir(biot-p-L)Cl] freeg | 2.6 | 0 | 8 | 444 |
| 19 | [Cp*Ir(4C-L)Cl] freeg | 80 | 0 | 42 | 104 |
| 20 | [Cp*Ir(4C-L)Cl]@prot-SNPs | 80 | 0 | 26 | 66 |
In line with the performance of the soluble ArM, the immobilized [Cp*Ir(biot-p-L)Cl]·S112A-K121A Sav@prot-SNP yielded the best results, with TON of 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 885 and almost full conversion (entry 11). Upon increasing the substrate concentration (200 mM), a TON of 46
885 and almost full conversion (entry 11). Upon increasing the substrate concentration (200 mM), a TON of 46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 747 was obtained using SNPs with [Cp*Ir(biot-p-L)Cl]·S112A-K121A Sav@prot-SNP (entry 14). Compared to the homogeneous ArMs (entries 5, 10 and 15 respectively), the TONs are significantly increased, at the cost of slight erosion in selectivity. We hypothesize that the increased rate results from the protective effect of the organosilane layer combined with the increased hydrophobicity around the active site. The preferential formation of enantioenriched salsolidine 1, depending on the Sav mutant, unambiguously demonstrates that the SNPs indeed contain the intact ArMs with preserved quaternary structure. Importantly, the SNPs can be recycled at least twice, at the cost of an erosion in conversion (entries 2, 3, 7, 8, 12 and 13), yet with virtually the same enantiomeric excess.
747 was obtained using SNPs with [Cp*Ir(biot-p-L)Cl]·S112A-K121A Sav@prot-SNP (entry 14). Compared to the homogeneous ArMs (entries 5, 10 and 15 respectively), the TONs are significantly increased, at the cost of slight erosion in selectivity. We hypothesize that the increased rate results from the protective effect of the organosilane layer combined with the increased hydrophobicity around the active site. The preferential formation of enantioenriched salsolidine 1, depending on the Sav mutant, unambiguously demonstrates that the SNPs indeed contain the intact ArMs with preserved quaternary structure. Importantly, the SNPs can be recycled at least twice, at the cost of an erosion in conversion (entries 2, 3, 7, 8, 12 and 13), yet with virtually the same enantiomeric excess.
Control experiments with native SNPs or SNPs without the presence of Sav (e.g. empty SNPs) did not show any activity towards NAD+ or imine reduction (entries 16 and 17). To address the synergistic effect of the silica layer and the streptavidin host on the catalytic performance of the iridium cofactor, a non-biotinylated [Cp*Ir(4C-L)Cl] analog was synthesized, immobilized and protected within SNPs ([Cp*Ir(4C-L)Cl]@prot-SNP). In contrast to the ArM, upon immobilization, the activity of [Cp*Ir(4C-L)Cl]@prot-SNP decreases upon mineralization within SNPs, (Table 2, entries 19 and 20).
We have also demonstrated the stability of enantioselectivity over time (see ESI†). The time-point assay showed only a slight erosion of enantioselectivity over the course of 7 days (decrease of 11%), which could be explained by disruption of the protective layer and leakage of the catalyst caused by friction in solution.20 However, the ICP-MS measurements of the reaction medium after the reaction revealed only traces of iridium. We thus argue that the decrease in activity is predominantly caused by loss of nanoparticles during centrifugation and washing cycles.
With biomedical applications23 in mind, we evaluated the regeneration of NAD+ by [Cp*Ir(biot-p-L)Cl]·S112A Sav@prot-SNP. For this purpose, the formation of NADH was monitored by absorbance spectroscopy at 340 nm (for detailed data see ESI†). The results confirm that the immobilization does not significantly affect the catalytic properties of the hybrid catalysts towards the NAD+ reduction. The protected catalyst [Cp*Ir(biot-p-L)Cl]·Sav@prot-SNP has a initial lower reaction rate, but overall slightly improved TONs compared to [Cp*Ir(biot-p-L)Cl]·Sav.
In order to test the compatibility of SNPs with complex cellular media, we performed experiments in the presence of E. coli cell lysates.
In stark contrast to the homogenous ArMs, which lose most of their activity in the presence of cellular debris solution, the SNPs partially maintained their catalytic activity. Up to 80 TONs were achieved with [Cp*Ir(biot-p-L)Cl]·S112A Sav@prot-SNPs and 171 TONs with [Cp*Ir(biot-p-L)Cl]·S112K Sav@prot-SNPs (for detailed data see ESI†). Other complex media were evaluated with [Cp*Ir(biot-p-L)Cl]·S112A-K121A Sav@prot-SNPs (Table 3).
| Entry | SNP | Cellular debrisa | eeb,c (%) | Conv.b (%) | TON |
|---|---|---|---|---|---|
Reactions were performed in a reaction buffer using 1 mg of SNPs in 100 μl reaction mixture with 20 mM substrate concentration at RT for 48 h.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a Reaction mixture contained 48 μl of crude cellular extract, organic solvent or stock solution of glutathione.b Enantiomeric excess and conversion were determined by HPLC analysis on a Chiralpak-IC column.c Positive ee values correspond to (R)-salsolidine 2 and negative ee values correspond to (S)-salsolidine 2.d Reaction performed with purified protein without any protection against the organic solvent. a Reaction mixture contained 48 μl of crude cellular extract, organic solvent or stock solution of glutathione.b Enantiomeric excess and conversion were determined by HPLC analysis on a Chiralpak-IC column.c Positive ee values correspond to (R)-salsolidine 2 and negative ee values correspond to (S)-salsolidine 2.d Reaction performed with purified protein without any protection against the organic solvent. |
|||||
| 1 | [Cp*Ir(biot-p-L)Cl]·S112A-K121A Sav@prot-SNP | Milk | 41 | 24 | 2569 |
| 2 | Yeast extract | 37 | 1 | 158 | |
| 3 | Urine pH 6 | 25 | 11 | 4554 | |
| 4 | Blood pH 7 | 35 | 1 | 392 | |
| 5 | Blood serum | 27 | 6 | 862 | |
| 6 | Empty vector CFE | 89 | 3 | 1873 | |
| 7 | Empty vector CL | 33 | 1 | 132 | |
| 8 | GSH 5 mM | 30 | 0.15 | 20 | |
| 9 | MeOH 50% | 14 | 30 | 4024 | |
| 10 | [Cp*Ir(biot-p-L)Cl]·S112A-K121A Sav purified proteind | MeOH 50% | 0 | 0 | 0 |
Up to 4500 TONs were obtained upon performing the imine reduction in urine. SNPs maintain their catalytic activity also in the presence of methanol (4000 TONs, accompanied by an erosion in ee), which results in protein denaturation without the presence of protective organosilane layer and a complete loss of activity (Table 3, entries 9 and 10).
The catalytic experiments reported herein reveal several noteworthy features:
(i) Immobilization of Sav isoforms on silica nanoparticles and their protection with organosilane layer yields catalysts with higher turnover number rates compared to the use of purified proteins.
(ii) Such nanoparticles can be recycled up to three times with only a slight erosion in selectivity.
(iii) Protected catalysts can withstand the presence of cellular debris without the need of diamide pretreatment.7
(iv) The protective layer also prevents the protein denaturation in the presence of chaotropic agents.
In summary, we have successfully developed an artificial biocatalyst consisting of a pianostool iridium cofactor within Sav immobilized in SNPs. These nanoparticles display remarkable catalytic activity with TON > 46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 using aqueous solutions and TON > 4000 in the presence of cellular debris. Presented SNPs offer interesting perspectives towards green chemistry as they operate under nearly physiological conditions and can be recycled with only a slight erosion of reactivity. Immobilization and protection of artificial metalloenzymes on silica nanoparticles represents a promising technique for activity enhancement of such hybrid catalysts. The next steps will include cellular uptake and engineering enzyme cascades incorporating natural and artificial metalloenzymes within SNPs.
000 using aqueous solutions and TON > 4000 in the presence of cellular debris. Presented SNPs offer interesting perspectives towards green chemistry as they operate under nearly physiological conditions and can be recycled with only a slight erosion of reactivity. Immobilization and protection of artificial metalloenzymes on silica nanoparticles represents a promising technique for activity enhancement of such hybrid catalysts. The next steps will include cellular uptake and engineering enzyme cascades incorporating natural and artificial metalloenzymes within SNPs.
This research was supported by the Swiss Nanoscience Institute (Project NanoZyme, DPA2238) and the NCCR Molecular Systems Engineering.
Notes and references
- H. Krässig, J. Schurz, R. G. Steadman, K. Schliefer, W. Albrecht, M. Mohring and H. Schlosser, Ullmann's encyclopedia of industrial chemistry, 2012, vol. 6, pp. 565–582 Search PubMed.
- O. Pàmies and J. E. Bäckvall, Chem. Rev., 2003, 103, 3247–3261 CrossRef PubMed.
- K. M. Polizzi, A. S. Bommarius, J. M. Broering and J. F. Chaparro-Riggers, Curr. Opin. Chem. Biol., 2007, 11, 220–225 CrossRef CAS PubMed.
- M. D. Truppo, H. Strotman and G. Hughes, ChemCatChem, 2012, 4, 1071–1074 CrossRef CAS.
- C. Letondor, N. Humbert and T. R. Ward, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 4683–4687 CrossRef CAS PubMed.
- T. R. Ward, Acc. Chem. Res., 2011, 44, 47–57 CrossRef CAS PubMed.
- Y. M. Wilson, M. Dürrenberger, E. S. Nogueira and T. R. Ward, J. Am. Chem. Soc., 2014, 136, 8928–8932 CrossRef CAS PubMed.
- M. E. Wilson and G. M. Whitesides, J. Am. Chem. Soc., 1978, 100, 306–307 CrossRef CAS.
- J. C. Lewis and P. R. Chen, ACS Catal., 2013, 3, 2954–2975 CrossRef CAS.
- F. Rosati and G. Roelfes, ChemCatChem, 2010, 2, 916–927 CrossRef CAS.
- P. J. Deuss, R. Denheeten, W. Laan and P. C. J. Kamer, Chem. – Eur. J., 2011, 17, 4680–4698 CrossRef CAS PubMed.
- F. Yu, V. M. Cangelosi, M. L. Zastrow, M. Tegoni, J. S. Plegaria, a. G. Tebo, C. S. Mocny, L. Ruckthong, H. Qayyum and V. L. Pecoraro, Chem. Rev., 2014, 114, 3495–3578 CrossRef CAS PubMed.
- T. K. Hyster and T. R. Ward, Angew. Chem., Int. Ed., 2016, 2–16 Search PubMed.
- T. Ueno, S. Abe, N. Yokoi and Y. Watanabe, Coord. Chem. Rev., 2007, 251, 2717–2731 CrossRef CAS.
- P. K. Sasmal, C. N. Streu and E. Meggers, Chem. Commun., 2013, 49, 1581–1587 RSC.
- M. Dürrenberger, T. Heinisch, Y. M. Wilson, T. Rossel, E. Nogueira, L. Knörr, A. Mutschler, K. Kersten, M. J. Zimbron, J. Pierron, T. Schirmer and T. R. Ward, Angew. Chem., Int. Ed., 2011, 50, 3026–3029 CrossRef PubMed.
- A. Cumbo, B. Lorber, P. F.-X. Corvini, W. Meier and P. Shahgaldian, Nat. Commun., 2013, 4, 1503–1507 CrossRef PubMed.
- S. Sykora, A. Cumbo, G. Belliot, P. Pothier, C. Arnal, Y. Dudal, P. F.-X. Corvini and P. Shahgaldian, Chem. Commun., 2015, 51, 2256–2258 RSC.
- P. Shahgaldian, R. Correro-Shahgaldian, A. Cumbo and P. F.-X. Corvini, WO/2015/014888, 2015.
- M. R. Correro, N. Moridi, H. Schützinger, S. Sykora, E. M. Amman, E. H. Peters, Y. Dudal, P. F.-X. Corvini and P. Shahgaldian, Angew. Chem., Int. Ed., 2016, 55, 6285–6289 CrossRef CAS PubMed.
- F. Schwizer, V. Köhler, M. Dürrenberger, L. Knörr and T. R. Ward, ACS Catal., 2013, 3, 1752–1755 CrossRef CAS.
- W. Stöber, A. Fink and E. Bohn, J. Colloid Interface Sci., 1968, 26, 62–69 CrossRef.
- S. Betanzos-Lara, Z. Liu, A. Habtemariam, A. M. Pizarro, B. Qamar and P. J. Sadler, Angew. Chem., Int. Ed., 2012, 51, 3897–3900 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details, methods, assays and spectra. See DOI: 10.1039/c6cc04604e |
| This journal is © The Royal Society of Chemistry 2016 |